Introduction
Cervical cancer (CC) is the fourth most common type
of cancer in women worldwide and the leading cause of
cancer-related death in some of the poorest countries in the world
(1). Recent global data estimated
527,624 new cases and 265,672 deaths due to CC during 2018
(2). The 5-year survival rate for
metastatic CC is 16.5% compared with 91.5% for localized CC
(3). CC is preventable via the
human papillomavirus (HPV) vaccination and cervical screening with
primary HPV testing followed by treatment of any detected
precancerous lesions (4).
Unfortunately, the majority of women in developing countries still
do not have access to CC prevention programs, resulting in
increased CC disease burden. To improve prevention of CC and cancer
treatment outcomes, there is an urgent need to consider exploring
an effective and simple means of diagnosing and treating CC.
Long non-coding RNAs (lncRNAs) are a class of
single-stranded RNAs, >200 nucleotides in length, which do not
possess protein coding capacity. However, they do contribute to the
modulation of tumor progression via regulation of biological
processes through chromatin remodeling, and transcriptional and
post-transcriptional processing (5,6).
Recent studies have reported that dysregulated expression of
lncRNAs are likely pervasive in human cancer types, and can drive
cancer development and progression of several types of cancer, such
as colorectal, breast and gastric cancer (7–9).
LINC00885, a novel oncogenic lncRNA type, was upregulated in breast
cancer cell lines (10). LINC00885
may also serve roles in preventing the progression and metastasis
of bladder cancer (11). However,
the relationship between LINC00885 and CC has not yet been
reported.
Forkhead box protein 3 (FOXP3)+ cells are
generally considered to suppress antitumor immune responses in
several types of cancer, such as gastrointestinal cancer and
gallbladder carcinoma (12,13). Immune dysregulation,
polyendocrinopathy, enteropathy and X-linked syndrome are caused by
different genetic defects in the FOXP3 gene (14). FOXP3 is a specific transcription
factor that is expressed on the surface of regulatory T cells
(Tregs), which serves important roles in the tumor microenvironment
and participates in tumor cell growth, invasion and metastasis, in
breast, gastric, thyroid and non-small cell lung cancer (15–18).
FOXP3 overexpression leads to severe immunodeficiency and
FOXP3+ Tregs are abundant in tumor infiltrates and
peripheral blood from patients with cancer, where it is often
associated with a poor prognosis (19). Importantly, FOXP3 is highly
expressed in CC, and able to facilitate proliferation and
invasiveness, resulting in the occurrence, development and
metastasis of CC, and FOXP3 serves an important role in lymph
angiogenesis of CC (20,21).
The present study aimed to examine the roles of
LINC00885 in CC in vitro. The results showed that LINC00885
can interact with FOXP3 in CC cells. Moreover, the heterologous
overexpression of LINC00885 in cultured CC cells promoted cell
proliferation, invasion and EMT. Therefore, LINC00885 may be a
potential therapeutic target for the treatment of CC.
Materials and methods
Expression of genes in cervical tumor
samples
Expression levels of LINC00885 and FOXP3 in CC tumor
samples in patients were examined using Gene Expression Profiling
Interactive Analysis 2 (GEPIA2; gepia2.cancer-pku.cn/#index), using
data obtained from The Cancer Genome Atlas (TCGA; cancer.gov/tcga)
and the Genotype Tissue Expression (GTEx) project (22–24).
Cell lines and culture
The human CC cell lines (C-33A, SiHa, HeLa and
CaSki) and normal cervical epithelial cell line (Ect1/E6E7) were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, and maintained in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/100 µg/ml streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2. Cells in the
logarithmic growth phase were harvested for further
experiments.
Cell transfection
For optimal short hairpin (sh)RNA transfection
efficiency, two shRNA sequences were designed to target the human
LINC00885 gene and one shRNA sequence was designed to target the
human FOXP3 gene. The LINC00885 shRNA-1 (sh-LINC00885#1) and
LINC00885 shRNA-2 (sh-LINC00885#2), FOXP3 shRNA (sh-FOXP3) and
control shRNA (sh-NC) were obtained from Shanghai GenePharma Co.,
Ltd. A LINC00885 overexpression plasmid, pcDNA3.1-LINC00885, was
commercially constructed by Shanghai GenePharma Co., Ltd., and an
empty pcDNA 3.1 vector (NC) was used as the control. Cells
(2×105 cells/well) were added to 6-well plates and
transfected with 50 nmol/l sh-LINC00885#1, sh-LINC00885#2,
sh-FOXP3, sh-NC, pcDNA3.1-LINC00885 or pcDNA3.1, or co-transfected
with sh-FOXP3 and pcDNA3.1 or pcDNA3.1-LINC00885 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
scientific, Inc.) according to the manufacturer's protocol. At 48 h
post-transfection, cells were harvested for subsequent
experiments.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from CC cell lines using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. For LINC00885
expression analysis, the RNA was reverse transcribed into cDNA
using a High Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Subsequently, qPCR was performed using the SYBR Premix Ex Taq™ II
kit (Thermo Fisher Scientific, Inc.) using an ABI Prism 7700
sequence detector (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The sequences of the PCR primers used were: LINC00885
forward, 5′-CAGGGTTGGTGCTATGAATGAC-3′ and reverse,
5′-GAAGATTGTCCATGTTGGCAGTAT-3′; and GAPDH forward,
5′-CCATCTTCCAGGAGCGAGAT-3′ and reverse, 5′-TGCTGATGATCTTGAGGCTG-3′.
The following thermocycling conditions were used for qPCR: Initial
denaturation at 95°C for 10 min; followed by 40 cycles of 94°C for
2 min, 60°C for 50 sec; a final extension at 60°C for 1 min. All
reactions were performed at least three times. GAPDH was used as
the house-keeping gene. The formula 2−∆∆Cq was used to
calculate relative gene expression (25).
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Beijing Solarbio
Science & Technology Co., Ltd.) assay was used to analyze cell
proliferation, according to the manufacturer's instructions.
Briefly, transfected CaSki and HeLa cells (1×104
cells/well) were seeded in 96-well plates for 0, 24, 48 or 72 h,
and subsequently 10 µl CCK-8 reagent was added to the medium and
further incubated in the dark at 37°C for 2 h. The absorbance of
every well was measured at 450 nm using a microplate reader.
EdU staining assay
EdU staining was used to detect proliferation.
Transfected CaSki and HeLa cells (4×104 cells/well) were
plated into a 96-well plate and incubated with 20 µM EdU (Thermo
Fisher Scientific, Inc.) for 3 h. Cells were analyzed under a
confocal microscope (magnification, ×200; Leica Microsystems GmbH).
DNA (blue) was stained using DAPI for 10 min at room temperature.
Green cells were the EdU/DAPI-positive cells.
Transwell assay
Transwell assays were used to measure the invasive
ability of the transfected cells. The 24-well Transwell chambers
(8-µm pores) pre-coated with Matrigel (BD Biosciences) overnight at
37°C were used to assess invasion. A total of 200 µl media with
transfected CaSki and HeLa cells (5×104 cells/ml) was
added to the upper chamber and 600 µl RPMI-1640 medium containing
10% FBS was added to the lower chamber. After incubation at 37°C
for 24 h, the cells that had not invaded were removed using cotton
swabs. After washing with PBS, the invaded cells were fixed in 4%
paraformaldehyde for 10 min at room temperature, and stained with
0.1% crystal violet for 15 min at room temperature. The number of
invaded cells was counted under an optical microscope
(magnification, ×100; Olympus Corporation) in five randomly
selected fields of view.
Dual-luciferase reporter assay
Based on the bioinformatics prediction tool PROMO
(alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3),
FOXP3 was identified as a transcription factor of LINC00885. CaSki
and HeLa cells were plated in 24-well plates (4×105
cells/well) and co-transfected with various plasmids as indicated
in the figures. The pGL3, pGL3-LINC00885 plasmids (Shanghai
GenePharma Co., Ltd.) were co-transfected with sh-FOXP3 or sh-NC
into cells using Lipofectamine 2000. Cells were collected 24 h
after transfection, and luciferase activity was analyzed using a
Dual-Luciferase Reporter assay kit (Promega Corporation). Relative
luciferase activity was normalized to Renilla luciferase
(control).
Chromatin immunoprecipitation
(ChIP)
ChIP assay was performed using the EZ-ChIP™
Chromatin immunoprecipitation kit (cat. no. 17-371; Sigma Aldrich;
Merck KGaA) according to the manufacturer's protocol. A total of
1×107 cells were cross-linked with 1% formaldehyde for
10 min at room temperature. The cell lysates were sonicated using a
10 sec on and 10 sec off mode for 12 cycles on the ice to obtain
chromatin fragments, which was then immunoprecipitated at 4°C
overnight using anti-FOXP3 (5 µg) antibody (cat. no. ab20034;
Abcam) and normal IgG complexes. Finally, the DNA was obtained via
phenol/chloroform extraction and ethanol precipitation, and RT-qPCR
was performed as aforementioned.
Western blotting
Total protein was extracted by ice-cold RIPA lysis
buffer (Beyotime Institute of Biotechnology). The concentration of
protein was detected using a BCA Protein Quantification kit
(Beyotime Institute of Biotechnology). A total of 30 µg protein was
loaded on a 10% SDS gel, resolved using SDS-PAGE and then
transferred to a PVDF membrane (Invitrogen; Thermo Fisher
Scientific, Inc.). After blocking with TBS with 0.1% Tween-20
(TBST; Beijing Solarbio Science & Technology Co., Ltd.)
containing 5% non-fat milk for 1 h at room temperature, the
membranes were incubated with primary antibodies against FOXP3
(1:1,000; cat. no. ab20034; Abcam), E-cadherin (1:1,000; cat. no.
ab40772; Abcam), Vimentin (1:1,000; cat. no. ab92547; Abcam) or
GAPDH (1:1,000; cat. no. ab8245; Abcam) overnight at 4°C. The
following day, the membranes were washed using TBST, and incubated
with the HRP-conjugated goat anti-rabbit immunoglobulin G (IgG;
1:20,000; cat. no. ZB-2301; OriGene Technologies, Inc.) or goat
anti-mouse IgG (1:20,000; cat. no. ZB-2305; OriGene Technologies,
Inc.) secondary antibodies for 1 h at room temperature.
Antigen-antibody complexes were detected using an enhanced
chemiluminescence reagent (Cytiva). The relative expression of
genes was analyzed using ImageJ (version 5.0; National Institutes
of Health) and normalized to GAPDH.
Statistical analysis
Results are presented as the mean ± standard
deviation, and all experiments were performed in triplicate. The
differences within groups were assessed using one-way ANOVA
followed by Tukey's post hoc test or an independent samples t-test.
Analysis was performed using SPSS (version 20; IBM Corp.) and
graphed using GraphPad Prism version 8.0 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
LINC00885 expression is upregulated in
CC tissues and cell lines
To examine the clinical significance of LINC00885,
the expression of LINC00885 in tissue samples from patients was
directly extracted from the GEPIA2 database together with the
graph, which showed that the expression of LINC00885 in CC tumor
tissue samples (n=306) were significantly higher compared with the
normal tissue samples (n=13) (Fig.
1A). Additionally, higher expression of LINC00885 was observed
in the four CC cell lines (C-33A, SiHa, HeLa and CaSki) compared
with the normal cervical epithelial cell line, Ect1/E6E7 (Fig. 1B), suggesting that LINC00885 may be
an oncogenic gene in CC.
 | Figure 1.Identification of LINC00885 based on
bioinformatics analysis using GEPIA2, and LINC00885 knockdown
inhibits the cell proliferation of CC cells. (A) Corresponding box
plots of the comparative expression of LINC00885 in CC tumor
samples (red) vs. normal tissue samples (grey) generated using
GEPIA2. *P<0.01 vs. normal. (B) Relative expression of LINC00885
using GAPDH as the loading control in C-33A, SiHa, HeLa and CaSki
CC cell lines, and Ect1/E6E7 normal cervical cells. **P<0.01,
***P<0.001 vs. Ect1/E6E7. (C) Interference efficiency in CaSki
and HeLa cells 48 h after transfection with two shRNAs against
LINC00885 (sh-LINC00885#1 and sh-LINC00885#2). (D) Following
knockdown of LINC00885, a Cell Counting Kit-8 assay was used to
detect changes in the cell proliferation of CaSki and HeLa cells.
(E) Knockdown of LINC00885 in CaSki and HeLa cells reduced their
proliferative capacities, as shown using an EdU staining assay.
***P<0.001 vs. control or sh-NC. GEPIA, Gene Expression
Profiling Interactive Analysis; CC, cervical cancer; sh, short
hairpin RNA; NC, negative control. |
Knockdown of LINC00885 reduces cell
proliferation of CC cells
To examine the role of LINC00885 in CC cell
progression, CaSki and HeLa cells were selected for the
loss-of-function assays and LINC00885 expression was significantly
reduced by sh-LINC00885#1 and sh-LINC00885#2 48 h after
transfection (Fig. 1C).
Subsequently, the CCK-8 assay results showed that these two
LINC00885 shRNAs significantly reduced the proliferative capacity
of CaSki and HeLa cells when compared with the si-NC-transfected
cells (Fig. 1D). As presented in
Fig. 1E, knockdown of LINC00885
significantly decreased the number of EdU-positive CC cells, which
suggested that the knockdown of LINC00885 reduced proliferation.
Together, these results suggest that LINC00885 accelerates the
malignant proliferation of CC cells.
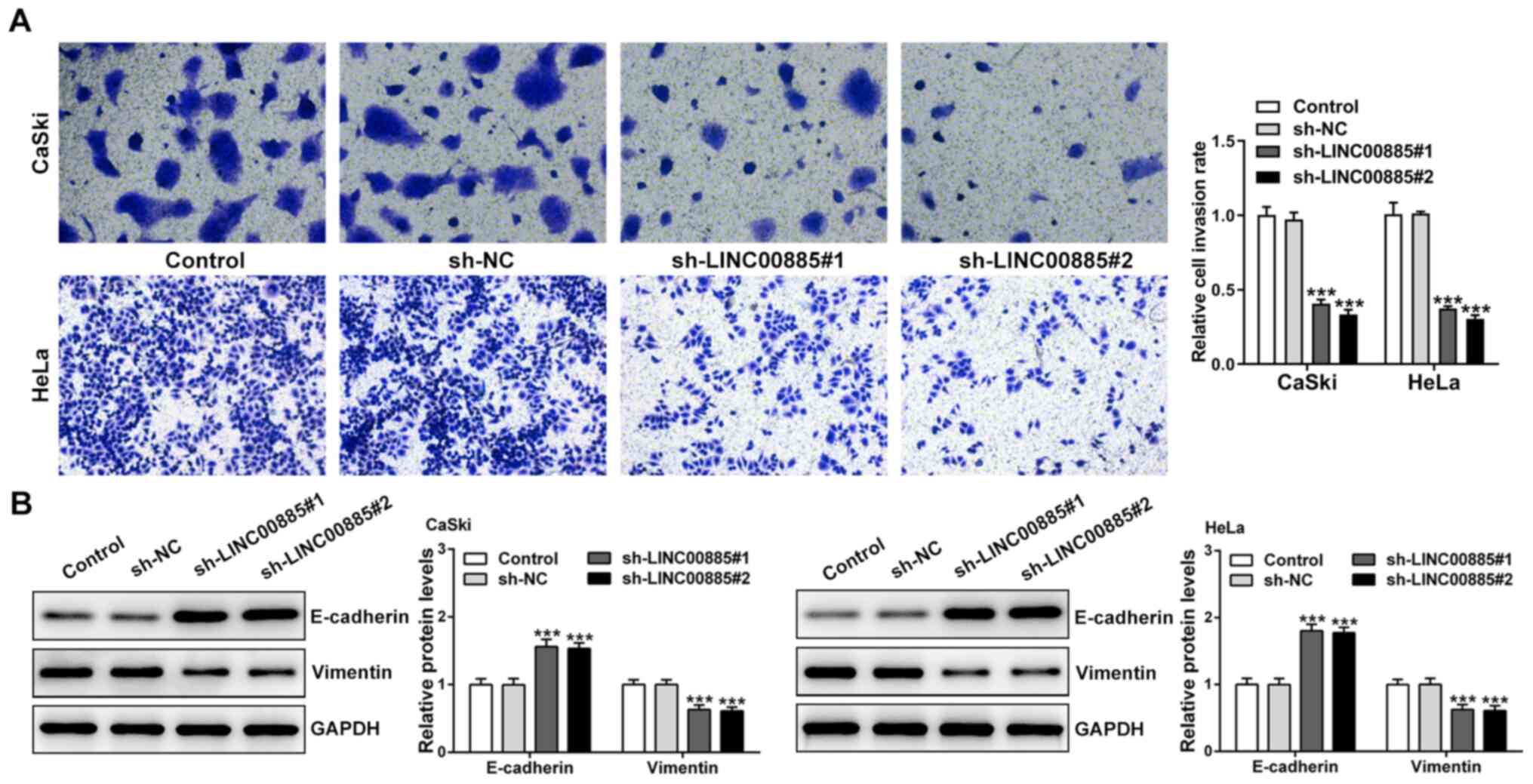
LINC00885 increases the invasive
capacity of CC cells and expression of EMT-related proteins
Transwell invasion assays showed that the knockdown
of LINC00885 reduced the invasive capacity of CC cells (Fig. 2A). In order to assess the effect of
LINC00885 on EMT in CC cells, the expression of E-cadherin
(epithelial marker) and Vimentin (mesenchymal marker) was evaluated
using western blotting. There was a significant increase in
E-cadherin expression and decrease in Vimentin expression following
knockdown of LINC00885, both in CaSki and HeLa cells (Fig. 2B). Collectively, the results showed
that LINC00885 increased invasion and EMT in CC cells.
FOXP3 directly regulates LINC00885 in
CC cells
Based on the prediction tool PROMO, FOXP3 was found
to be a transcription factor of LINC00885 (Fig. 3A). First, FOXP3 expression was
assessed in the CC tissues from the GEPIA2 database. Showing a
similar pattern to LINC00885, FOXP3 expression was higher in tumor
tissues compared with the normal tissues, indicating its potential
oncogenic role in CC cells (Fig.
3B). In addition, FOXP3 expression was higher in the four CC
cell lines compared with Ect1/E6E7 cells (Fig. 3C). To examine the biological
functions of FOXP3 in CC cells, specific small shRNA was used to
knock down FOXP3 expression in two CC cell lines, CaSki and HeLa
cells. The transfection efficiency was evaluated 48 h
post-transfection by western blotting (Fig. 3D). The knockdown of FOXP3 expression
significantly reduced LINC00885 expression in CaSki and HeLa cells
(Fig. 3E). As indicated in Fig. 3F, the knockdown of FOXP3
significantly reduced the luciferase activity in CaSki and HeLa
cells co-transfected with pGL3-LINC00885, whereas the knockdown of
FOXP3 had no effect on luciferase activity when the cells were
co-transfected with pGL3-NC, suggesting that FOXP3 bound directly
to the LINC00885 promoter. ChIP was used to validate the potential
endogenous interaction between LINC00885 and FOXP3. The results
showed that the LINC00885 promoter was preferentially enriched in
the anti-FOXP3 group compared with the IgG control group (Fig. 3G).
Effects of FOXP3 knockdown on the
proliferation of CC cells are reversed by LINC00885
overexpression
To verify whether LINC00885 mediated-regulation of
the proliferation of CC cells was dependent on FOXP3, LINC00885 was
overexpressed in CaSki and HeLa cells using pcDNA-LINC00885
(Fig. 4A). FOXP3 knockdown
significantly inhibited the proliferation of CaSki and HeLa cells,
and LINC00885 partially reversed such alterations caused by
sh-FOXP3 (Fig. 4B). Moreover, as
shown in Fig. 4C, the knockdown of
FOXP3 significantly reduced the number of EdU-positive (green)
CaSki and HeLa cells, and this was restored by pc-LINC00885
transfection. These results showed that FOXP3 directly binds to the
promoter of LINC00885, thus indirectly promoting the proliferation
of CC cells.
Effects of FOXP3 knockdown on the
invasion and EMT of CC cells is reversed by LINC00885
overexpression
First, CaSki and HeLa cells were divided into four
groups: sh-NC, sh-FOXP3, sh-FOXP3 + pcDNA3.1 and sh-FOXP3 +
pc-LINC00885. FOXP3 knockdown alone attenuated the invasive
abilities of CaSki and HeLa cells, and the addition of pc-LINC00885
reversed the effects of sh-FOXP3 (Fig.
5A). Likewise, the expression levels of EMT-related factors
were also measured by western blotting. As shown in Fig. 5B, the expression levels of
E-cadherin were increased, whereas Vimentin expression was
decreased after transfection with sh-FOXP3 compared with sh-NC.
Overexpression of LINC00885 reduced E-cadherin expression and
increased Vimentin expression in both CaSki and HeLa cells.
Together, these results demonstrated that LINC00885 leads to EMT of
CC cells via modulation of FOXP3.
Discussion
CC is one of the most common types of cancer in
women worldwide with high morbidity and mortality rates, and
continues to be a serious public health problem in the developing
world, including in China (26).
Due to its large population and the geographical and socioeconomic
inequities, China has a high burden of CC and considerable
disparities in the capacity to detect and manage the disease
amongst different regions (26).
For patients with CC, there are two types of metastasis:
Hematogenous metastasis and lymphatic metastasis. Patients with
hematogenous metastasis have a higher risk of death compared with
those with lymphatic metastasis, and the metastatic sites include
the lungs, liver, bone and brain (27). Thus, the identification of novel
suitable prognostic biomarkers is critical to improve the detection
of CC.
LINC00885 is a newly discovered lncRNA with few
relevant studies, and its abnormal expression has been observed
only in human breast and bladder cancer (10,11).
To the best of our knowledge, the present study was the first to
show the increased expression of LINC00885 in CC and its crucial
role in the acquisition of a malignant phenotype in CC cells. In
the present study, based on the analysis of data obtained from TCGA
and GTEx, the expression of LINC00885 in CC tissues was notably
higher compared with the normal cervical tissues. Additionally, the
expression of LINC00885 in CC cells was consistent with TCGA and
GTEx data. These findings suggested that LINC00885 may act as a
carcinogenic lncRNA involved in the pathophysiology of CC. In order
to further identify whether LINC00885 contributes to the
proliferation, invasion and EMT of CC, loss-of-function experiments
were performed. The proliferative and invasive abilities, and
extent of EMT of CC CaSki and HeLa cells were notably decreased in
cells transfected with two different LINC00885 shRNAs, which showed
that LINC00885 served a crucial role in maintaining the malignant
phenotype of CC. Studies on other lncRNAs have found similar
results in CC, such as HOTAIR (28), MALATl (29) and EBIC (30). In brief, overexpression of HOTAIR
can be used as a predictor for poor prognosis of CC and suppression
of lncRNA HOTAIR can reduce the proliferation, migration and
invasion of CC cells and decrease the expression of EMT-related
proteins (28). Downregulation of
lncRNA-MALATl expression can reduce the proliferative and invasive
capacities of CC cells and was correlated with HPV in cervical
cancer (29). Furthermore, the
lncRNA-EBIC-mediated increase in CC cell infiltration may be
related to inhibition of E-cadherin expression (30).
FOXP3 is a specific biomarker commonly used to
identify Tregs; Tregs suppress the antitumor immune response, and
thereby promote tumor progression. The roles of FOXP3 in cervical
cancer has been previously demonstrated (20,21).
Thus, it was not surprising that FOXP3 participated in the
acquisition of a malignant phenotype in CC cells, including
proliferation, invasion and EMT, which can be suppressed by the
knockdown of FOXP3. Following the knockdown of FOXP3, the ectopic
overexpression of LINC00885 partially restored the malignant
phenotype of CC cells, which also highlighted a potential
association between LINC00885 and FOXP3. Furthermore, FOXP3 is a
transcription factor of LINC00885, and this was verified in the
present study based on the results of the dual-luciferase reporter
and ChIP assays. Therefore, it was hypothesized that LINC00885
promotes CC development via regulation of FOXP3 expression.
Although further investigation is required, the results of the
present study showed that LINC00885 and FOXP3 may function as a
tumor promoter in CC.
In conclusion, FOXP3 regulated the effects of
LINC00885 on the proliferation, invasion and EMT of CC cells.
LINC00885 and FOXP3 may thus serve as biomarkers for the early
diagnosis of CC and a potential therapeutic target for reversing
the malignant phenotype of CC. Finally, the combined results
provided a novel perspective of FOXP3 and LINC00885 function in
papillary CC tumorigenesis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JX and LL conceived and designed the study. YL, HT
and LZ performed the experiments. YL and HT analyzed the data. YL
and LL wrote the manuscript. JX revised the manuscript. All authors
read and approved the final manuscript. YL, JX and LL are
responsible for confirming the authenticity of the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shrestha AD, Neupane D, Vedsted P and
Kallestrup P: Cervical cancer prevalence, incidence and mortality
in low and middle income countries: A systematic review. Asian Pac
J Cancer Prev. 19:319–324. 2018.PubMed/NCBI
|
|
3
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Canfell K: Towards the global elimination
of cervical cancer. Papillomavirus Res. 8:1001702019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tano K and Akimitsu N: Long non-coding
RNAs in cancer progression. Front Genet. 3:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian
J, Zhang X and Fang JY: A long non-coding RNA signature to improve
prognosis prediction of colorectal cancer. Oncotarget. 5:2230–2242.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Wang W, Xia P, Wan L, Zhang L, Yu L,
Wang L, Chen X, Xiao Y and Xu C: Identification of a five-lncRNA
signature for predicting the risk of tumor recurrence in patients
with breast cancer. Int J Cancer. 143:2150–2160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zaheed O, Samson J and Dean K: A
bioinformatics approach to identify novel long, non-coding RNAs in
breast cancer cell lines from an existing RNA-sequencing dataset.
Noncoding RNA Res. 5:48–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Liu Y, Zhang X, Liu J and Wang P:
Transcriptomic analysis of high-throughput sequencing about
circRNA, lncRNA and mRNA in bladder cancer. Gene. 677:189–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Y, Liao H, Zhang Y, Yuan R, Wang F,
Gao Y, Wang P and Du Z: Prognostic value of tumor-infiltrating
FoxP3+ T cells in gastrointestinal cancers: A meta analysis. PLoS
One. 9:e943762014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Huang Y and Qin M:
Tumour-infiltrating FoxP3+ and IL-17-producing T cells affect the
progression and prognosis of gallbladder carcinoma after surgery.
Scand J Immunol. 78:516–522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bennett CL, Christie J, Ramsdell F,
Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT,
Chance PF and Ochs HD: The immune dysregulation,
polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused
by mutations of FOXP3. Nat Genet. 27:20–21. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng GL, Li L, Guo YW, Yu P, Yin XJ, Wang
S and Liu CP: CD8(+) cytotoxic and FoxP3(+) regulatory T
lymphocytes serve as prognostic factors in breast cancer. Am J
Transl Res. 11:5039–5053. 2019.PubMed/NCBI
|
|
16
|
Li F, Sun Y, Huang J, Xu W, Liu J and Yuan
Z: CD4/CD8 + T cells, DC subsets, Foxp3, and IDO expression are
predictive indictors of gastric cancer prognosis. Cancer Med.
8:7330–7344. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang S, Wu J, Ren J, Vlantis AC, Li MY,
Liu SYW, Ng EKW, Chan ABW, Luo DC, Liu Z, et al: MicroRNA-125b
interacts with Foxp3 to induce autophagy in thyroid cancer. Mol
Ther. 26:2295–2303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang S, Liu Y, Li MY, Ng CSH, Yang SL,
Wang S, Zou C, Dong Y, Du J, Long X, et al: FOXP3 promotes tumor
growth and metastasis by activating Wnt/beta-catenin signaling
pathway and EMT in non-small cell lung cancer. Mol Cancer.
16:1242017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka A and Sakaguchi S: Targeting Treg
cells in cancer immunotherapy. Eur J Immunol. 49:1140–1146.
2019.PubMed/NCBI
|
|
20
|
Luo Q, Zhang S, Wei H, Pang X and Zhang H:
Roles of Foxp3 in the occurrence and development of cervical
cancer. Int J Clin Exp Pathol. 8:8717–8730. 2015.PubMed/NCBI
|
|
21
|
Tang J, Yang Z, Wang Z, Li Z, Li H, Yin J,
Deng M, Zhu W and Zeng C: Foxp3 is correlated with VEGF-C
expression and lymphangiogenesis in cervical cancer. World J Surg
Oncol. 15:1732017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Jensen MA and Zenklusen JC: A
practical guide to The Cancer Genome Atlas (TCGA). Methods Mol
Biol. 1418:111–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
GTEx Consortium: The Genotype-Tissue
Expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Di J, Rutherford S and Chu C: Review of
the cervical cancer burden and population-based cervical cancer
screening in China. Asian Pac J Cancer Prev. 16:7401–7407. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Wu X and Cheng X: Advances in
diagnosis and treatment of metastatic cervical cancer. J Gynecol
Oncol. 27:e432016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim
SW and Kim YT: Long non-coding RNA HOTAIR is associated with human
cervical cancer progression. Int J Oncol. 46:521–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie
P, Zhou G and Li G: The role of MALAT1 correlates with HPV in
cervical cancer. Oncol Lett. 7:2135–2141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar : PubMed/NCBI
|