Introduction
Esophageal cancer (EC) is one of the most aggressive
and lethal types of malignant tumor, and is considered the sixth
leading cause of cancer-related mortality, and the eighth most
common cancer worldwide (1).
Esophagectomy resection remains the major treatment strategy for
patients with EC that are at the early stage of the disease
(2). However, in order to improve
prognosis, ~half of these patients required chemotherapy due to
systemic or local recurrence (3).
During chemotherapy, drug resistance is a major obstacle. Cisplatin
was discovered in 1845 and is a chemotherapy medication widely used
to treat various types of cancer, including EC (4). Tumor resistance and low sensitivity to
cisplatin frequently occurs in EC, resulting in ineffective
treatment and poor prognosis. Therefore, it is imperative to
identify suitable biomarkers that could be used to overcome the
potential resistance of patients with EC in chemotherapy.
MicroRNAs (miRNAs/miRs) constitute a class of
endogenous, small non-coding RNAs that are involved in regulating
the expression of target mRNAs (5).
Previously, miRNAs were reported to be associated with the response
of chemotherapy in a number of cancer types (6–8). For
instance, miR-106b-3p was reported to act as a potent tumor
promoter participating in tumor progression, development and
sensitivity to chemotherapeutic drugs (9–11). In
EC cells, it was also reported that miR-106b could promote cell
proliferation, migration and invasion by targeting Smad7 (12). Furthermore, it was also demonstrated
in another study that miR-106b could regulate the chemosensitivity
of lung cancer cells to cisplatin (13). However, the exact mechanism of the
contribution of miR-106b-3p in regulating cisplatin sensitivity to
EC remains unknown.
Indeed, miRNAs regulate cellular processes or cancer
development by modulating hundreds of target genes and signaling
pathways (14,15). Recently, a number of studies have
reported the promising target genes of miR-106-3p concerning
cellular networks (16,17). By using the TargetScan online tool,
thousands of potential targets of miR-106-3p were verified,
including protein-glutamine γ-glutamyltransferase E (TGM3). As
commonly known, TGM3 is a tumor suppressor gene in various cancer
types, such as human neck and head cancer and colorectal cancer
(18,19). However, the relationship between
miR-106-3p and TGM3 in regulating the development of EC remains
unknown. Hence, in the present study, the effects of miR-106b-3p on
the sensitivity of KYSE30 cells to cisplatin were investigated by
targeting TGM3, along with the potential molecular mechanism of
action.
Materials and methods
Specimens
A total of 30 pairs of esophageal squamous cell
carcinoma (ESCC) and non-tumor adjacent tissues (>5 cm away from
tumor tissues) were obtained from Baoji Central Hospital (Shaanxi,
China) between June 2015 and November 2017. All patients were
diagnosed with ESCC by a pathological evaluation, and they had not
received biotherapy, chemotherapy or any other treatment prior to
the initiation of the study. Ethical approval was granted by the
Ethics Committee of the Baoji Central Hospital and written informed
consent was received from each patient in accordance with the
institutional guidelines.
Cell line and transfection
The KYSE30 human esophageal cancer cell line (The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences) and the human squamous epithelial cell line Het-1A (BeNa
Culture Collection; Beijing Beina Chunglian Biotechnology Research
Institute) were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc.) at 37°C in the presence of
5% CO2 and penicillin-streptomycin (Sigma-Aldrich; Merck
KGaA). miR-106b-3p mimics, inhibitor and scramble (NC) were
purchased from Shanghai GenePharma Co., Ltd. KYSE30 cells were
seeded into 6-well plates at an initial density of 2×105
cells/well. Cells were transfected with 40 nM NC or miR-106b-3p
inhibitor or miR-106b-3p mimics sequences using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. After 24 h
transfection, subsequent experimentation was conducted and the
transfection efficiency of miR-106b-3p was assessed. The sequences
were as follows: miR-106b-3p mimics, 5′-TAAAGTGCTGACAGTGCAGAT−3′;
miR-106b-3p inhibitor, 5′-AUCUGCACUGUCAGCACUUUA−3′; and NC,
5′-CAGUACUUUUGUGUAGUACAA−3′.
Small interfering (si)RNA, siRNA-1 TGM3 and siRNA-2
TMG3, were designed and purchased from Shanghai GenePharma Co.,
Ltd. KYSE30 cells were seeded into a 6-well plate at a density of
2×105 cells/well. Subsequently, 40 nM siRNA-1 TGM3 and
siRNA-2 TMG3 sequences were transfected into cells with
Lipofectamine 2000 reagent, according to the manufacturer's
instructions. siRNA-1 TGM3 sequence,
5′-TATGAATTCTGTACGGGAGGCCACCAGCGC−3′; siRNA-2 TGM3 sequence,
5′-TATGAATTCTGTACGGGAGGCCACCAGCGC−3′.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.).
Subsequently, cDNA was synthesized using the miScript Reverse
Transcription kit at room temperature for 10 min, and qPCR was
performed using the miScript SYBR-Green PCR kit (both purchased
from Qiagen, Inc.). The U6 small nuclear RNA was used for
normalization. Determination of the relative levels of TGM3
was performed using the TaqMan™ PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.), while glyceraldehyde phosphate
dehydrogenase (GAPDH) functioned as an internal control. The
relative expression levels were calculated according to the
2−ΔΔCq method (20).
Cell proliferation assay
The transfected cells were seeded into a 96-well
plate at a density of 1×104 cells/well and cultivated
for 24 h at 37°C in the presence of 5% CO2. As performed
previously, cells were treated with 4 µmol/l cisplatin for an
additional 24 h (21). Cell
viability was detected using Cell Counting Kit-8 reagent (CCK-8;
Dojindo Molecular Technologies, Inc.), according to the
manufacturer's instructions, and the absorbance was measured at 490
nm with a microplate reader.
Flow cytometry assay
The transfected cells were seeded into a 96-well
plate at a density of 1×105 cells/well. Medium
containing 4 µmol/l cisplatin was added for 48 h at 37°C, in the
presence of 5% CO2. Following trypsinization and washing
with PBS (Thermo Fisher Scientific, Inc.), the cells were double
stained with FITC Annexin V and propidium iodide (PI) for 10 min in
the dark at room temperature. Finally, the apoptotic rate at the
early and late period was determined via a FACScan flow cytometer
and analyzed with CellQuest software version 5.2.1 (both from BD
Biosciences).
Western blot analysis
The transfected cells were isolated and protein was
extracted using RIPA buffer (Beijing Solarbio Science &
Technology Co., Ltd.) containing Protease Inhibitor Cocktail
(Bimake). The concentrations of proteins were measured using BCA
reagent (Solarbio Life Sciences) and 20 µg/lane proteins were
separated by 10% SDS-PAGE (Thermo Fisher Scientific, Inc.) and
transferred to PVDF membranes (Beijing Solarbio Science &
Technology Co., Ltd.). The membranes were blocked with 5% non-fat
milk for 50 min at room temperature and the primary antibodies were
added to the membranes at 37°C overnight. The primary antibodies
were as follows: Rabbit anti-TGM3 (cat. no. NBP1- 86950;
Bio-Techne), rabbit anti-Bcl-2 (cat. no. IMG-5685; Bio-Techne),
rabbit anti-Bax (cat. no. AF820; Bio-Techne), rabbit anti-caspase 3
(cat. no. AF835; Bio-Techne) and rabbit anti-GAPDH (cat. no.
IMG-5143A; Bio-Techne). Subsequently, the HRP-conjugated secondary
antibody IgG H&L (1:1,000; cat. no. ab7090; Abcam) was added
and cultured for another 2 h at room temperature. Finally, the
signals were detected using the electrochemiluminescence assay (BD
Pharmingen; BD Biosciences). GAPDH was used as the endogenous
reference.
Luciferase reporter assay
TargetScan 7.2 software (targetscan.org) was used to
predict the potential binding sequences of miR-106b-3p and TGM3
according to a previous study (22). Het-1A cells were cultured in a
24-well plate at a density of 1×104 cells/well.
Afterwards, the full length of TGM3 was amplified from the cDNA of
Het-1A cells, and inserted into the pGL3-Basic vector (Promega
Corporation) in order to construct wild-type (WT) TGM3.
Subsequently, Het-1A cells were co-transfected with WT and mutant
TGM3 along with NC and miR-106b-3p mimics sequences using
Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) at
37°C, in the presence of 5% CO2. Following 48 h of cell
culture after transfection, luciferase activity was determined
using the dual-luciferase reporter gene assay kit (Promega
Corporation) and normalized to Renilla luciferase enzyme
activity, according to the manufacturer's protocol.
Statistical analysis
SPSS version 22.0 (IBM Corp.) was used to analyze
the data. All data are presented as the mean ± standard deviation.
The differences between groups were analyzed using the Student's
t-test or one-way ANOVA followed by Newman-Keuls post hoc test.
Pearson's correlation analysis was performed to measure the
correlation between miR-106b-3p and TGM3 expression levels.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Dysregulation of miR-106b-3p and TGM3
in ESCC tissues
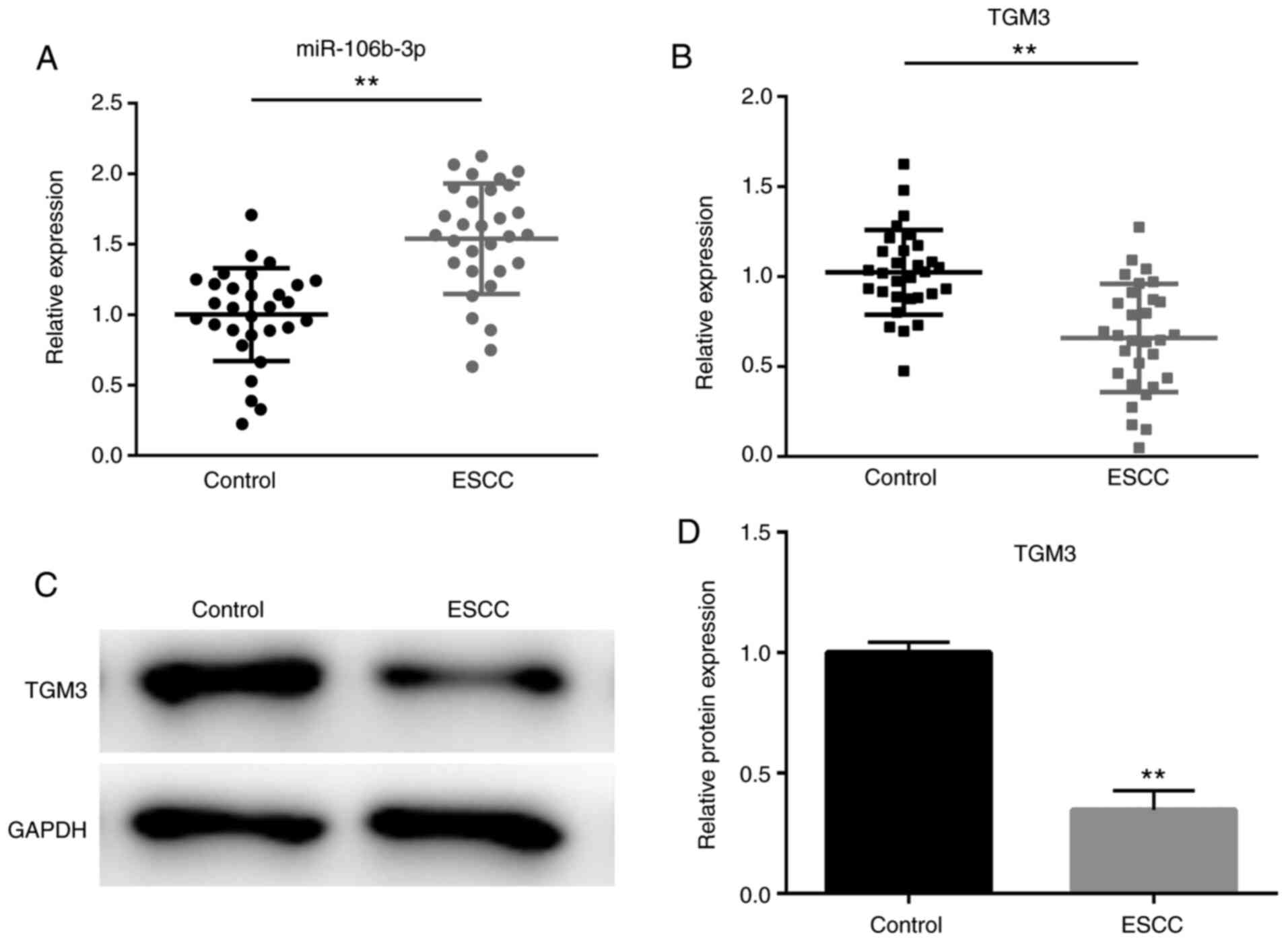
As shown in Fig. 1A,
the expression levels of miR-106b-p were significantly increased in
the ESCC tissues compared with the non-tumor adjacent tissues
(P<0.01); whereas the mRNA and protein expression levels of TGM3
were significantly downregulated in ESCC tissues compared with
those in non-tumor adjacent tissues (P<0.01; Fig. 1B-D).
TGM3 is a downstream target of
miR-106b-3p
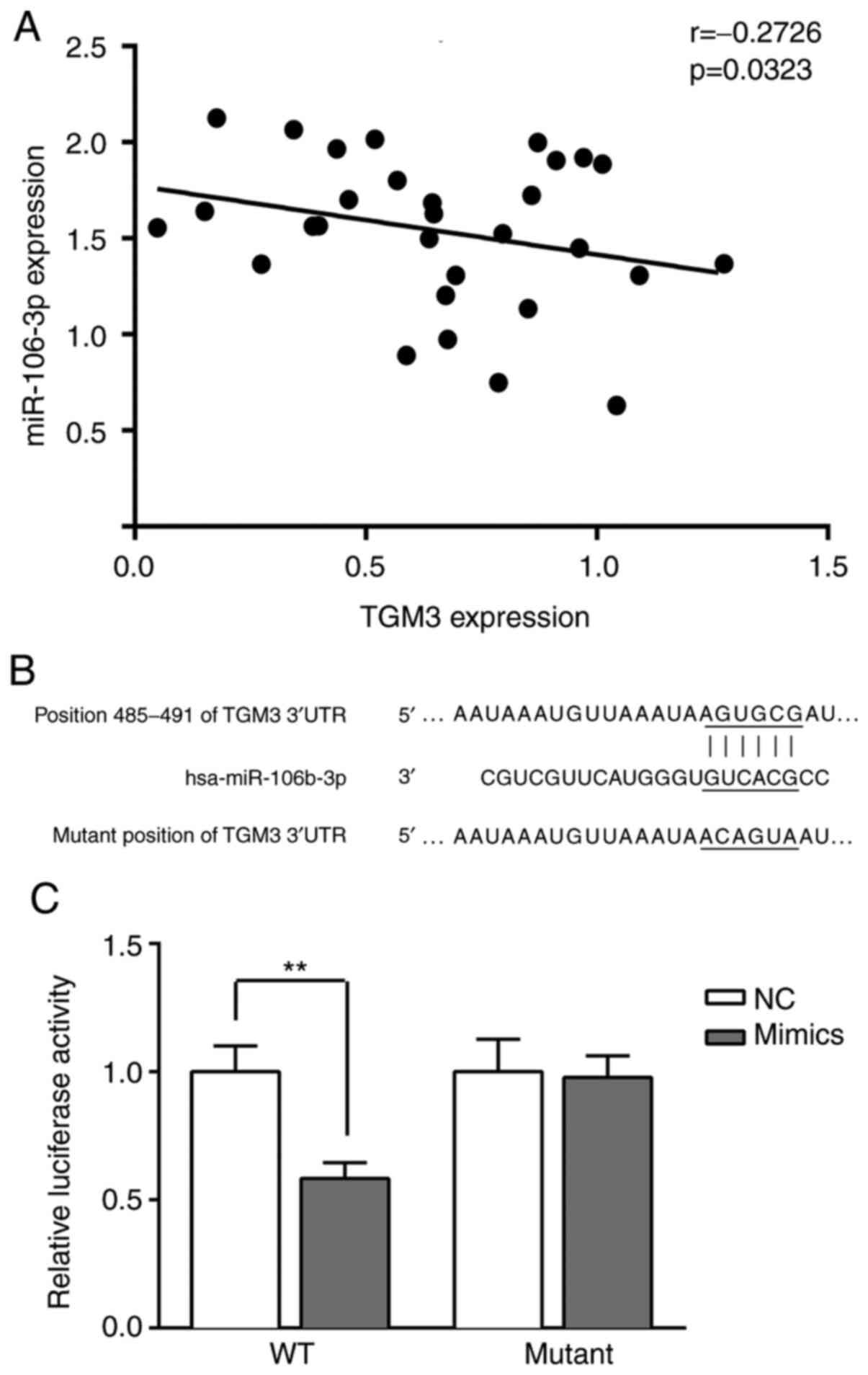
As shown in Fig. 2A,
TGM3 expression was negatively correlated with miR-106-3p
(r=−0.2726, P=−0.0323). Furthermore, the sequences of TGM3
3′-untranslated region (UTR) contained the putative miR-106b-3p
binding sites as predicted by TargetScan (Fig. 2B). Following transfection with WT
and mutant TGM3 3′-UTR plasmids, luciferase activity was
significantly decreased in the cells transfected with miR-106b-3p
mimics (P<0.01; Fig. 2C).
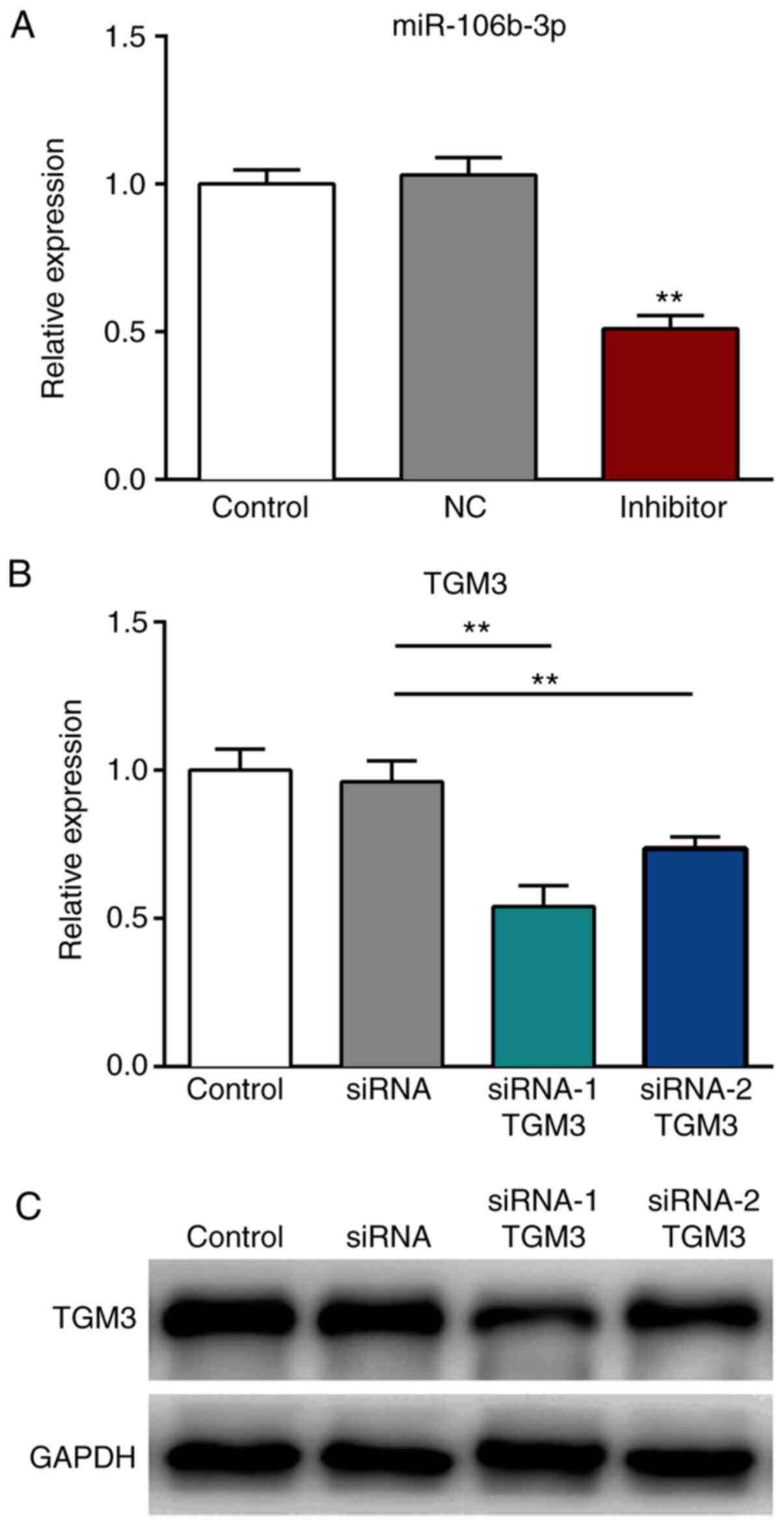
Transfection efficiency
The expression levels of miR-106b-3p were
significantly decreased following transfection with the miR-106b-3p
inhibitor, compared with those transfected with the NC (P<0.01;
Fig. 3A). Furthermore, the mRNA and
protein expression levels of TMG3 were significantly reduced
following transfection with siRNA-1 TMG3 or siRNA-2 TMG3 sequences,
compared with those noted in the siRNA group (P<0.01; Fig. 3B and C).
Downregulation of miR-106b-3p
suppresses ESCC cell viability following treatment with
cisplatin
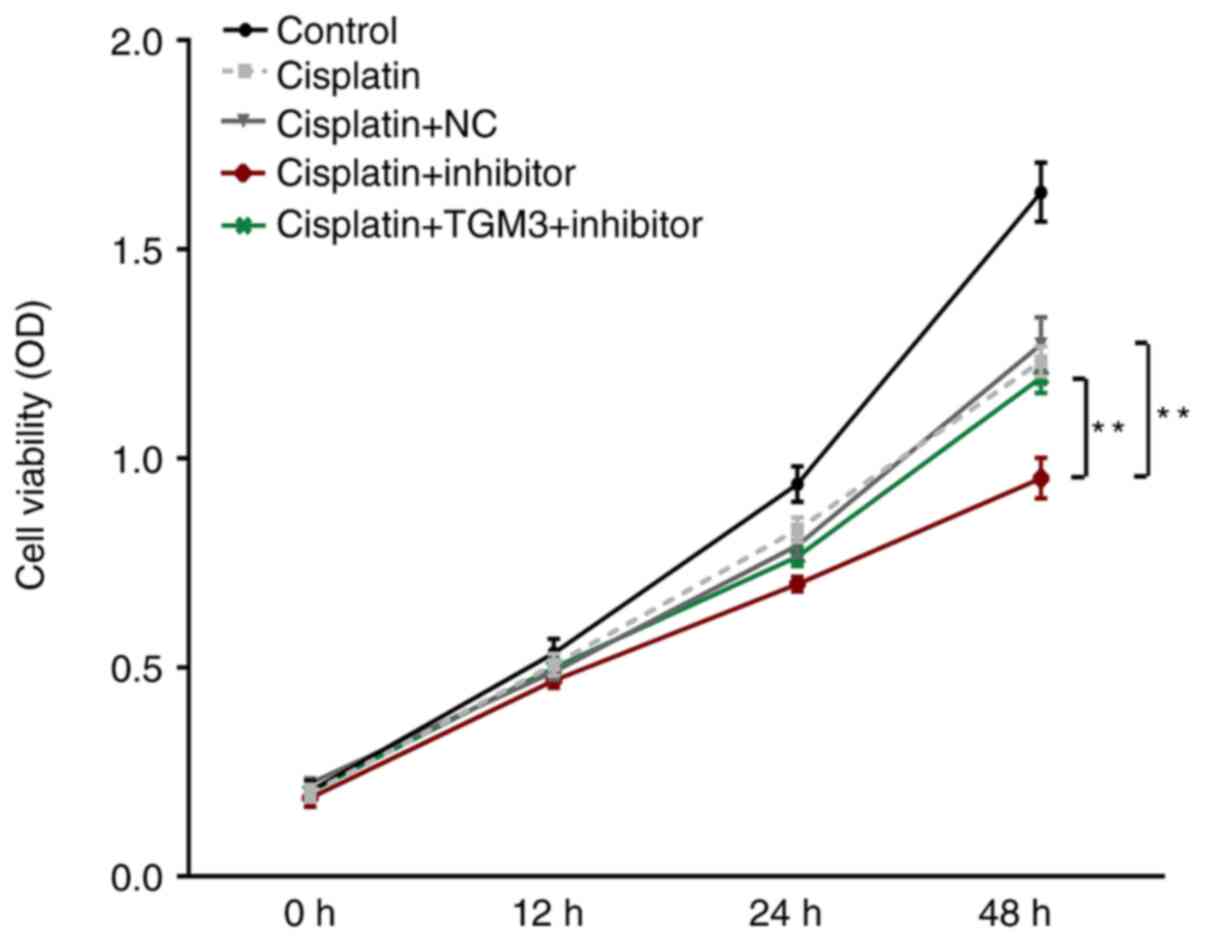
Following treatment with cisplatin, cell viability
was significantly suppressed by transfection with the miR-106b-3p
inhibitor (Fig. 4). However, this
suppressive effect could be reversed by co-transfection with TMG3
siRNA (P<0.01).
Downregulation of miR-106b-3p promotes
ESCC cell apoptosis following treatment with cisplatin
The induction rate of apoptosis was significantly
increased by transfection with the miR-106b-3p inhibitor. However,
this effect was reversed by co-transfection with TMG3 siRNA
(P<0.01; Fig. 5A-F).
Effects of miR-106b-3p on the
expression levels of apoptosis-related proteins
Following treatment of the cells with cisplatin, the
expression levels of Bcl-2 were decreased, whereas the expression
levels of TGM3, Bax and caspase-3 were increased following
transfection with the miR-106b-3p inhibitor. This variation could
be reversed by co-transfection with TMG3 siRNA (Fig. 6).
Discussion
TGM3 is a member of the Ca2+-dependent enzyme family
and is hypothesized to be involved in the formation of the
cornified cell envelope and shape determination (23,24).
Previously, TGM3 was reported to be involved in human head and neck
cancer development (25,26). A number of previous studies revealed
that dysregulation of TGM3 was associated with tumorigenesis and
development of a variety of human cancer types, including basal
cell carcinoma, laryngeal carcinoma, oral squamous cell carcinoma
and ESCC (27–29). It was also found that TGM3
expression was significantly reduced in ESCC tissues (30). In addition, it was reported by
another study that TGM3 could suppress tumor growth via the NF-κB
signaling pathway in EC (31). More
importantly, TGM3 was identified as a potential prognostic
indicator in ESCC, suggesting that TGM3 may be a novel target in
ESCC treatment (32). In the
present study, TGM3 was downregulated in EC tissues, which was in
accordance with the results reported in previous studies. However,
the underlying molecular mechanisms of TMG3 in ESCC cisplatin
sensitivity still remains unclear.
Acquired drug resistance and low sensitivity to
chemotherapy have become major obstacles of successful cancer
treatment. Growing evidence has highlighted the important roles of
certain proteins in regulating sensitivity of cancer cells to
chemotherapeutic agents. Accumulating evidence has reported that
various miRNAs are involved in regulating cisplatin
chemosensitivity in human cancer types, including ESCC. For
instance, miR-218, miR-145, miR-338-5p and miR-125a-5p (33–36).
Jiao et al (11) reported
that miR-106b could regulate 5-fluorouracil resistance by targeting
zinc finger and BTB domain-containing protein 7A in
cholangiocarcinoma. Yu et al (13) proposed that miR-106b could enhance
the sensitivity of A549/DDP cells to cisplatin by targeting
polycystic kidney disease-2. Fang et al (37) revealed that miR-106b played a
crucial role in causing gemcitabine resistance of pancreatic cancer
as well. The present study further confirmed that inhibition of
miR-106b-3p via transfection could increase chemosensitivity of
KYSE30 cells to cisplatin in vitro, most likely by targeting
TGM3. To the best of our knowledge, this is the first study to
investigate the underlying mechanism of miR-106b-3p in regulating
cisplatin sensitivity of EC cell lines via TGM3 targeting.
Additional work must focus on the detailed molecular mechanisms by
investigating how miR-106b-3p and TGM3 affect chemoresistance in
vivo. Moreover, Bcl-2, Bax and Caspase-3 were chosen as the
three typical apoptosis-related factors to verify the effects of
TGM3 and miR-106b-3p on apoptosis. In the future, we will conduct
immunohistochemical staining of apoptotic markers.
In conclusion, the present study validated that
downregulation of miR-106b-3p may increase cisplatin sensitivity in
KYSE30 cell lines by targeting TGM3. Therefore, miR-106b-3p may
function as a promising sensitizer of cisplatin therapy in patients
with EC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YoZ, YuZ, XL and YS drafted the manuscript. NW, MC
and ZY contributed to the manuscript revision. YoZ, YuZ, XL and YS
designed the study. XL, NW, MC and ZY performed the experiments.
YoZ, XL, YS, NW, MC and ZY contributed to the data acquisition and
supervision. YuZ, XL, MC and ZY contributed to data analysis and
interpretation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval was granted by the Ethics Committee
of the Baoji Central Hospital (approval no. 2015010398) and written
informed consent was received from each patient in accordance with
the institutional guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim JJ, Park JK and Moon SW: Usefulness of
positron emission tomography-computed tomography in pre-operative
evaluation of intra-thoracic esophageal cancer. Thorac Cancer.
6:687–694. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miyata H, Yamasaki M, Kurokawa Y,
Takiguchi S, Nakajima K, Fujiwara Y, Konishi K, Mori M and Doki Y:
Survival factors in patients with recurrence after curative
resection of esophageal squamous cell carcinomas. Ann Surg Oncol.
18:3353–3361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akutsu Y and Matsubara H: Chemotherapy and
surgery for T4 esophageal cancer in Japan. Surg Today.
45:1360–1365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martens-Uzunova ES, Olvedy M and Jenster
G: Beyond microRNA-novel RNAs derived from small non-coding RNA and
their implication in cancer. Cancer Lett. 340:201–211. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu C, Shan Z, Li C and Yang L: miR-129
regulates cisplatin-resistance in human gastric cancer cells by
targeting P-gp. Biomed Pharmacother. 86:450–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mutlu M, Raza U, Saatci O, Eyupoglu E,
Yurdusev E and Sahin O: miR-200c: A versatile watchdog in cancer
progression, EMT, and drug resistance. J Mol Med (Berl).
94:629–644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lang B, Shang C and Meng L: Targeted
silencing of S100A8 gene by miR-24 increase chemotherapy
sensitivity of endometrial carcinoma cells to paclitaxel. Med Sci
Monit. 22:1953–1958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun C, Yao X, Jiang Q and Sun X: miR-106b
targets DAB2 to promote hepatocellular carcinoma cell proliferation
and metastasis. Oncol Lett. 16:3063–3069. 2018.PubMed/NCBI
|
|
10
|
Bu W, Wang Y and Min X: microRNA-106b
promotes the proliferation, migration and invasion of
retinoblastoma cells by inhibiting the expression of ZBTB4 protein.
Exp Ther Med. 16:4537–4545. 2018.PubMed/NCBI
|
|
11
|
Jiao D, Yan Y, Shui S, Wu G, Ren J, Wang Y
and Han X: miR-106b regulates the 5-fluorouracil resistance by
targeting Zbtb7a in cholangiocarcinoma. Oncotarget. 8:52913–52922.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai F, Liu T, Zheng S, Liu Q, Yang C, Zhou
J, Chen Y, Shehidin I and Lu X: miR-106b promotes migration and
invasion through enhancing EMT via downregulation of Smad 7 in
Kazakh's esophageal squamous cell carcinoma. Tumor Biol.
37:14959–14604. 2016. View Article : Google Scholar
|
|
13
|
Yu S, Qin X, Chen T, Zhou L, Xu X and Feng
J: microRNA-106b-5p regulates cisplatin chemosensitivity by
targeting polycystic kidney disease-2 in non-small-cell lung
cancer. Anticancer Drugs. 28:852–860. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao GD, Zhang YF, Chen P and Ren XB:
MicroRNA-544 promotes colorectal cancer progression by targeting
forkhead box O1. Oncol Lett. 15:991–997. 2018.PubMed/NCBI
|
|
15
|
Long XH, Shi Y, Ye P, Guo J, Zhou Q and
Tang YT: MicroRNA-99a suppresses breast cancer progression by
targeting FGFR3. Front Oncol. 9:14732019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni SJ, Weng WW, Xu MD, Wang QF, Tan C, Sun
H, Wang L, Huang D, Du X and Sheng WQ: miR-106b-5p inhibits the
invasion and metastasis of colorectal cancer by targeting CTSA.
Onco Targets Ther. 11:3835–3845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou YT, Tian WH, Zhang M, Ren TH, Sun GR,
Jiang RR, Han RL, Kang XT and Yan FB: Transcriptom analysis
revealed regulation of dexamethasone induced microRNAs in chicken
thymus. J Cell Biochem. 120:6570–6579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu XB, Cao W, Wang X, Zhang JJ, Lv ZJ, Qin
X, Wu YD and Chen WT: TGM3, a candidate tumor suppressor gene,
contributes to human head and neck cancer. Mol Cancer. 12:1512013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng Y, Ji D, Huang Y, Ji B, Zhang Y, Li
J, Peng W, Zhang C, Zhang D, Sun Y and Xu Z: TGM3 functions as a
tumor suppressor by repressing epithelial-to-mesenchymal transition
and the PI3K/AKT signaling pathway in colorectal cancer. Oncol Rep.
43:864–876. 2020.PubMed/NCBI
|
|
20
|
Livak KJ and Schimittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J, Wang L, Du X, Sun Q, Wang Y, Li M,
Zang W, Liu K and Zhao G: α-solanine enhances the chemosensitivity
of esophageal cancer cells by inducing microRNA-138 expression.
Oncol Rep. 39:1163–1172. 2018.PubMed/NCBI
|
|
22
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steven AC and Steinert PM: Protein
composition of cornified cell envelopes of epidermal keratinocytes.
J Cell Sci. 107:693–700. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo A, Kong J, Hu G, Liew CC, Xiong M,
Wang X, Ji J, Wang T, Zhi H, Wu M and Liu Z: Discovery of
CA2+-relevant and differentiation-associated genes downregulated in
esophageal squamous cell carcinoma using cDNA microarray.
Oncotarget. 23:1291–1299. 2004.PubMed/NCBI
|
|
25
|
Wu X, Cao W, Wang X, Zhang J, Lv Z, Qin X,
Wu Y and Chen W: TGM3, a candidate tumor suppressor gene,
contributes to human head and neck cancer. Mol Cancer. 12:1512013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X, Wang R, Jiao J, Li S, Yu J, Yin Z,
Zhou L and Gong Z: Transglutaminase 3 contributes to malignant
transformation of oral leukoplakia to cancer. Int J Biochem Cell
Biol. 104:34–42. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He G, Zhao Z, Fu W, Sun X, Xu Z and Sun K:
Study on the loss of heterozygosity and expression of
transglutaminase 3 gene in laryngeal carcinoma. Zhonghua Yi Xue Yi
Chuan Xue Za Zhi. 19:120–123. 2002.(In Chinese). PubMed/NCBI
|
|
28
|
Negishi A, Masuda M, Ono M, Honda K,
Shitashige M, Satow R, Sakuma T, Kuwabara H, Nakanishi Y, Kanai K,
et al: Quantitative proteomics using formalin-fixed
paraffin-embedded tissues of oral squamous cell carcinoma. Cancer
Sci. 100:1605–1611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stacey SN, Sulem P, Gudbjartsson DF,
Jonasdottir A, Thorleifsson G, Gudjonsson SA, Masson G, Gudmundsson
J, Sigurgeirsson B, Benediktsdottir KR, et al: Germline sequence
variants in TGM3 and RGS22 confer risk of basal cell carcinoma. Hum
Mol Genet. 23:3045–3053. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu W, Yu ZC, Cao WF, Ding F and Liu ZH:
Functional studies of a novel oncogene TGM3 in human esophageal
squamous cell carcinoma. World J Gastroenterol. 12:3929–3932. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li W, Zhang Z, Zhao W and Han N:
Transglutaminase 3 protein modulated human esophageal cancer cell
growth by targeting the NF-κB signaling pathway. Oncol Rep.
36:1723–1730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uemura N, Nakanishi Y, Kato H, Saito S,
Nagino M, Hirohashi S and Kondo T: Transglutaminase 3 as a
prognostic biomarker in esophageal cancer revealed by proteomics.
Int J Cancer. 124:2106–2115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian H, Hou L, Xiong YM, Huang JX, She YJ,
Bi XB and Song XR: miR-218 suppresses tumor growth and enhances the
chemosensitivity of esophageal squamous cell carcinoma to
cisplatin. Oncol Rep. 33:981–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng TL, Li DP, He ZF and Zhao S: miR-145
sensitizes esophageal squamous cell carcinoma to cisplatin through
directly inhibiting PI3K/AKT signaling pathway. Cancer Cell Int.
19:2502019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin WC, Chen LH, Hsieh YC, Yang PW, Lai
LC, Chuang EY, Lee JM and Tsai HM: miR-338-5p inhibits cell
proliferation, colony formation, migration and cisplatinresistance
in esophageal squamous cancer cells by targeting FERMT2.
Carcinogenesis. 40:883–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Y, Ma K, Yang S, Zhang X, Wang F,
Zhang X, Liu H and Fan Q: MicroRNA-125a-5p enhances the sensitivity
of esophageal squamous cell carcinoma cells to cisplatin by
suppressing the activation of the STAT3 signaling pathway. Int J
Oncol. 53:644–658. 2018.PubMed/NCBI
|
|
37
|
Fang Y, Zhou W, Rong Y, Kuang T, Xu X, Wu
W, Wang D and Lou W: Exosomal miRNA-106b from cancer-associated
fibroblast promotes gemcitabine resistance in pancreatic cancer.
Exp Cell Res. 383:1115432019. View Article : Google Scholar : PubMed/NCBI
|