Introduction
Hepatocellular carcinoma (HCC) is a common type of
cancer that originates from hepatocytes and accounts for 70–90% of
primary liver cancer cases worldwide (1). Approximately 500,000 cases of HCC are
diagnosed globally every year, and HCC is the fifth most common
type of cancer in men and the eighth most common in women (2). If HCC is not treated, patients succumb
quickly, and the global 5-year survival rate is only 5% (3–5).
Therefore, it is necessary to elucidate the specific mechanism
underlying the occurrence and development of HCC, and to provide a
scientific basis for the diagnosis and treatment of HCC.
Cell division cycle 20 (CDC20) is a cell cycle
checkpoint regulator (6). CDC20 and
another regulator E-cadherin can directly bind and activate the
anaphase-promoting complex (APC), which has an important role in
the process of cells entering and exiting mitosis (7). Recently, an increasing number of
studies have shown that CDC20 is upregulated in various types of
human malignant tumor, including pancreatic ductal adenocarcinoma,
breast cancer, glioblastoma and gastric cancer; in addition, CDC20
may be closely associated with the poor prognosis of various types
of cancer (8–11). A previous study reported that
overexpression of CDC20 may promote the resistance of
castration-resistant prostate cancer cell lines to docetaxel in a
Bim-dependent manner (12). Zhang
et al (13) reported that
reducing the expression of CDC20 may inhibit the expression of
CD44+ prostate cancer stem cells. Moreover, CDC20 has
been shown to promote the degradation of AXIN1, reduce the
phosphorylation of β-catenin and promote the nuclear translocation
of β-catenin, thus enhancing the self-renewal ability of
CD44+ prostate stem cells (13). However, to the best of our
knowledge, the association of CDC20 with HCC has only been
determined through bioinformatics analysis, and no research has
been conducted on the association of CDC20 with cell function and
related molecular signaling pathways in HCC (13). Therefore, the present study aimed to
assess the specific mechanism underlying the effects of CDC20 on
HCC.
Epithelial-mesenchymal transition (EMT) refers to
the process by which cells transition from an epithelial phenotype
to a mesenchymal phenotype under specific physiological or
pathological conditions. Notably, EMT has been confirmed to play an
important role in HCC (14). The
process of EMT causes loss of expression of E-cadherin, claudin,
occludin and other connecting molecules in epithelial cells,
destroys cell polarity and promotes some lytic enzymes that are
involved in degradation of the extracellular matrix and basement
membrane (15). Increased
expression of matrix metalloproteinases, which occurs during EMT,
destroys the histological barrier of tumor cell invasion,
facilitating the separation of tumor cells from the primary tumor,
invasion and metastasis (16).
These findings indicated that EMT may serve an important role in
promoting HCC metastasis.
The present study aimed to investigate the effect of
CDC20 on the poor prognosis of patients with HCC and the biological
functions of HCC cells. In addition, the molecular mechanism by
which CDC20 affects malignant progression of HCC through EMT was
investigated, in order to provide potential strategies for the
prognosis and treatment of patients with HCC.
Materials and methods
Patients and tumor specimens
A total of 71 HCC and paired adjacent tissue
specimens were collected from Department of Hepatocellular Surgery,
Affiliated Hospital of North Sichuan Medical College (Nanchong,
China) between January 2010 and May 2013. The mean age of the
patients was 61 years (age range, 30–77 years), and the cohort
consisted of 30 men and 41 women (Table
I). Patients with HCC were followed up for 90 months after
surgery, in order to evaluate survival rate (patients were followed
up by phone every 4 months, and the total follow-up period was 90
months). All procedures involving human participants were approved
by the Ethics Committee of Affiliated Hospital of North Sichuan
Medical College and were conducted in accordance with the
Declaration of Helsinki. The corresponding clinicopathological data
were obtained from Affiliated Hospital of North Sichuan Medical
College. All patients who participated in the experiment provided
written informed consent.
 | Table I.Association between CDC20 expression
and clinicopathological features of patients with hepatocellular
carcinoma. |
Table I.
Association between CDC20 expression
and clinicopathological features of patients with hepatocellular
carcinoma.
|
|
| CDC20
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases | Low (n=29) | High (n=42) | P-value |
|---|
| Age, years |
|
|
| 0.801 |
|
<50 | 33 | 14 | 19 |
|
|
≥50 | 38 | 15 | 23 |
|
| Sex |
|
|
| 0.901 |
|
Male | 30 | 12 | 18 |
|
|
Female | 41 | 17 | 24 |
|
| TNM stage |
|
|
| 0.163 |
|
I/II | 37 | 18 | 19 |
|
|
III/IV | 34 | 11 | 23 |
|
| Tumor size, cm |
|
|
| 0.003 |
| ≤5 | 29 | 18 | 11 |
|
|
>5 | 42 | 11 | 31 |
|
| HBsAg |
|
|
| 0.233 |
|
Positive | 45 | 16 | 29 |
|
|
Negative | 26 | 13 | 13 |
|
| AFP, ng/ml
(17) |
|
|
| 0.629 |
|
≤20 | 27 | 12 | 15 |
|
|
>20 | 44 | 17 | 27 |
|
| Liver
cirrhosis |
|
|
| 0.541 |
|
Present | 47 | 18 | 29 |
|
|
Absent | 24 | 11 | 13 |
|
| Number of
tumors |
|
|
| 0.002 |
|
Single | 36 | 21 | 15 |
|
|
Multiple (≥2) | 35 | 8 | 27 |
|
| Vascular
invasion |
|
|
| 0.030 |
|
Present | 33 | 9 | 24 |
|
|
Absent | 38 | 20 | 18 |
|
The detailed inclusion criteria were as follows: i)
HCC was confirmed by postoperative pathology, and the
histopathological diagnosis was clear; ii) patients were diagnosed
with HCC for the first time without distant metastasis; iii)
patients had not received any treatment prior to surgery; iv) no
other serious malignant disease had been diagnosed; v) the
clinical, pathological and surgical data were complete; vi) the
follow-up information was complete and available.
Immunohistochemical staining
A total of 71 HCC tissues were fixed in 10% formalin
for 12 h at room temperature, embedded in paraffin and cut into
4-µm sections. The tissue sections were then used to generate
tissue microarray cores (1.5-mm diameter). The microarrays were
deparaffinized in xylene I for 15 min and xylene II for 15 min at
room temperature, and were rehydrated in a graded ethanol series
(100, 95, 80 and 75% ethanol, 5 min each). Subsequently, the
microarrays were incubated with 3% H2O2 for
30 min at 37°C and with 5% goat serum (Origene Technologies, Inc.)
for 15 min at 37°C to block non-specific binding. The microarrays
were then incubated with a monoclonal anti-CDC20 antibody (1:2,000;
cat. no. ab215908; Abcam) at 4°C overnight. Subsequently, the
sections were incubated with a secondary biotin-labeled IgG
antibody (1:100; cat. no. SAP-9100; Origene Technologies, Inc.) at
37°C for 30 min. The visualization signal was detected using
3,3′-diaminobenzidine (Boster Biological Technology) at room
temperature for 10 sec. Slides were visualized using a light
microscope (magnification, ×100; Zeiss AG).
The immunostaining results were analyzed using the
following scoring parameters: Staining intensity [according to the
color development degree of the positive marker (range, 0–3): No
staining, negative (score, 0); light yellow staining, weak (score,
1); brown-yellow staining, moderate (score, 2); and brown black
staining, strong (score, 3)] and percentage of positive cells
(range, 0–4: 0, <5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; and 4,
76–100%). Total score was determined by adding the staining
intensity score to the percentage of positive cells score. Slides
with a total score <4 were defined as low CDC20 expression,
whereas slides with a score ≥4 were defined as high CDC20
expression (17).
Cell lines and transfection
Hep3B, MHCC-97H, MHCC-97L and Huh-7 cell lines, and
the THLE2 cell line were obtained from the Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. The HCC cell
lines (Hep3B, MHCC-97H, MHCC-97L and Huh-7) were cultured in DMEM
supplemented with 10% FBS, whereas the normal liver cell line
(THLE2) was cultured in Bronchial Epithelial Cell Growth Medium
supplemented with 10% FBS (all from Gibco; Thermo Fisher
Scientific, Inc.); all cells were maintained in a humidified
incubator containing 5% CO2 at 37°C.
Hep3B and Huh-7 cell lines (2×105) were
transfected with 50 nM small interfering (si)RNA against CDC20
(si-CDC20) (Guangzhou RiboBio Co., Ltd.). The sequences for the
siRNAs were as follows: si-CDC20 forward,
5′-GGAUUGGAGUUCUGGGAAUTT-3′ and reverse,
5′-AUUCCCAGAACUCCAAUCCTT-3′; and siRNA-negative control (si-NC;
scrambled siRNA) forward, 5′-UUCUUCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The cells were transiently transfected
using Lipofectamine® 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc) and the transfection was maintained for
>24 h. Subsequent experiments were performed 72 h after
transfection.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Briefly, total RNA was extracted from HCC tissues,
adjacent tissues and cell lines using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RT to cDNA was performed using a
PrimeScript RT reagent (Takara Bio, Inc.), and qPCR was performed
using SYBR Premix Ex Taq II (Takara Bio, Inc.) and a LightCycler
system (Roche Diagnostics). The temperature protocol for RT was as
follows: 25°C for 10 min, followed by 45°C for 30 min and 80°C for
30 min. The primer sequences were as follows: CDC20 forward,
5′-GACCACTCCTAGCAAACCTGG-3′ and reverse, 5′-GGGCGTCTGGCTGTTTTCA-3′;
β-actin forward, 5′-GGATGCAGAAGGAGATCACTG-3′ and reverse,
5′-CGATCCACACGGAGTACTTG-3′. The following thermocycling conditions
were used for qPCR: Initial denaturation at 95°C for 10 min,
followed by 40 cycles at 90°C for 30 sec and 60°C for 30 sec, and a
final extension step at 72°C for 10 min. β-actin was used as the
internal reference gene, and the mRNA expression levels of CDC20
were analyzed using the 2−ΔΔCq method (18).
Western blotting
Total protein was extracted from cell lines
(1×106) and tissues using a 100:1 mixed solution of RIPA
buffer (Beyotime Institute of Biotechnology) and PMSF (Beyotime
Institute of Biotechnology). The protein concentrations were
quantified using the Bradford protein assay (Bio-Rad Laboratories,
Inc.). Equal concentrations of protein (30 µg/lane) were separated
by SDS-PAGE on a 10% gel and transferred to PVDF membranes. The
membranes were then blocked with 5% nonfat dried milk at room
temperature for 1 h and were probed at 4°C overnight with primary
antibodies against CDC20 (1:2,000; cat. no. ab183479;), N-cadherin
(1:5,000; cat. no. ab76011), vimentin (1:5,000; cat. no. ab92547),
E-cadherin (1:100; cat. no. ab40772), Ki-67 (1:5,000; cat. no.
ab92742) and β-actin (1:5,000; cat. no. ab8227) (all from Abcam).
After washing, the membranes were incubated with the appropriate
HRP-conjugated secondary antibodies (1:5,000; cat. no. ab6721;
Abcam) at room temperature for 1 h. Finally, immunoreactive protein
bands were visualized using an enhanced chemiluminescence solution
(EMD Millipore) and a ChemiDoc Imaging system (Bio-Rad
Laboratories, Inc.). β-actin was used as the internal control.
Protein expression was semi-quantified using Quantity One version
4.6 software (Bio-Rad Laboratories, Inc.).
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA) (19) database (http://gepia.cancer-pku.cn/index.html)
was used to analyze the expression levels of CDC20 in unpaired
non-tumor liver tissue samples and HCC tissue samples. The
threshold settings were set to P<0.01, fold change ≥1 (19).
Cell immunofluorescence
Hep3B and Huh-7 cells were seeded (1×105
cells) and cultured on coverslips. After si-CDC20 transfection,
Hep3B and Huh-7 cells were fixed with 4% paraformaldehyde at room
temperature for 30 min, and permeabilized with 0.25% Triton X-100
solution at room temperature for 15 min. Subsequently, Hep3B and
Huh-7 cells were washed with PBS and then blocked with 5% bovine
serum albumin (OriGene Technologies, Inc.) at room temperature for
1 h. The coverslips were incubated with anti-CDC20 (1:500; cat. no.
ab215908; Abcam) overnight at 4°C. After washing with PBS, cells
were incubated with an appropriate goat anti-rabbit secondary
antibody (1:100; cat. no. ZF-0311; OriGene Technologies, Inc.) at
37°C for 30 min and DAPI. The slides were imaged using an inverted
fluorescence microscope and results were recorded.
Cell Counting Kit-8 (CCK-8) assay
Hep3B and Huh-7 cells (2×103 cells/well)
were transfected with si-CDC20 or si-NC for 48 h and cultured in a
96-well plate for 24, 48 and 72 h. The CCK-8 assay (Dojindo
Molecular Technologies, Inc.) was used to detect cell proliferation
according to the manufacturer's protocol.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Hep3B and Huh-7 cells (1×103/well) were
transfected with si-CDC20 or si-NC for 48 h and cultured in a
96-well plate. Hep3B and Huh-7 cells were incubated with 50 µM EdU,
100 µl 1X ApolloR reaction cocktail (cat. no. 100T; Guangzhou
RiboBio Co., Ltd.) and 100 µl 1X Hoechst 33342 for 30 min at 37°C.
Hep3B and Huh-7 cell proliferation was analyzed by counting the
mean number of cells in three fields for each sample using a
fluorescence microscope (magnification, ×100).
Cell migration and invasion
assays
After si-CDC20 transfection, Hep3B and Huh-7 cells
were resuspended in high-glucose DMEM containing 1% FBS and seeded
into the upper Transwell chamber at a density of 1×105
cells/well. Transwell chambers (Corning, Inc.) were used to detect
cell invasion and migration. For migration assays, 500 µl
high-glucose DMEM containing 10% FBS was added to the lower
chamber. After incubation for 12 h at 37°C, the Transwell chambers
were fixed with 4% methanol at room temperature for 30 min and
stained with 0.1% crystal violet for 20 min at room temperature.
For the invasion assay, the inserts were precoated with Matrigel (1
mg/ml). After incubation for 16 h at 37°C, the Transwell chambers
were fixed with 4% paraformaldehyde at room temperature for 30 min
and stained with 0.5% crystal violet at room temperature for 20
min. Finally, invasive and migratory cells were counted under an
inverted optical microscope (Olympus DP70 light microscope; Olympus
Corporation; magnification, ×200).
Statistical analysis
All experiments were performed in triplicate.
Statistical analyses were performed using SPSS version 22.0
software (IBM Corp.) and GraphPad Prism version 8.0 software
(GraphPad Software, Inc.). The significant difference in CDC20
expression between HCC tissues and adjacent normal tissues was
assessed using paired t-test. The relationship between CDC20
expression and clinicopathological characteristics was analyzed by
χ2 test. Overall survival analysis was performed using
the Kaplan-Meier method and the log-rank test. Univariate and
multivariate Cox regression analyses were used to analyze the
prognostic significance of CDC20. Statistical differences among
multiple groups were analyzed by one-way ANOVA, followed by Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
CDC20 is highly expressed in HCC
The present study analyzed the expression levels of
CDC20 in HCC using the GEPIA database; the results demonstrated
that the mRNA expression levels of CDC20 were significantly higher
in HCC tissues compared with those in paired adjacent tissues
(P<0.05; Fig. 1A). This finding
was confirmed in collected HCC samples; the mRNA expression levels
of CDC20 were higher in the HCC samples compared with those in the
paired adjacent tissues (P<0.05; Fig. 1B). Furthermore, the expression
levels of CDC20 were higher in HCC cell lines compared with those
in a normal liver cell line (THLE2) (P<0.05; Fig. 1C).
CDC20 is associated with
clinicopathological features of patients with HCC
Immunohistochemistry was used to detect the protein
expression levels of CDC20 in HCC; the results indicated that high
CDC20 expression was observed in 59.2% of HCC samples (41/71)
(Fig. 2A). Furthermore, the
association between CDC20 expression and the clinicopathological
characteristics of patients with HCC was assessed. High CDC20
expression was revealed to be positively associated with tumor size
(P=0.003), number of tumors (P=0.002) and vascular invasion
(P=0.030) (Table I).
CDC20 overexpression is associated
with poor prognosis in HCC
The association between CDC20 expression and the
poor prognosis of patients with HCC was analyzed using the 71
collected clinical HCC samples; the results revealed that high
CDC20 expression was associated with significantly shorter overall
patient survival compared with low CDC20 expression (P<0.01;
Fig. 2B). In addition, univariate
analysis indicated that tumor size (HR=1.863; P=0.011),
multiplicity (HR=1.366; P=0.036) and CDC20 expression (HR=1.735;
P=0.011) were significantly associated with overall survival in HCC
(Table II). Multivariate analysis
also suggested that tumor size (HR=1.903; P=0.017), multiplicity
(HR=1.997; P=0.031) and CDC20 expression level (HR=2.036; P=0.007)
were independent prognostic factors for overall survival in HCC
(Table II).
 | Table II.Univariate and multivariate analyses
of risk factors for overall survival in patients with
hepatocellular carcinoma. |
Table II.
Univariate and multivariate analyses
of risk factors for overall survival in patients with
hepatocellular carcinoma.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
| 0.769
(0.423–1.304) | 0.534 |
|
|
|
<50 | 33 |
|
|
|
|
|
≥50 | 38 |
|
|
|
|
| Sex |
| 0.506
(0.347–0.863) | 0.364 |
|
|
|
Male | 30 |
|
|
|
|
|
Female | 41 |
|
|
|
|
| TNM stage |
| 1.338
(0.450–4.338) | 0.673 |
|
|
|
I/II | 37 |
|
|
|
|
|
III/IV | 34 |
|
|
|
|
| Tumor size, cm |
| 1.863
(1.035–3.713) | 0.011 | 1.903
(1.156–3.066) | 0.017 |
| ≤5 | 29 |
|
|
|
|
|
>5 | 42 |
|
|
|
|
| HBsAg |
| 1.304
(0.408–4.368) | 0.364 |
|
|
|
Positive | 45 |
|
|
|
|
|
Negative | 26 |
|
|
|
|
| AFP, ng/ml
(17) |
| 0.783
(0.425–1.557) | 0.403 |
|
|
|
≤20 | 27 |
|
|
|
|
|
>20 | 44 |
|
|
|
|
| Liver
cirrhosis |
| 0.933
(0.489–1.806) | 0.869 |
|
|
|
Presence | 47 |
|
|
|
|
|
Absence | 24 |
|
|
|
|
| Number of
tumors |
| 1.366
(1.108–3.761) | 0.036 | 1.997
(1.297–6.330) | 0.031 |
|
Single | 36 |
|
|
|
|
|
Multiple (≥2) | 35 |
|
|
|
|
| Vascular
invasion |
| 0.673
(0.339–1.208) | 0.574 |
|
|
|
Present | 33 |
|
|
|
|
|
Absent | 38 |
|
|
|
|
| CDC20
expression |
| 1.735
(1.196–3.667) | 0.011 | 2.036
(1.296–6.647) | 0.007 |
|
High | 29 |
|
|
|
|
|
Low | 42 |
|
|
|
|
Knockdown of CDC20 inhibits the
proliferation of Hep3B and Huh-7 cells
The present study used si-CDC20 to silence CDC20
expression. Hep3B and Huh-7 cells were used, as the expression
levels of CDC20 were significantly different; the expression levels
of CDC20 in Huh-7 were relatively high, whereas those in Hep3B
cells were relatively low. Post-transfection with si-CDC20, the
expression levels of CDC20 were significantly decreased in Hep3B
and Huh-7 cells (P<0.05; Fig. 3A and
B). Furthermore, cellular immunofluorescence confirmed the
reduction in the expression levels of CDC20 in Hep3B and Huh-7
cells transfected with si-CDC20 (Fig.
3C and D). The results of the CCK-8 assay revealed that
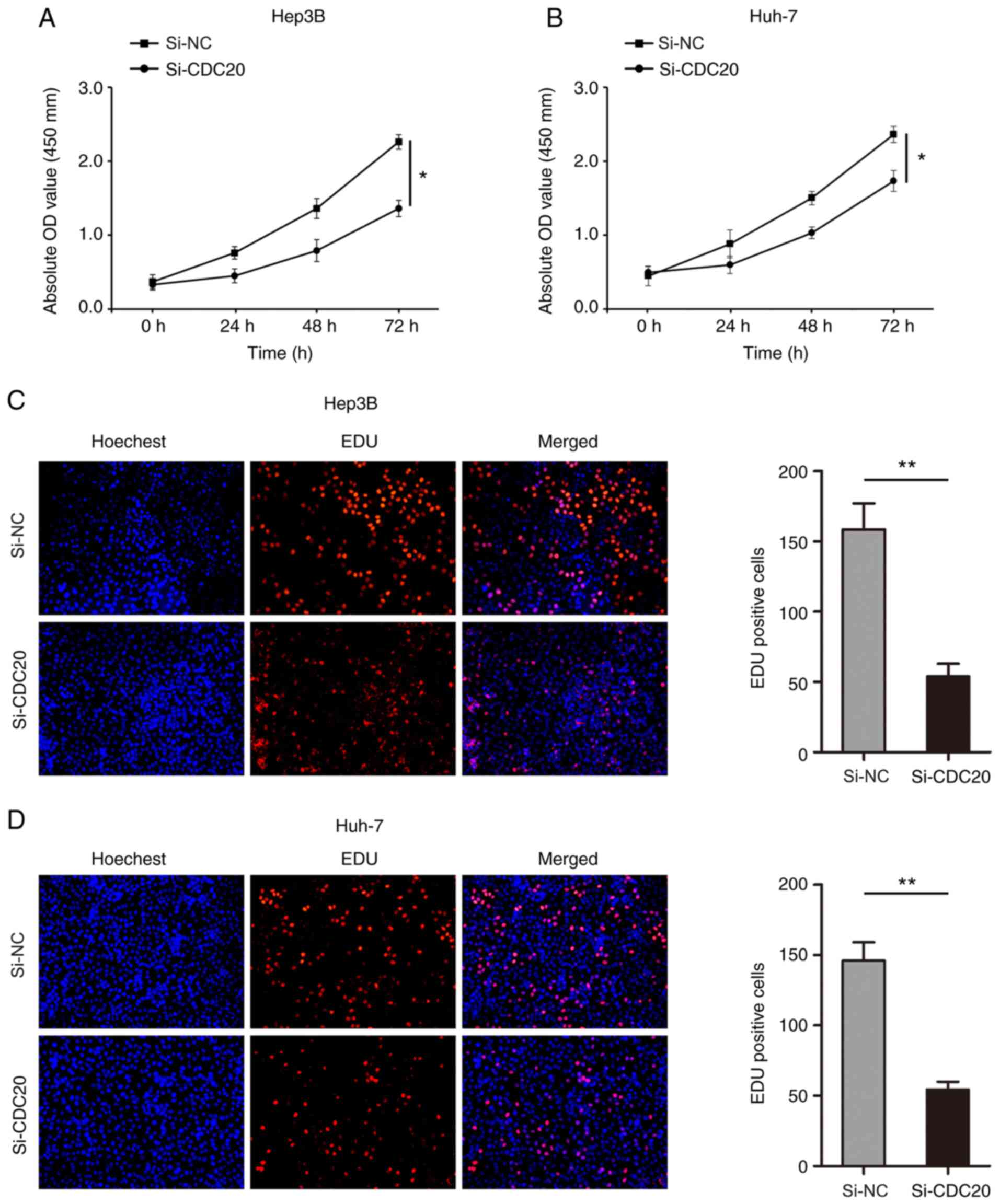
knockdown of CDC20 significantly inhibited the proliferation of
Hep3B and Huh-7 cells (P<0.05; Fig.
4A and B). In addition, the number of Hep3B and Huh-7
EdU-positive cells was significantly decreased in the si-CDC20
group compared with that in the si-NC group (P<0.01; Fig. 4C and D).
Knockdown of CDC20 inhibits the
migration and invasion of Hep3B and Huh-7 cells
The present study examined the effect of CDC20 on
the migration and invasion of Hep3B and Huh-7 cells through
Transwell assays. The results revealed that knockdown of CDC20
inhibited Hep3B and Huh-7 cell migration and invasion compared with
those in the si-NC group (P<0.05; Fig. 5A and B).
Knockdown of CDC20 inhibits the
expression of EMT-related proteins
The present study demonstrated that the
proliferation, migration and invasion of HCC cells was reduced when
expression of CDC20 was decreased. Therefore, whether the changes
were related to alterations in EMT- and proliferation-associated
proteins was assessed. The results of western blotting revealed
that CDC20 silencing decreased N-cadherin, vimentin and Ki-67
expression levels, and increased E-cadherin expression levels
compared with those in the si-NC group (P<0.05; Fig. 6A and B).
Discussion
The present study confirmed that CDC20 was highly
expressed in HCC, and its high expression was significantly
associated with the poor prognosis of patients with HCC. In
addition, it was revealed that CDC20 could regulate the malignant
biological behavior of HCC through EMT, which lays the foundation
for further research into the specific mechanism underlying the
effects of CDC20 on HCC.
CDC20 is a regulator of the APC, which can
accelerate mitotic exit and interact with the spindle assembly
checkpoint (SAC) (20). The SAC
protein aggregates at the centromere in the prometaphase stage of
mitosis and induces conformational changes of Mad2, which helps it
bind to CDC20 and BubRl to form the mitotic checkpoint complex, an
APC inhibitor (20). The high
expression of CDC20 in tumor tissues has been reported to promote
the malignant progression of tumors (21). Conversely, inhibition of CDC20
activity may regulate cell division cycle and accelerate cell
apoptosis (21). A previous study
demonstrated that decreasing the expression of CDC20 inhibited the
migration of pancreatic cancer cells and metastatic breast cancer
cells, and triterpene mixture extracted from the mushroom Poria
cocos, purified triterpene dehydropropionic acid and
polyvalerate C could significantly inhibit the expression of CDC20
to regulate the malignant biological behavior of tumor cells
(22). In estrogen
receptor-positive breast cancer, CDC20 mRNA was reported to be
highly expressed, and high CDC20 expression was closely related to
tumor size and poor tumor grade (23). High mRNA expression levels of CDC20
were also significantly associated with poor prognosis. Notably,
the high expression of CDC20 has been reported to be closely
associated with the adverse effects of endocrine therapy in
patients receiving hormone therapy; CDC20 is therefore considered
an independent predictor of adverse clinical outcomes after
endocrine therapy (23). A previous
study also demonstrated that CDC20 expression was high and the
expression of BIM was decreased in glioma cell lines resistant to
temozolomide (24). Furthermore,
inhibition of CDC20 expression could inhibit the EMT
characteristics of drug-resistant cell lines and regulate the
malignant growth of glioma drug-resistant cell lines (24). CDC20 has also been revealed to be
highly expressed in primary cutaneous squamous cell carcinoma and
may promote the malignant biological behavior of tumor cells
through the Wnt/β-catenin signaling pathway (25). In addition, CDC20 is a known key
downstream gene of the MDM2-p53 signaling pathway in diffuse large
B-cell lymphoma, and has been shown to regulate tumor cell
proliferation, apoptosis and cell cycle changes (26). A previous study reported that CDC20
was very lowly expressed in normal liver tissues, and to the best
of our knowledge, there are no reports of CDC20 expression in other
liver diseases (1). Notably, the
results of the present study demonstrated that CDC20 was highly
expressed in HCC, and its high expression was significantly
associated with the poor prognosis of patients with HCC. Our future
studies aim to collect information on the expression of CDC20 in
HCC from a public database, and further analyze the relationship
between the expression of CDC20 and the poor prognosis of HCC.
One of the important biological characteristics of
HCC is its high levels of metastasis, which can lead to poor
prognosis of patients (27). At
present, although the mechanism of tumor metastasis is not fully
understood, existing studies have revealed that tumor metastasis is
a complex process involving multiple factors and stages, which
depends on the interaction between tumor cells and internal
environmental factors, such as promoting tumor cell growth,
invasion, migration and angiogenesis (28). A large number of studies have
demonstrated that EMT may have an important role in tumor
metastasis. EMT is a process during which epithelial cells lose
polarity, tight junctions and adhesion junctions under the action
of certain factors, and thus gain infiltrative and migratory
abilities, and acquire interstitial cell morphology and
characteristics (29–31). The loss of an epithelial phenotype
and the acquisition of stromal characteristics are the main
features of EMT. EMT includes the following: i) Decreased
expression of cell adhesion molecules, which leads to the loss of
epithelial cell intercellular adhesion, and ii) the keratin
cytoskeleton is transformed into a vimentin cytoskeleton, and cells
transform into spindle cells. In addition, EMT sometimes affects
cell function and morphology (30,31).
This phenotypic transformation enables tumor cells to eliminate
intercellular adhesion and thus become more aggressive.
The EMT process has been reported to be associated
with changes in the expression levels of slug, snail and twist,
which can regulate the malignant proliferation of HCC (32). Previous studies have reported that
inhibition of CDC20 expression in cisplatin-resistant osteosarcoma
cells significantly reversed the EMT phenotype and altered the
expression of EMT biomarkers (24,33).
In estrogen receptor-positive luminal A breast cancer cells, EMT
changes have been shown to serve an important regulatory role in
breast cancer migration, invasion and metastasis (34). In the present study, CDC20 silencing
decreased the expression levels of N-cadherin, vimentin and Ki-67,
and increased E-cadherin expression suggesting that CDC20
expression may promote the metastasis of HCC through EMT. However,
how this mechanism is achieved and the further regulatory mechanism
require in-depth research. The present study demonstrated that
CDC20 could regulate the malignant biological behavior of HCC
through EMT, and these findings may provide a scientific and
objective basis for further exploring the role of CDC20 in HCC from
the perspective of EMT. Although changes in the expression level of
CDC20 may cause changes in EMT, it is still unclear which specific
molecule it acts on, and its specific regulatory mechanism is
unclear. In addition, our future studies aim to analyze the
mechanism of CDC20 in other liver diseases.
Research on CDC20 in HCC has mainly been conducted
through bioinformatics analysis, and studies exploring the specific
mechanism by which CDC20 affects HCC via cell function analysis are
rare. The present study explored the role of CDC20 in HCC from the
perspective of cell function and aimed to determine the specific
molecular mechanism of CDC20. In addition, the relationship between
CDC20 and the characteristics of patients with HCC was assessed.
The present research is innovative and comprehensively analyzed the
role of CDC20 in HCC. In conclusion, the present results indicated
that the expression of CDC20 was significantly increased in HCC
cells, and CDC20 overexpression was associated with poor HCC
prognosis. Furthermore, CDC20 may promote proliferation, migration
and invasion of HCC cells and could affect the biological function
of HCC cells through EMT.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Project of
Department of Education, Sichuan Provincial (grant no. 16TDD00025),
the Scientific Research Project of Affiliated Hospital of North
Sichuan Medical College (grant no. 2020ZD001), the Scientific
Research Project of Affiliated Hospital of North Sichuan Medical
College (grant no. 2020JC035), Pre-research of State-level Project
of North Sichuan Medical College (grant no. CBY19-YZ17) and the
Popularization and Application Project of Sichuan Health Commission
(grant no. 20PJ149).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GY, GW, YX and JL conceived and designed the
experiments. GY, GW, YX and JS conducted experiments. GY, WL, TT
and JL performed data analysis and wrote the paper. GY, GW, YX and
JL confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures involving human participants were
approved by the Ethics Committee of Affiliated Hospital of North
Sichuan Medical College, and complied with the 1964 Helsinki
Declaration and its later amendments or comparable ethical
standards. The participants signed an extensive informed consent
form after being informed about the benefits and risks associated
with the present study.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greten TF, Wang XW and Korangy F: Current
concepts of immune based treatments for patients with HCC: From
basic science to novel treatment approaches. Gut. 64:842–848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kapanidou M, Curtis NL and Bolanos-Garcia
VM: Cdc20: At the Crossroads between chromosome segregation and
mitotic exit. Trends Biochem Sci. 42:193–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schrock MS, Stromberg BR, Scarberry L and
Summers MK: APC/C ubiquitin ligase: Functions and mechanisms in
tumorigenesis. Semin Cancer Biol. 67:80–91. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong S, Huang F, Zhang H and Chen Q:
Overexpression of BUB1B, CCNA2, CDC20, and CDK1 in tumor tissues
predicts poor survival in pancreatic ductal adenocarcinoma. Biosci
Rep. Feb 26–2019.doi: 10.1042/BSR20182306. View Article : Google Scholar
|
|
9
|
Parmar MB, K C RB, Löbenberg R and Uludağ
H: Additive polyplexes to undertake siRNA therapy against CDC20 and
Survivin in breast cancer cells. Biomacromolecules. 19:4193–4206.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De K, Grubb TM, Zalenski AA, Pfaff KE, Pal
D, Majumder S, Summers MK and Venere M: Hyperphosphorylation of
CDH1 in glioblastoma cancer stem cells attenuates APC/C
CDH1 activity and pharmacologic inhibition of APC/C
CDH1/CDC20 compromises viability. Mol Cancer Res.
17:1519–1530. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim Y, Choi JW, Lee JH and Kim YS: Spindle
assembly checkpoint MAD2 and CDC20 overexpressions and cell-in-cell
formation in gastric cancer and its precursor lesions. Hum Pathol.
85:174–183. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li K, Mao Y, Lu L, Hu C, Wang D, Si-Tu J,
Lu M, Peng S, Qiu J and Gao X: Silencing of CDC20 suppresses
metastatic castration-resistant prostate cancer growth and enhances
chemosensitivity to docetaxel. Int J Oncol. 49:1679–1685. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Huang H, Liu A, Li J, Liu C, Sun
B, Chen L, Gao Y, Xu D and Su C: Cell division cycle 20 (CDC20)
drives prostate cancer progression via stabilization of β-catenin
in cancer stem-like cells. EBioMedicine. 42:397–440. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Giannelli G, Koudelkova P, Dituri F and
Mikulits W: Role of epithelial to mesenchymal transition in
hepatocellular carcinoma. J Hepatol. 65:798–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Diepenbruck M and Christofori G:
Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no,
maybe? Curr Opin Cell Biol. 43:7–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu H, Zhang W, Wu ZR, Liu Y, Shi Y, Gong
J, Shen W and Liu C: MiR-29c-3p regulates DNMT3B and LATS1
methylation to inhibit tumor progression in hepatocellular
carcinoma. Cell Death Dis. 10:482019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Z, He M, Shah AA and Wan Y: Insights
into APC/C: From cellular function to diseases and therapeutics.
Cell Div. 11:92016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McLean JR, Chaix D, Ohi MD and Gould KL:
State of the APC/C: Organization, function, and structure. Crit Rev
Biochem Mol Biol. 46:118–136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng S, Castillo V and Sliva D: CDC20
associated with cancer metastasis and novel mushroom-derived CDC20
inhibitors with antimetastatic activity. Int J Oncol. 54:2250–2256.
2019.PubMed/NCBI
|
|
23
|
Alfarsi LH, Ansari RE, Craze ML, Toss MS,
Masisi B, Ellis IO, Rakha EA and Green AR: CDC20 expression in
oestrogen receptor positive breast cancer predicts poor prognosis
and lack of response to endocrine therapy. Breast Cancer Res Treat.
178:535–544. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Zhou F, Li Y, Li Q, Wu Z, Yu L,
Yuan F, Liu J, Tian Y, Cao Y, et al: Cdc20 overexpression is
involved in temozolomide-resistant glioma cells with
epithelial-mesenchymal transition. Cell Cycle. 16:2355–2365. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chu Z, Zhang X, Li Q, Hu G, Lian CG and
Geng S: CDC20 contributes to the development of human cutaneous
squamous cell carcinoma through the Wnt/β-catenin signaling
pathway. Int J Oncol. 54:1534–1544. 2019.PubMed/NCBI
|
|
26
|
Sun C, Li M, Feng Y, Sun F, Zhang L, Xu Y,
Lu S, Zhu J, Huang J, Wang J, et al: MDM2-P53 signaling
pathway-mediated upregulation of CDC20 promotes progression of
human diffuse large B-cell lymphoma. Onco Targets Ther.
13:10475–10487. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Couri T and Pillai A: Goals and targets
for personalized therapy for HCC. Hepatol Int. 13:125–137. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yeung KT and Yang J:
Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol.
11:28–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu M, Xue H, Wang Y, Shen Q, Jiang Q,
Zhang X, Li K, Jia M, Jia J, Xu J and Tian Y: MiR-345 inhibits
tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT
pathway in hepatocellular carcinoma. Int J Oncol. 50:975–983. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu E, Gao Y, Zhang B, Xia T, Zhang Z and
Shang G: Upregulation of cell division cycle 20 in cisplatin
resistance-induced epithelial-mesenchymal transition in
osteosarcoma cells. Am J Transl Res. 12:1309–1318. 2020.PubMed/NCBI
|
|
34
|
Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, Mo
Q, Liao L and Xu J: Twist1 promotes breast cancer invasion and
metastasis by silencing Foxa1 expression. Oncogene. 36:1157–1166.
2017. View Article : Google Scholar : PubMed/NCBI
|