Introduction
Colorectal cancer (CRC) is the fourth most deadly
cancer in the world with nearly 900,000 deaths annually, accounting
for about 10% of all annually diagnosed cancers and cancer-related
deaths (1). The occurrence of CRC
is a multi-step process. Invasion and metastasis are the main
causes of morbidity and mortality, and ~1/3 of patients with CRC
eventually develop metastatic disease (2). Therefore, early diagnosis can directly
affect or even determine the survival time of patients with CRC.
The pathogenesis of CRC is complex, involving the regulation of
numerous molecular pathways including Wnt/β-catenin, p53,
TGF-β/SMAD, NF-κB and Notch signaling pathways (3). Thus, studies into the molecular
mechanisms of CRC will aid in the development of novel molecular
diagnostic tools and targeted treatment methods, and thus improve
the survival of patients.
MicroRNAs (miRNAs/miRs) are a large class of highly
conserved endogenous non-coding single-stranded small RNAs, 18–25
nucleotides in length (4). At
present, >28,600 miRNAs have been identified, of which >2,600
mature miRNAs have been found in humans (5). miRNAs bind to the 3′untranslated
region of target gene mRNA through incomplete base pairing and form
an RNA-induced silencing complex, which promotes the degradation of
RNA or inhibits the translation of proteins, thereby regulating the
expression of the downstream target genes (6). Furthermore, miRNAs have been found to
regulate gene expression in numerous biological processes in cells,
including proliferation, differentiation, metabolism and apoptosis
(7).
The gene of miR-483 is located in the second intron
region of insulin-like growth factor receptor 2 on chromosome 11
(8). Since it was first cloned in
the human embryonic liver in 2005, researchers have identified that
the expression pattern of miR-483 is inconsistent among different
types of tumors. For instance, large number of studies have
reported that miR-483 was not only involved in the occurrence of
relatively benign diseases, such as type 2 diabetes,
osteoarthritis, ischemic heart disease and polycystic ovary
syndrome, but was also involved in the occurrence of malignant
tumors, including nephroblastoma, lung cancer, adrenocortical
carcinoma and numerous digestive system tumors (9–16).
Moreover, miR-483 serves a role in multiple biological functions of
tumor cells, acting on target mRNAs that are closely associated
with the occurrence and development of tumors (17).
EI24 autophagy associated transmembrane protein
(EI24) is an early, rapidly induced gene involved in p53-mediated
apoptosis (18). Furthermore, it
serves an important role in inhibiting cell proliferation and
activating autophagy, as well as exerts tumor-suppressive activity
(19). The human EI24 gene is
located on chromosome 11q23, where heterozygote deficiency occurs
in a variety of malignant tumors, leading to the decrease or
disappearance of its anti-cancer function (20). Abnormalities in EI24 expression are
closely associated with the occurrence and progression of tumors
(21–23). In addition, our previous study
confirmed EI24 as the target molecule of miR-483, using reporter
gene detection (24).
The present study aimed to investigate the effects
of miR-483 and EI24 on the malignant phenotype of CRC, the
regulation of EI24 expression by miR-483 and its significance in
CRC.
Materials and methods
Cell culture and clinical samples
The normal colorectal epithelial cell line, NCM460,
and the CRC cell lines, Caco-2, RKO, LoVo and HCT 116, were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 1% penicillin-streptomycin (HyClone;
Cytiva) and 10% FBS (HyClone; Cytiva) in a cell incubator at 37°C
under a humidified atmosphere of 5% CO2.
A total of 183 cases of CRC and adjacent normal
mucosa tissue specimens were collected from Xijing Hospital of The
Fourth Military Medical University (age range, 24–91 years) between
January 1, 2014 and December 31, 2015; the date of last complete
follow-up was December 31, 2019. All specimens were diagnosed as
CRC by postoperative pathological examination, and the clinical
features of the specimens are presented in the Table SI. All tissue samples used in this
study were immediately frozen in liquid nitrogen and stored at
−80°C. The use of patient data was approved by the patient and
his/her family members, and the study was approved by the Ethics
Committee of Xijing Hospital, The Fourth Military Medical
University (approval no. KY20203211-1). All operations were
performed in accordance with the Helsinki Declaration and good
clinical practice guidelines (25).
Reverse transcription-quantitative
(RT-q)PCR
PCR primers and RT primers were designed and
synthesized by Guangzhou RiboBio Co., Ltd., and RNA was extracted
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocols. The quality of RNA was detected
via the ultraviolet absorption method. The purity of RNA was
determined using 10% denaturing agarose gel electrophoresis.
Isolated RNAs were reverse transcription using the Promega M-MLV
kit (cat. no. M1705; Promega Corporation) according to the
manufacturer's protocols. The cDNA was used to perform RT-qPCR on
LightCycler 480 Real-time PCR system(Roche, USA) using the SYBR
Master Mix (cat. no. DRR041B; Takara Bio). Amplification was
performed at 95°C for 30 sec, followed by 40 cycles of 95°C for 5
sec and 60°C for 30 sec. U6 was used as an endogenous control for
miRNA detection, and GAPDH was used as the RT-qPCR control of EI24.
The following primers were used in the study: U6 forward,
5′-CGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCA-3′; GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′; and EI24 forward,
5′-AATGCACCAGCGGTTGTCTAA-3′ and reverse,
5′-GATAGAGAAAAGGCAGCCACTGA-3′; miR-483 5′-AAGACGGGAGGAAAGAAGGGA-3′.
The relative RNA expression level was calculated using the
2−∆∆Cq method (26).
Cell transfection
The human CRC cell line Caco-2 was cultured in
RPMI-1640 complete medium containing 1% penicillin-streptomycin
(10,000 U/ml; cat. no. 15140122; Gibco; Thermo Fisher Scientific,
Inc.) and 10% FBS at 37°C with 100% humidity and 5% CO2.
To generate the hsa-miR-483 lentiviral expression plasmid, the pre
hsa-miR-483 sequence (GGAAAGGACGAAACACCGGCTGATGGCACCTGCCCTTTGG) was
synthesized and cloned into lentiviral expression vector GV309
(Shanghai GeneChem Co., Ltd.). A scrambled sequence
(TTCTCCGAACGTGTCACGT) was created as a negative control construct.
The miR-483 lentivirus (LV-miR-483) and negative control (LV-NC)
virus were GFP (Green fluorescent protein)-tagged. After being
confirmed by DNA sequencing, 293 cells (Cell Resource Center,
Institute of Basic Medicine, Chinese Academy of Medical Sciences)
were co-transfected with the vector plasmid and the packing
plasmids of pHelper 1.0 and pHelper 2.0 (Shanghai GeneChem Co.,
Ltd.). To obtain stable lentivirus infected cell lines, Caco-2
cells were plated at 30% confluence and lentivirus vector
(1×109 TU/ml) containing 2 mg/ml polybrene (Shanghai
GeneChem Co., Ltd.) was transfected into serum-free medium under
the conditions of 37°C, 100% humidity and 5% CO2. After
16 h, fresh complete medium was used instead of culture medium. The
transfection efficiency was observed via fluorescence microscope
(magnification, ×100) 72 h later. After transfection with 48 h,
RT-qPCR was used to verify whether the transfection was successful.
miR-483 inhibitor was used to construct miR-483 low expression CRC
cells. 100 pmol miR-483 inhibitor (cat. no. miR20002173-1-5;
Guangzhou RiboBio Co., Ltd.) or inhibitor NC (cat. no.
miR20002173-1-5; Guangzhou RiboBio Co., Ltd.) was added into 250 µl
Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.), 5 µl
Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.) reagent was diluted and mixed with 250 µl
Opti-MEM at room temperature for 5 min. Lipofectamine®
was mixed with miR-483 inhibitor or inhibitor NC at room
temperature for 20 min. The complexes of inhibitor or inhibitor NC
were added into Caco-2 cells respectively. The cells were incubated
at 37°C, 100% humidity and 5% CO2. After transfection
for 48 h, RT-qPCR was used to verify whether the transfection was
successful.
MTT assay
Logarithmic growth stage cells were inoculated into
96-well plates with 3,000 cells per well, using a microsample gun,
and then cultured in a cell culture box under the conditions of
37°C, 100% humidity and 5% CO2. The following day, 20 µl
sterile MTT solution (5 mg/ml; Beijing Dingguo Changsheng
Biotechnology, Co., Ltd.) was added to the wells. After 4 h, the
culture solution in each well was absorbed and removed. DMSO (100
µl; Sigma-Aldrich; Merck KGaA) was added to each well to dissolve
the formazan particles. An oscillator was used for oscillation at
37°C for 5 min at a frequency of 20 rpm, and the optical density
value was measured at 490/570 nm using an enzyme labeling
instrument.
Cell cycle assay
Cells in the logarithmic growth stage
(1×106 cells/well) were resuspended in a cell
suspension, centrifuged several times (room temperature, 200 × g, 5
min) and immobilized with 75% ethanol. Cells were suspended in cell
staining solution containing 0.01% RNase and 0.5% propidium iodide
(Shanghai Ruji Biological Technology Development Co., Ltd.) and
stained at 4°C for 20 min. The cellular DNA content was measured
via EPICS XL flow cytometer (Beckman Coulter, Inc.) at 488 nm
excitation wavelength, and the proliferation index (PI) was
calculated by FlowJo software (v. 7.6.1; FlowJo LLC), PI=(S +
G2)/(S + G2 + G1).
Colony formation assay
Logarithmic growth stage cells (70%) were inoculated
into 6-well plates with a microsample gun and cultured in a cell
incubator for 10 days. The cell culture fluid was changed every 3
days, and the cell state was closely monitored. Cell colonies were
imaged (fluorescence microscope; magnification, ×100) before the
experiment was terminated. Cells were fixed with 4%
paraformaldehyde (Sinopharm Chemical Reagent Co., Ltd.) at room
temperature for 30 min and stained with Giemsa solution (Beijing
Dingguo Changsheng Biotechnology, Co., Ltd.). The cells were washed
and dried repeatedly with ddH2O. Imaged were captured,
and the number of colonies >1 mm were counted.
Migration and invasion assays
Cell invasion and migration assays were performed
using Transwell chambers. In the cell migration assay,
1.0×105 transfected cells were inoculated into the upper
chamber, and 600 µl 30% FBS-containing medium was added to the
lower chamber. The cells were cultured in a 37°C incubator for 48
h. The non-migrated cells in the chamber were carefully removed
with a cotton swab, fixed with 4% paraformaldehyde (Sinopharm
Chemical Co., Ltd.) at 37°C for 15 min and stained with crystal
violet (Beyotime Institute of Biotechnology) after drying,
following which they were observed and counted under a microscope
(light microscope; magnification, ×100). For the cell invasion
assay, a Matrigel-coated membrane (Becton, Dickinson and Company)
was used (pre-coated at 4°C for 10 min).
Tumorigenicity assays in nude
mice
A total of 12 4-week-old male BALB/C nude mice
weighing ~20 g were randomly divided into two groups with six mice
in each group. Lentiviral-transduced Caco-2 cells (the
concentration of cell suspension was 2×107/ml) were
subcutaneously injected in nude mice, and the number of cells used
per injection was 5×105 cells/mouse. The nude mice were
reared at 21±2°C, relative humidity 30–70%, 12-h light/dark cycle
and normal diet. The tumor diameter was measured every 3 days to
monitor the growth rate of tumor. The maximum (L) and minimum
length (W) of tumor were measured with slide caliper. The tumor
volume was calculated using the following formula: Volume = 1/2
(LxW2) formula. At 28 days after injection, the nude
mice were anesthetized with sodium pentobarbital (40 mg/kg) and
euthanized via cervical dislocation. The tumors were collected and
then weighed. All animal experiments were approved and supervised
by the Animal Care Committee of The Fourth Military Medical
University (approval no. IACUC-20200402), and were conducted
according to international standards for animal welfare (27).
Western blot analysis
Cells were lysed in lysis buffer (Beyotime Institute
of Biotechnology). Following centrifugation (4°C, 13,000 × g, 15
min), the supernatant was collected, and the protein concentration
determined using the BCA method (Thermo Fisher Scientific, Inc.).
The mass of protein loaded per lane was 20 µg. The protein was
separated using 10% SDS-PAGE and transferred to a PVDF membrane
(EMD Millipore). After protein transfer and blocking (blocking
reagent was TBST solution containing 5% skimmed milk and 0.1%
Tween-20), the membrane was incubated with primary antibody (rabbit
monoclonal antibody; cat. no. ab130957; Abcam) overnight at 4°C,
and horseradish peroxidase (HRP)-conjugated secondary antibody
(1:5,000 dilution, cat. no. 31466; Invitrogen; Thermo Fisher
Scientific, Inc.) for 1 h at room temperature and visualized using
the ECL reagent (EMD Millipore). GAPDH (mouse monoclonal antibody;
cat. no. ab8245; Abcam) and β-actin (rabbit monoclonal antibody;
cat. no. SAB5500001; Sigma-Aldrich; Merck KGaA) were used as
controls. The bands were measured using an X-ray film and were
quantified using ImageJ software (v 1.6.0, NIH).
Immunohistochemistry
The 3 µm thick frozen tissue slices were prepared
and rewarmed at room temperature, blocked with 5% goat serum at
room temperature for 30 min (Beyotime Institute of Biotechnology)
and anti-EI24 antibody (1:200 dilution, cat. no. ab130957; Abcam)
was added. The samples were incubated overnight at 4°C. Sections
were then washed with PBS (Gibco; Thermo Fisher Scientific, Inc.),
following which the secondary antibody (1:200 dilution; cat. no.
31466; Invitrogen; Thermo Fisher Scientific, Inc.) was added and
incubated in a 37°C incubator for 30 min. Following rinsing with
PBS, the tissue microarray was placed in a chromogenic substrate
solution (Tiangen Biotech Co., Ltd.) for 10 min and then tap water
was used to fully rinse the samples in order to stop the
chromogenic process. After 1 min of re-dyeing with hematoxylin at
room temperature (Beyotime Institute of Biotechnology), the steps
of dehydration, transparency and sealing were conducted, and the
results were determined using a microscope (light microscope;
magnification, ×100).
Tissue microarray immunohistochemical
staining
After the aforementioned staining, two pathologists
independently interpreted the tissue microarray, including the
staining intensity and size of the stained area. The coloring
intensity score was as follows: Colorless was scored 0; light
yellow scored 1; brown-yellow scored 2; and brown scored 3. The
staining area score was 0 for <5%, 1 for 5–25%, 2 for 26–50%, 3
for 51–75% and 4 for 76–100%. If the scores of the two pathologists
were different, the average of the two was taken. In this study,
the final score of staining intensity was the product of two parts:
0 was negative (−), 1–4 was weak positive (+), 5–8 was positive
(++) and 9–12 was strong positive (+++).
Statistical analysis
In the cell experiment, Tukey's post hoc tests and
one-way ANOVA were used to compare the differences between the
groups. EI24 and miR-483 expression was analyzed using the Wilcoxon
signed rank test in CRC tissues and adjacent normal tissues.
χ2 tests were used to analyze the association between
the expression level of EI24 and clinical parameters. Kaplan-Meier
and logarithmic rank tests were used to compare survival rates, and
the Bonferroni correction was applied for pairwise adjustment. Data
are presented as mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Triplicate repeats were performed for each experiment. All
statistical analyses were performed using SPSS 19.0 software (IBM
Corp.).
Results
miR-483 is upregulated in CRC tissues
and cells
The current study designed and synthesized specific
miRNA primers, and then used RT-qPCR to detect the expression level
of miR-483 in CRC cells and corresponding normal colorectal cells.
The expression level of miR-483 in the CRC cell lines Caco-2, RKO
and HCT 116 was significantly higher compared with that in normal
colorectal cells; however, there was no significant difference in
its expression in LoVo cells (Fig.
1A). Moreover, the expression level of miR-483 in CRC tissues
was significantly higher compared with in the adjacent normal
tissues (Fig. 1B). Thus, it was
suggested that miR-483 may serve a regulatory role in the
occurrence and development of CRC.
Overexpression of miR-483 promotes the
proliferation, invasion and migration of CRC cells
Caco-2 cell lines with a relatively low expression
of miR-483 were selected, and lentivirus expression vectors were
used to establish a stable overexpression cell model. The
proliferative level of Caco-2 cells in the groups transfected with
LV-miR-483 or LV-NC were positive for green immunofluorescence
(Fig. 2A). The expression level of
miR-483 in these two groups was also detected via RT-PCR. The PCR
amplification curves and results revealed that the expression level
of miR-483 in the LV-miR-483 transfected Caco-2 cells was
significantly higher compared with that in the NC group (Fig. 2A). This observation indicated that
LV-miR-483 and LV-NC stable Caco-2 cell lines were successfully
established, providing credibility to the subsequent functional
experiments.
 | Figure 2.Overexpression of miR-483 promotes
the proliferation, invasion and migration of CRC cells. (A) The
proliferative level of Caco-2 cells in the two groups transfected
with LV-miR-483 and LV-NC was observed by immunofluorescence. The
PCR amplification curves and results demonstrated that the
expression level of miR-483 in the OE group was significantly
higher compared with that in the NC group. (B) MTT assay results
revealed that the proliferation rate of the OE group was
significantly higher than that of the NC group. (C) Flow cytometry
was used to analyze cell cycle and detect the percentage of OE
group and NC group cells in G0/G1, S and
G2/M phases, suggesting that upregulated miR-483 may be
involved in the regulation of the CRC cell cycle. (D) Colony
formation assay indicated that the number of cell clones in
OE-Caco-2 cells was significantly higher than that in NC group.
Transwell (E) invasion and (F) migration assays were used to
elucidate the invasion and migration of cells. All error bars
represent the SEM of ≥3 independent experiments. The results showed
that the mobility and invasive rate of the OE-Caco-2 group were
significantly higher than those of NC group. (G) Nude mice
tumorigenicity assays detected the effect of miR-483 OE on tumor
formation in vivo, and the results demonstrated that the
tumor volume and tumor weight in the OE group were significantly
larger compared with the NC group. *P<0.05 vs. NC-Caco-2.
LV-miR-483, miR-483 lentivirus; LV-NC, negative control virus; CRC,
colorectal cancer; miR, microRNA; OE, overexpression; OD, optical
density. |
MTT assay showed that the proliferation of CRC cells
was accelerated, leading to greater malignancy (Fig. 2B). Compared with the LV-NC group,
the LV-miR-483 group was associated with a lower probability of G1
arrest (Fig. 2C). These results
demonstrated that overexpression of miR-483 promoted the
proliferation of CRC cells, and participated in the regulation of
the cell cycle. Overexpression of miR-483 also increased the
proliferation and invasiveness of CRC cells, as shown by colony
formation and Transwell assays (Fig. 2D
and E). Further experiments revealed that overexpression of
miR-483 increased the migration of Caco-2 cells (Fig. 2F), suggesting that miR-483 increased
the possibility of distant metastasis of CRC.
In order to further clarify the effect of miR-483
overexpression on CRC cells, tumorigenicity assays were conducted
in nude mice. After subcutaneous implantation of Caco-2 cells in
nude mice, tumor volume and weight in the miR-483 overexpression
group were significantly higher compared with those in the NC group
(Fig. 2G).
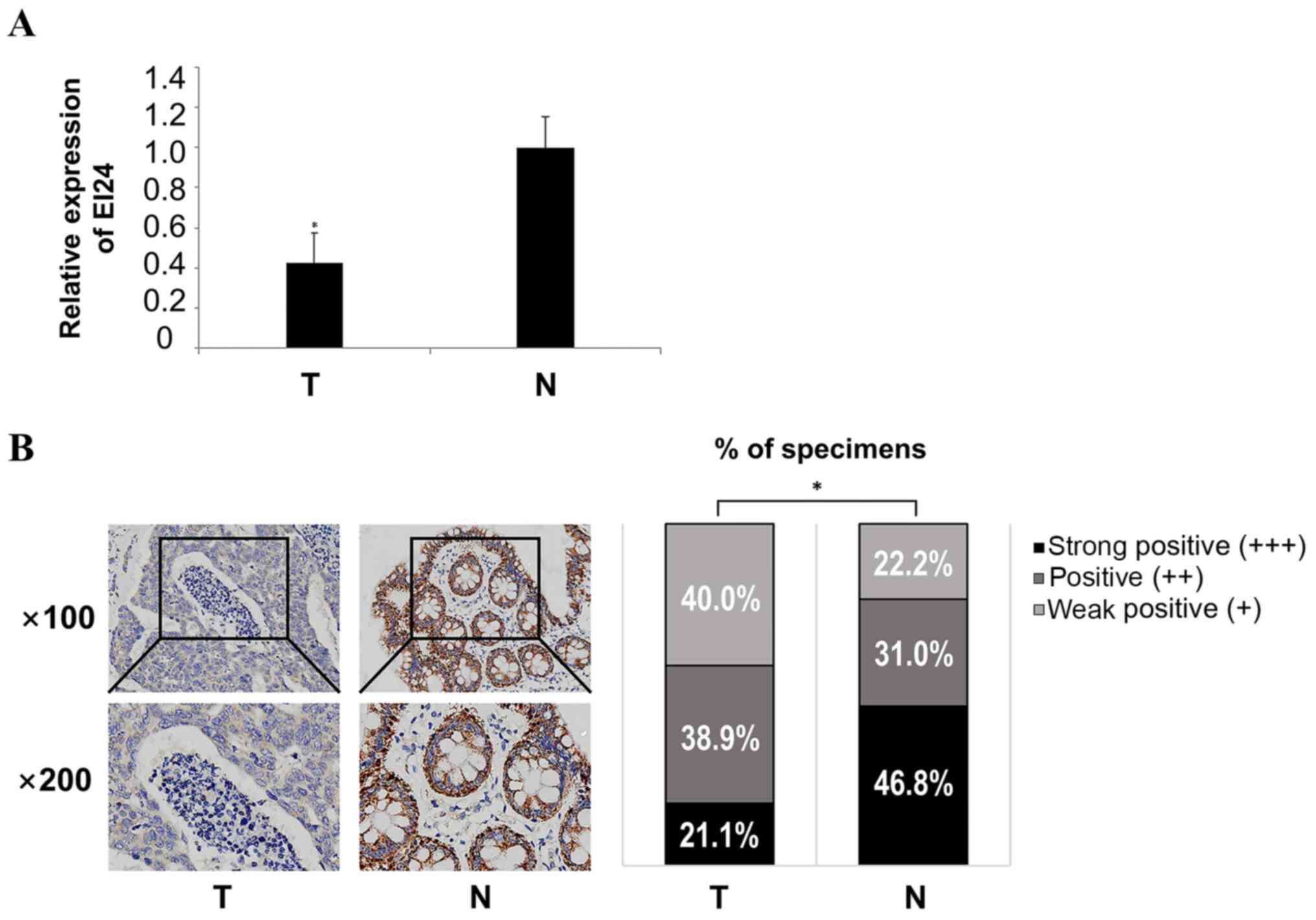
EI24 is downregulated in CRC
tissues
The RT-qPCR results demonstrated that the expression
levels of EI24 in CRC tissues were significantly lower compared
with those in adjacent normal tissues (Fig. 3A). The expression intensity of EI24
in cancer and adjacent normal tissues was analyzed using
immunohistochemistry, and the results indicated that the expression
level of EI24 in adjacent normal tissues was significantly
upregulated compared with CRC tissues (Fig. 3B). These experiments suggested that
EI24 was weakly expressed in CRC tissues, suggesting that EI24
serves a role as a tumor suppressor gene in CRC and may be a
prognostic molecule for CRC.
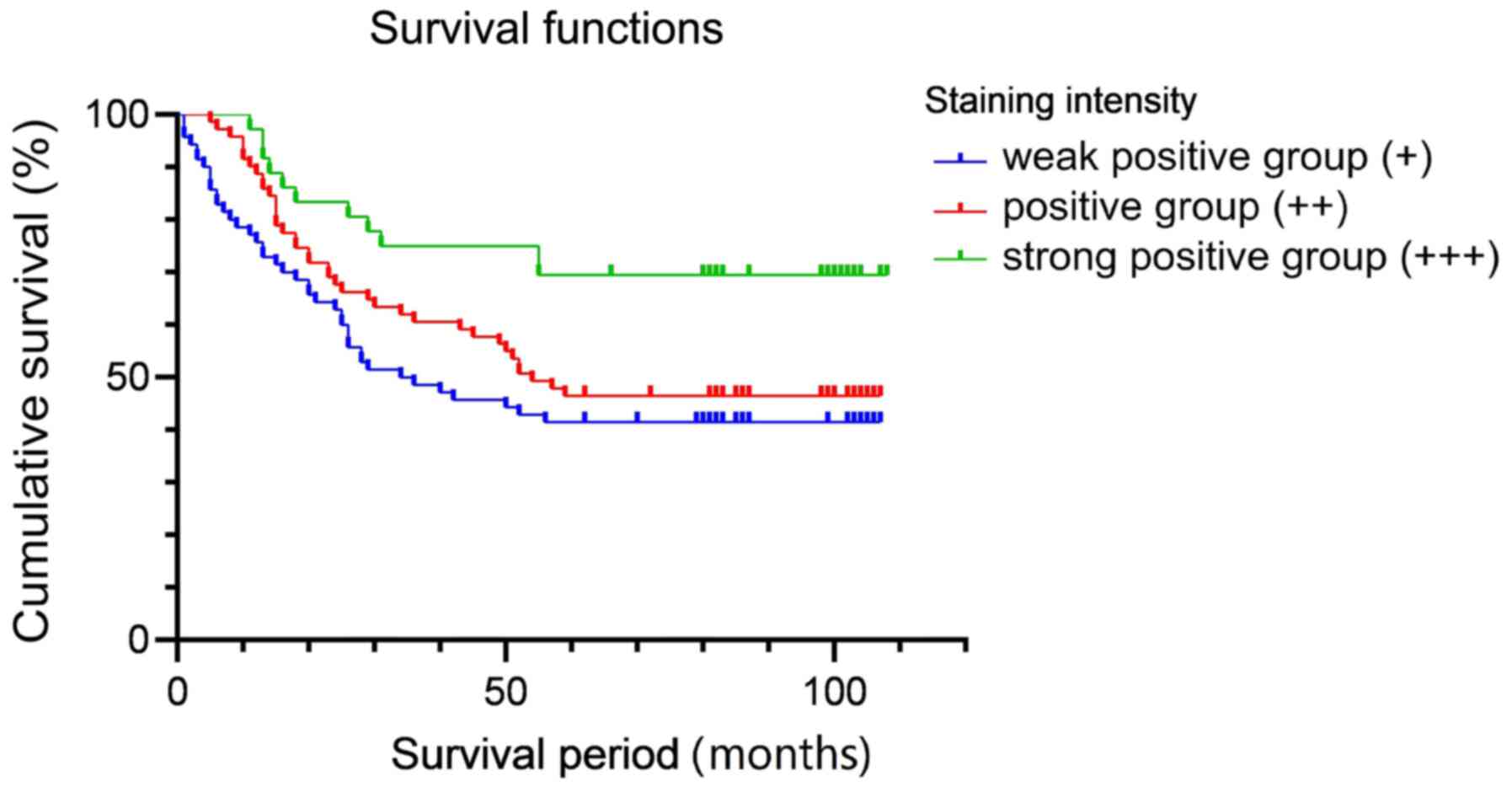
EI24 expression is associated with
patient prognosis
In order to clarify the relationship between the
expression level of EI24 and the prognosis of CRC, 183 patients
with CRC were recruited. Based on the expression level of EI24 in
the immunohistochemical staining of pathological sections, the
patients were divided into a high-expression group and a
low-expression group. The general data pertaining to the patients
are shown in Table SI.
Kaplan-Meier and logarithmic rank tests were used for survival
analysis. The result demonstrated that there was significant
difference between strong positive group (+++) and weak positive
group (+; P=0.005), and there was also significant difference
between strong positive group (+++) and positive group (++;
P=0.03), but there was no significant difference between + and ++
groups (P=0.296). After Bonferroni correction, only +++ and +
groups had a significant difference (P<0.05; Fig. 4). The results of the survival
analysis indicated that the prognosis of patients with high
expression of EI24 was improved compared with that of patients with
a low expression of EI24.
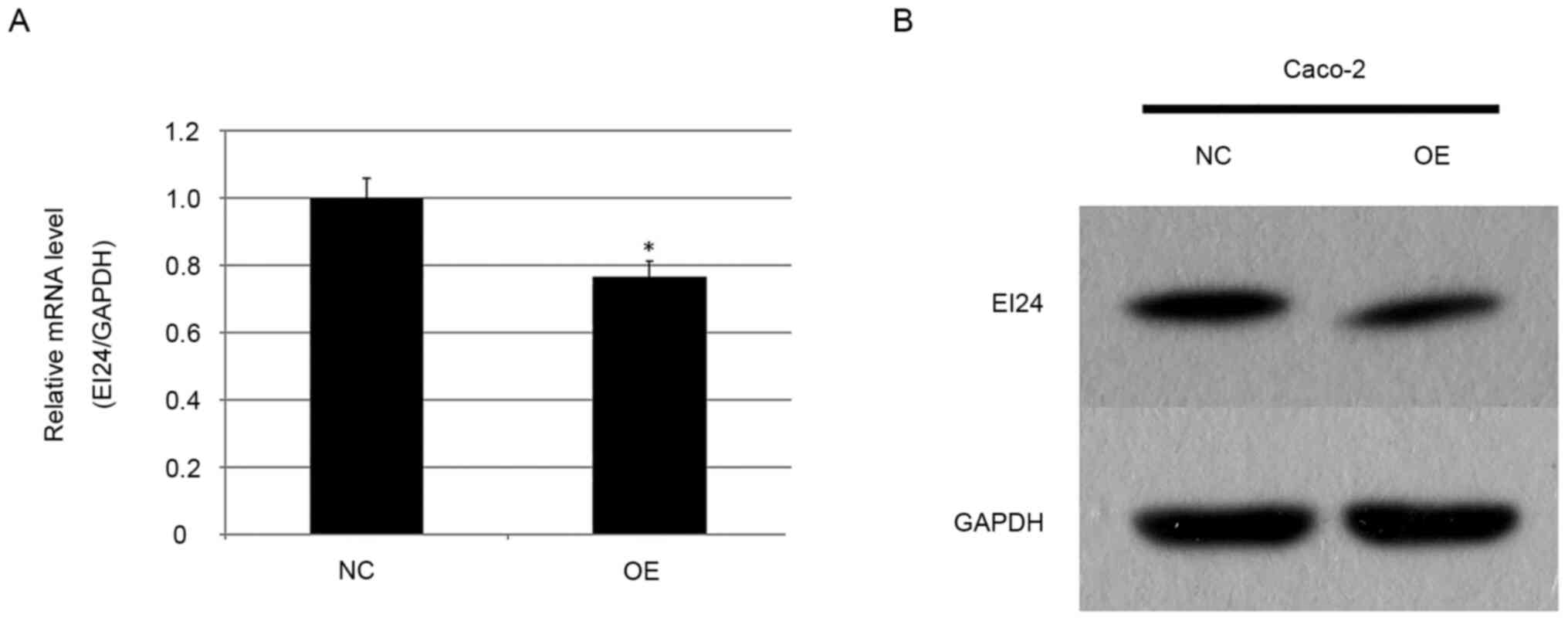
miR-483 expression is negatively
associated with EI24 levels in CRC tissues
Our previous studies reported that the expression
level of miR-483 was increased in esophageal squamous cell
carcinoma, and that upregulation of miR-483 could promote the
development of esophageal cancer (24,28).
Furthermore, a reporter gene test confirmed that EI24 was the
target molecule of miR-483 (21).
In the current experiment, the expression level of EI24 in miR-483
overexpressing stable Caco-2 cells and in the NC group was detected
using RT-qPCR. It was found that the expression level of EI24 in
the overexpression group was 0.766 times higher compared with that
in the NC group (P<0.05; Fig.
5A). Western blotting was also used to detect the expression
level of EI24 in the miR-483 overexpressing CRC cell line (Fig. 5B), and it was identified that EI24
expression in the overexpression group was notably lower compared
with that in the NC group, suggesting that upregulation of miR-483
may inhibit the expression of EI24 protein. These results are
consistent with our previous studies (24,28),
suggesting a targeting relationship between miR-483 and EI24.
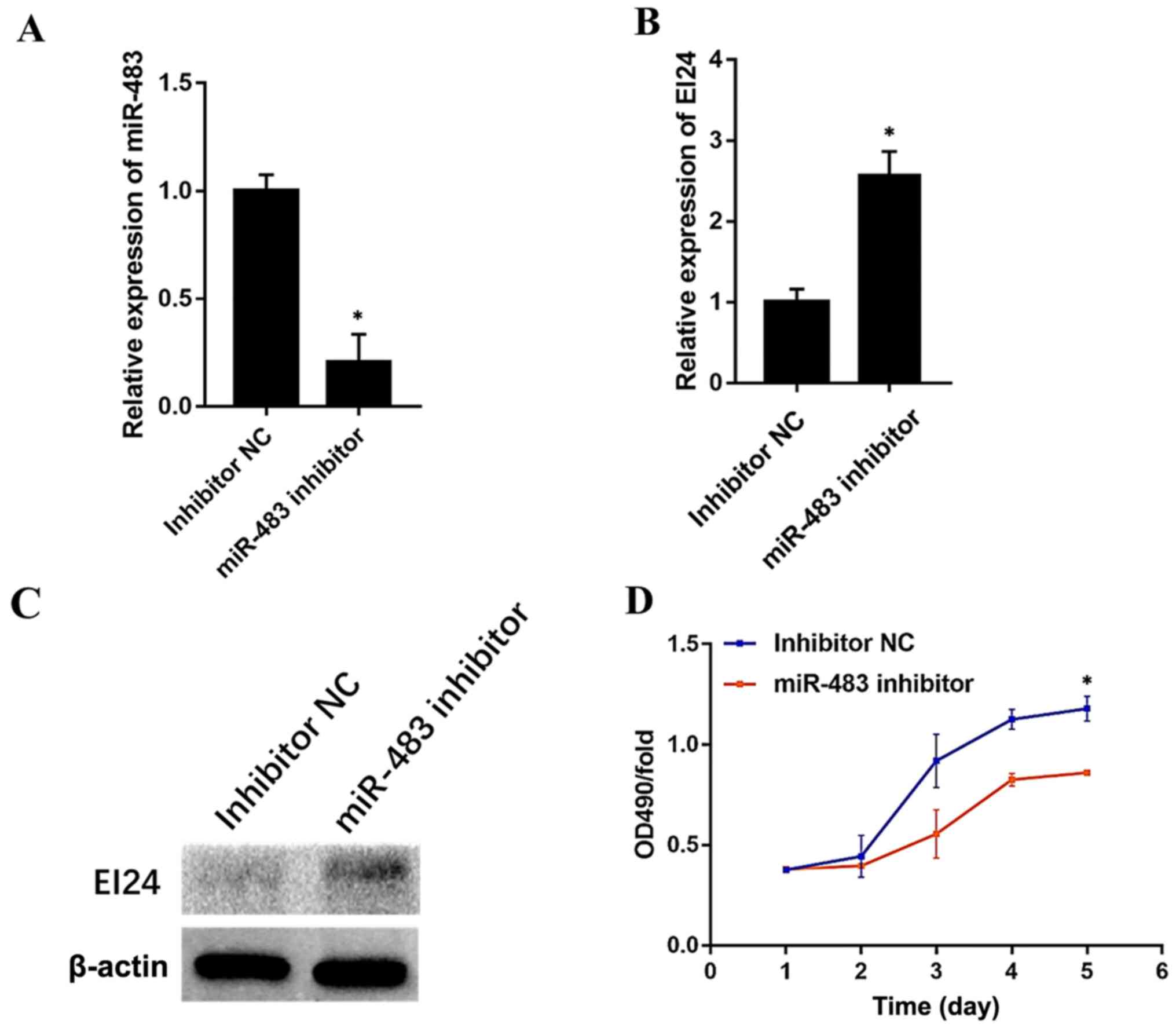
Endogenous miR-483 modulates EI24
expression and CRC cell proliferation
To investigate whether endogenous miR-483 regulates
EI24 expression, the effect of an miR-483 inhibitor in Caco-2
cells, which relatively expresses high levels of miR-483, was
determined. miR-483 inhibitor (cat. no. miR20002173-1-5; Guangzhou
RiboBio Co., Ltd.) and the NC (cat. no. miR20002173-1-5; Guangzhou
RiboBio Co., Ltd.) were used for this experiment. Compared with the
NC group, the expression level of miR-483 was significantly
decreased in the cells transfected with inhibitor (Fig. 6A). Furthermore, the transfection of
the miR-483 inhibitor markedly enhanced EI24 expression both at the
mRNA and protein levels (Fig. 6B and
C). Moreover, CRC cell proliferation was significantly
increased after miR-483 inhibitor transfection (Fig. 6D). These findings may suggest that
miR-483 endogenously modulates EI24 expression and CRC cell
proliferation.
Discussion
Invasion and metastasis are important
characteristics of CRC and are one of the main causes of mortality
in patients with CRC (1). The
invasion and proliferation of cancer cells are continuous and
dynamic biological processes involving multiple steps and factors
(29). The process consists of
several relatively independent steps, including separation and
exfoliation of cancer cells, adhesion between cancer cells and the
cell matrix, invasion and movement of cancer cells, invasion and
penetration of vascular walls, the presence and survival of cancer
cells in the circulatory system, penetration of the hemorrhagic
wall by cancer cells, implantation of cancer cells in distal
organs, and the proliferation and metastasis of cancer cells after
neovascularization (30). Although
a variety of CRC-related molecules have been identified, the signal
regulatory networks involved in the invasion and metastasis of CRC
are complex, and the specific mechanisms, molecular markers and
targets of action remain to be fully determined. Therefore, it is
important to investigate the key molecules and molecular mechanisms
of CRC metastasis, and to identify effective intervention targets.
Current studies have shown that the mechanisms of CRC metastasis
involve the epithelial-mesenchymal transformation (EMT)-related
signaling pathway, Wnt/β-catenin signaling pathway, Notch signaling
pathway, TGF-β signaling pathway, tumor-related genes, including
p53, and numerous miRNA molecules (31–36).
Invasive and metastatic behaviors are important
biological features of malignant tumors and are also important
factors leading to the progression of tumors and the outcome of
treatment (37). The invasion and
metastasis of tumors are a dynamic process, involving the
interaction of multiple factors associated with the tumor cells
themselves, and are also closely related to their microenvironment,
forming complex signaling pathways (38). A large number of studies have
reported that miR-483 was involved in the invasion and metastasis
of tumors. For instance, a study on esophageal cancer showed that
miR-483-5p was positively correlated with lymph node metastasis of
esophageal cancer (39). In
addition to the digestive system, a study of lung cancer suggested
that miR-483-5p was activated by the Wnt/β-catenin signaling
pathway, and promoted metastasis by directly acting on two
metastasis inhibitors, GDP dissociation inhibitor 1 (RhoGDI1) and
activated leukocyte adhesion molecule (ALCAM). The downregulation
of RhoGDI1 can enhance snail family transcriptional repressor 1
gene expression, thereby promoting the EMT (14,40).
Moreover, the expression level of miR-483-5p was positively
correlated with the expression level of the β-catenin protein but
was negatively correlated with the expression levels of RhoGDI1 and
ALCAM. Therefore, miR-483-5p was not only the key factor involved
in activating β-catenin to promote metastasis but was also a
negative regulator of the metastasis inhibitors RhoGDI1 and ALCAM
(14). The present results
demonstrated that miR-483 was highly expressed in CRC cells. In
vivo and in vitro experiments revealed that miR-483
served a regulatory role in the proliferation of CRC cells, and
overexpression of miR-483 promoted the invasion and migration of
CRC cells. After completing the current study, additional MTT
assay, plate colony formation assay, Transwell invasion experiment,
cell cycle assay and nude mouse tumorigenesis tests were performed
in an RKO cell line, and the results were consistent with those of
Caco-2 cell line (data not shown).
EI24 is downregulated in malignant tumors of the
digestive tract, including pancreatic ductal adenocarcinoma and
esophageal cancer, as well as in breast, lung and skin cancer types
(24). Zang et al (18) reported that the mRNA expression
level of EI24 in pancreatic ductal adenocarcinoma tissue was
downregulated compared with adjacent normal tissues, and the
downregulation trend was associated with the degree of
differentiation of cancer cells. Furthermore, overexpression of
EI24 reduced the expression level of the oncogene c-Myc by
activating c-Myc autophagic lysosomes, thereby inhibiting the
proliferation of cancer cells and promoting cell cycle stagnation.
Thus, these authors proposed the hypothesis that the
EI24/Beclin-1/p62/c-Myc pathway mediated the progression of
pancreatic ductal adenocarcinoma, and it was suggested that EI24
served an important role in anti-oncogenesis (18). Moreover, Li et al (19) revealed that the expression level of
miR-455-3p was increased in triple negative breast cancer, which
promoted the proliferation, invasion and migration of cancer cells.
The authors further predicted, and verified, that EI24 was a target
gene. In addition, it was found the knockdown of EI24 expression
using small interfering RNA could enhance the invasion and
migration of cancer cells, and miR-455-3p could enhance the
invasion and migration of cancer cells. Thus, targeting the tumor
suppressor gene EI24 promoted the invasion and migration of tumors
(19). The aforementioned studies
showed that EI24 expression was significantly decreased in tumor
tissues, potentially inhibiting the occurrence and metastasis of
tumors. In the present study, it was identified that the expression
level of EI24 in CRC tissues was lower compared with that in
adjacent normal tissues. Moreover, the survival analysis of CRC
patients revealed that patients with high EI24 expression had a n
improved prognosis.
Both miR-483 and EI24 are involved in important
biological processes, including tumor cell proliferation, invasion
and migration, and they overlap in the process of tumor metastasis
(28). Our previous study revealed
that the expression level of miR-483 was increased in esophageal
cancer (24). Furthermore,
upregulation of miR-483 promotes cancer cell proliferation,
migration and other malignant phenotypes, whereas downregulation of
miR-483 could inhibit malignant phenotypes (41). These results suggested that miR-483
could serve a role in promoting esophageal cancer. Bioinformatics
analyses and genome-wide expression profile chips were combined to
predict that EI24 may be the downstream target gene of miR-483, and
that miR-483 may promote the metastasis and progression of cancer
cells by inhibiting the expression of EI24 (24). Therefore, miR-483 may promote the
metastasis of CRC by regulating the expression level of EI24. The
present study conducted experiments using RT-qPCR and western
blotting. These experiments demonstrated that miR-483 could inhibit
the expression level of EI24, which suggested the existence of a
miR-483/EI24 signaling pathway. miR-483 appears to be co-expressed
with its host gene IGF2 and promote the proliferation of CRC cells
by inhibiting its downstream target gene DLC-1 (42). These results suggest that the
miR-483/EI24 signaling pathway may serve an important role in the
metastasis of CRC, representing a possible alternative for the
future treatment of CRC.
There were several limitations to the current study.
Recent studies have reported that the occurrence and progression of
CRC are associated a series of oncogenes and tumor suppressor gene
mutations, including RAS, BRAF, PIK3CA and TP53 gene, amongst
others (40,43,44).
However, such tests were not performed at this time. Therefore,
future studies will detect and analyze the expression of related
genes in CRC with abnormal expression of miR-483. In addition,
relevant experiments will be performed in other CRC cell lines, to
obtain more complete results. Overexpression and gene knockout
tests should also be performed to assess the intrinsic association
between miR-483 and EI24. The sample size of clinical data should
be expanded, and patient-related genetic testing results should be
collected. Further studies should focus on the investigation of the
upstream and downstream pathways of miR-483, and the mechanisms via
which miR-483 regulates the cellular proliferation and metastasis
of CRC.
In conclusion, the present study demonstrated that
miR-483 could regulate the expression level of EI24 by targeting
EI24, as well as promoted the proliferation, migration and invasion
of CRC cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ and HZ are responsible for confirming the
authenticity of the raw data. WZ, WY, JY and HZ participated in the
design and performance of the experiments. LD, XW and YL
contributed to the data analysis. LN, SX and RZ contributed to the
collection of samples and clinical data analyses. JY and LH
participated in the conception and design of manuscripts,
coordinated the acquisition of clinical specimens and interpretated
the data results. All authors read and approved the final
manuscript, and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in the present study
involving human participants were approved by the Ethic Committee
of Xijing Hospital of The Fourth Military Medical University/Air
Force Military Medical University (Xi'an, China; approval no.
KY20203211-1). Written informed consent for the publication of any
associated data and accompanying images was obtained from all
patients prior to surgery. All animal experiments were approved and
supervised by the Animal Care Committee of The Fourth Military
Medical University (approval no. IACUC-20200402), according to
international standards for animal welfare.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA/miR
|
microRNA
|
|
EI24
|
EI24 autophagy associated
transmembrane protein
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang Q, Hou C, Huang D, Zhuang C, Jiang W,
Geng Z, Wang X and Hu L: miR-455-5p functions as a potential
oncogene by targeting galectin-9 in colon cancer. Oncol Lett.
13:1958–1964. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou W, Zhou X, Liu JQ, Zhang YJ and Hong
L: High expression of miR-21 in tissue correlated with the poor
survival of patients with esophageal cancer: A pilot study using
the meta-analysis. J Prev Med Care. 1:9–15. 2016. View Article : Google Scholar
|
|
5
|
Chen X, Zhang DH and You ZH: A
heterogeneous label propagation approach to explore the potential
associations between miRNA and disease. J Transl Med. 16:3482018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma J, Hong L, Chen Z, Nie Y and Fan D:
Epigenetic regulation of microRNAs in gastric cancer. Dig Dis Sci.
59:716–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou W, Yang W, Ma J, Zhang H, Li Z, Zhang
L, Liu J, Han Z, Wang H and Hong L: Role of miR-483 in digestive
tract cancers: From basic research to clinical value. J Cancer.
9:407–414. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferland-McCollough D, Fernandez-Twinn DS,
Cannell IG, David H, Warner M, Vaag AA, Bork-Jensen J, Brøns C,
Gant TW, Willis AE, et al: Programming of adipose tissue miR-483-3p
and GDF-3 expression by maternal diet in type 2 diabetes. Cell
Death Differ. 19:1003–1012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Zhang H, Sun Q, Wang Y, Yang J,
Yang J, Zhang T, Luo S, Wang L, Jiang Y, et al: Intra-articular
delivery of Antago-miR-483-5p inhibits osteoarthritis by modulating
matrilin 3 and tissue inhibitor of metalloproteinase 2. Mol Ther.
25:715–727. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiao Y, Ma N, Wang X, Hui Y, Li F, Xiang
Y, Zhou J, Zou C, Jin J, Lv G, et al: miR-483-5p controls
angiogenesis in vitro and targets serum response factor. FEBS Lett.
585:3095–3100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi L, Liu S, Zhao WQ and Shi JZ:
miR-483-5p and miR-486-5p are down-regulated in cumulus cells of
metaphase II oocytes from women with polycystic ovary syndrome.
Reprod Biomed Online. 31:565–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu X, Li Z, Chan MT and Wu WK: The roles
of microRNAs in Wilms' tumors. Tumour Biol. 37:1445–1450. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen
J, Zhang Y, Lai P, Fan X, Zhou X, et al: miR-483-5p promotes
invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1
and ALCAM. Cancer Res. 74:3031–3042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patterson EE, Holloway AK, Weng J, Fojo T
and Kebebew E: MicroRNA profiling of adrenocortical tumors reveals
miR-483 as a marker of malignancy. Cancer. 117:1630–1639. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Si Y, Zhang H, Ning T, Bai M, Wang Y, Yang
H, Wang X, Li J, Ying G and Ba Y: miR-26a/b inhibit tumor growth
and angiogenesis by targeting the HGF-VEGF axis in gastric
carcinoma. Cell Physiol Biochem. 42:1670–1683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xue L, Nan J, Dong L, Zhang C, Li H, Na R,
He H and Wang Y: Upregulated miR-483-5p expression as a prognostic
biomarker for esophageal squamous cell carcinoma. Cancer Biomark.
19:193–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zang Y, Zhu L, Li T, Wang Q, Li J, Qian Y,
Wei L, Xie M, Tang WH, Liu X, et al: EI24 suppresses tumorigenesis
in pancreatic cancer via regulating c-Myc. Gastroenterol Res Pract.
2018:26265452018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Z, Meng Q, Pan A, Wu X and Li L:
MicroRNA-455-3p promotes invasion and migration in triple negative
breast cancer by targeting tumor suppressor EI24. Oncotarget.
8:19455–19466. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi JM, Jang JY, Choi YR, Kim HR, Cho BC
and Lee HW: Reduced expression of EI24 confers resistance to
gefitinib through IGF-1R signaling in PC9 NSCLC cells. Lung Cancer.
90:175–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nam TW, Park SY, Lee JH, Roh JI and Lee
HW: Effect of EI24 expression on the tumorigenesis of
ApcMin/+ colorectal cancer mouse model. Biochem Biophys
Res Commun. 514:1087–1092. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi JM, Devkota S, Sung YH and Lee HW:
EI24 regulates epithelial-to-mesenchymal transition and tumor
progression by suppressing TRAF2-mediated NF-κB activity.
Oncotarget. 4:2383–2396. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mazumder Indra D, Mitra S, Singh RK, Dutta
S, Roy A, Mondal RK, Basu PS, Roychoudhury S and Panda CK:
Inactivation of CHEK1 and EI24 is associated with the development
of invasive cervical carcinoma: Clinical and prognostic
implications. Int J Cancer. 129:1859–1871. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma J, Hong L, Xu G, Hao J, Wang R, Guo H,
Liu J, Zhang Y, Nie Y and Fan D: miR-483-3p plays an oncogenic role
in esophageal squamous cell carcinoma by targeting tumor suppressor
EI24. Cell Biol Int. 40:448–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mentz RJ, Hernandez AF, Berdan LG, Rorick
T, O'Brien EC, Ibarra JC, Curtis LH and Peterson ED: Good clinical
practice guidance and pragmatic clinical trials: Balancing the best
of both worlds. Circulation. 133:872–880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
MacArthur Clark JA and Sun D: Guidelines
for the ethical review of laboratory animal welfare People's
Republic of China National Standard GB/T 3589218 [Issued 6 February
2018 Effective from 1 September 2018]. Animal Model Exp Med.
3:103–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duan L, Ma J, Yang W, Cao L, Wang X, Niu
L, Li Y, Zhou W, Zhang Y, Liu J, et al: EI24 inhibits cell
proliferation and drug resistance of esophageal squamous cell
carcinoma. Front Oncol. 10:15702020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Malki A, ElRuz RA, Gupta I, Allouch A,
Vranic S and Al Moustafa AE: Molecular mechanisms of colon cancer
progression and metastasis: Recent insights and advancements. Int J
Mol Sci. 22:1302020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leber MF and Efferth T: Molecular
principles of cancer invasion and metastasis (review). Int J Oncol.
34:881–895. 2009.PubMed/NCBI
|
|
31
|
Vu T and Datta PK: Regulation of EMT in
colorectal cancer: A culprit in metastasis. Cancers (Basel).
9:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haase G, Gavert N, Brabletz T and
Ben-Ze'ev A: The Wnt target gene L1 in colon cancer invasion and
metastasis. Cancers (Basel). 8:482016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weidle UH, Birzele F and Krüger A:
Molecular targets and pathways involved in liver metastasis of
colorectal cancer. Clin Exp Metastasis. 32:623–635. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu X, Ji Q, Fan Z and Li Q: Cellular
signaling pathways implicated in metastasis of colorectal cancer
and the associated targeted agents. Future Oncol. 11:2911–2922.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen
H, Xia D, Xu E, Lai M, Wu Y and Zhang H: Mutations of key driver
genes in colorectal cancer progression and metastasis. Cancer
Metastasis Rev. 37:173–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang S, Tan X, Huang Z, Chen Z, Lin P and
Fu SW: MicroRNA biomarkers in colorectal cancer liver metastasis. J
Cancer. 9:3867–3873. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Niu L, Yang W, Duan L, Wang X, Li Y, Xu C,
Liu C, Zhang Y, Zhou W, Liu J, et al: Biological implications and
clinical potential of metastasis-related miRNA in colorectal
cancer. Mol Ther Nucleic Acids. 23:42–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Allgayer H, Leupold JH and Patil N:
Defining the ‘Metastasome’: Perspectives from the genome and
molecular landscape in colorectal cancer for metastasis evolution
and clinical consequences. Semin Cancer Biol. 60:1–13. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Y and Hong L: Prediction value of
miR-483 and miR-214 in prognosis and multidrug resistance of
esophageal squamous cell carcinoma. Genet Test Mol Biomarkers.
17:470–474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang C, Wang X, Su Z, Fei H, Liu X and Pan
Q: miR-25 promotes hepatocellular carcinoma cell growth, migration
and invasion by inhibiting RhoGDI1. Oncotarget. 6:36231–36244.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiao Y, Guo Q, Jiang TJ, Yuan Y, Yang L,
Wang GW and Xiao WF: miR-483-3p regulates osteogenic
differentiation of bone marrow mesenchymal stem cells by targeting
STAT1. Mol Med Rep. 20:4558–4566. 2019.PubMed/NCBI
|
|
42
|
Cui H, Liu Y, Jiang J, Liu Y, Yang Z, Wu
S, Cao W, Cui IH and Yu C: IGF2-derived miR-483 mediated
oncofunction by suppressing DLC-1 and associated with colorectal
cancer. Oncotarget. 7:48456–48466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Løes IM, Immervoll H, Sorbye H, Angelsen
JH, Horn A, Knappskog S and Lønning PE: Impact of KRAS, BRAF,
PIK3CA, TP53 status and intraindividual mutation heterogeneity on
outcome after liver resection for colorectal cancer metastases. Int
J Cancer. 139:647–656. 2016. View Article : Google Scholar
|
|
44
|
Kawaguchi Y, Kopetz S, Newhook TE, De
Bellis M, Chun YS, Tzeng CD, Aloia TA and Vauthey JN: Mutation
status of RAS, TP53, and SMAD4 is superior to mutation status of
RAS alone for predicting prognosis after resection of colorectal
liver metastases. Clin Cancer Res. 25:5843–5851. 2019. View Article : Google Scholar : PubMed/NCBI
|