Introduction
Hepatocellular carcinoma (HCC) is a common digestive
tract malignant tumor with poor prognosis in China, and is the
fourth most common cause of cancer-related mortality worldwide,
accounting for 7% of all cancer-related deaths (1,2). The
primary treatment strategy used for HCC is hepatectomy. However,
>80% of HCC cases are diagnosed at an advanced stage (3,4), only
<5% of cases are candidates for surgical resection and the
recurrence rate is >70% within 5 years after surgery (5). At present, there is a lack of
effective treatments for advanced HCC, particularly for patients
following failure of sorafenib treatment (6,7).
Therefore, the design of targeted therapies for HCC has been a
research hotspot.
Circular RNAs (circRNAs) are a class of endogenous
non-coding RNAs that are produced by an exon and/or intron sequence
of the original transcription by reverse splicing (8). Unlike classical linear RNAs with 5′
and 3′ ends, circRNAs produce covalent closed-loop constructs that
prevent degradation by RNA exonuclease or RNase R, which results in
increased stability of circRNAs compared with that of linear mRNAs
(9). The expression of circRNAs in
humans is tissue-specific, and various circRNAs have been reported
to play important roles in HCC (10). A previous study demonstrated that
circRNA dedicator of cytokinesis 1 (DOCK1) is highly expressed in
thyroid cancer tissues (11).
circRNA DOCK1 inhibits the expression of microRNA (miRNA/miR)-124
in thyroid cancer cells, blocks the signal transduction of the
Janus kinase/signal transducer and activator of
transcription/adenosine 5′-monophosphate-activated protein kinase
signaling pathway and participates in the occurrence of thyroid
cancer via downregulating miR-124 (11). circRNA DOCK1 has been reported to
promote the progression of bladder cancer by regulating the
circDOCK1/hsa-miR-132-3p/Sox5 signaling pathway (12). Moreover, circRNA DOCK1 can inhibit
miR-196a-5p-induced apoptosis by targeting baculoviral IAP repeat
containing 3 in oral squamous cell carcinoma (13). However, to the best of our
knowledge, the role of circRNA DOCK1 in HCC has not been previously
reported.
It was previously reported that hsa-circ-u0085131
upregulates autophagy-related 7 via sponge adsorption to enhance
the resistance of non-small cell lung cancer cells to cisplatin,
resulting in autophagy (14).
miR-654-5p downregulation has been observed in colorectal cancer
cells, and miR-654-5p overexpression can inhibit colorectal cancer
cell proliferation, invasion and migration by targeting
hematopoietic lineage cell-specific protein (HCLS1)-associated
protein X-1 (15). miR-654-5p has
been reported to inhibit the occurrence of ovarian cancer through
regulating the MYC, WNT and AKT signaling pathways (16). Furthermore, miR-654-5p
downregulation can lead to the upregulation of transmembrane
protein 52B in gastric cancer, which was found to promote cell
invasion and metastasis both in vivo and in vitro
(17). Previous studies have also
demonstrated that SMAD2 is associated with the regression of HCC,
whereas inhibition of SMAD2 can suppress the regression of HCC
(18–20).
Therefore, the present study aimed to investigate
the role of circRNA DOCK1 in HCC and determine whether it affects
cell proliferation, invasion and migration by regulating the
miR-654-5p/SMAD2 axis.
Materials and methods
Cell culture
A human normal hepatocyte cell line (HHL-5) and HCC
cell lines (MHCC97-H, SK-Hep-1, Huh-7 and Hep3b) were purchased
from BioVector NTCC, Inc. HHL-5 and HCC cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin at
37°C with 5% CO2 and saturated humidity. After
resuscitation, cells of the 4 to 10th generation were used for
subsequent experiments.
Plasmids and cell transfection
For the stable overexpression of circ-SMAD2,
sequences were constructed into pcDNA3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.). The plasmids of short hairpin RNA
(shRNA)-negative control (NC) and shRNA-DOCK1#1/2 were commercially
synthesized by Shanghai GenePharma Co., Ltd. Other plasmids were
obtained from Guangzhou RiboBio Co., Ltd., as follows: mimic-NC
(cat. no. miR1N0000001-1-5), miR-654-5p mimic (cat. no.
miR10003330-1-5), NC inhibitor (cat. no. miR2N0000001-1-5) and
miR-654-5p inhibitor (cat. no. miR20003330-1-5).
Hep3b cells in the logarithmic growth phase were
selected and inoculated (4×105 cells) in a 6-cm culture
dish. Following culture for 24 h, Hep3b cells were transfected with
shRNA-NC (50 nM), shRNA-DOCK1#1/2 (50 nM), mimic-NC (50 nM),
miR-654-5p mimic (50 nM), NC inhibitor (50 nM), miR-654-5p
inhibitor (50 nM), pcDNA3.1-NC (50 nM) or pcDNA3.1-SMAD2 (50 nM),
or co-transfected with shRNA-DOCK1 + NC inhibitor, shRNA-DOCK1 +
miR-654-5p inhibitor, shRNA-DOCK1 + pcDNA3.1-NC or shRNA-DOCK1 +
pcDNA3.1-SMAD2 using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol at 37°C for 24 h. Hep3b cells without any treatment were
used as the blank control group. At 24 h post-transfection,
subsequent experiments were performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
A PARIS™ kit (Thermo Fisher Scientific, Inc.) was
used according to the manufacturer's protocols for cell
fractionation. Briefly, Hep3b cells (1×105) were mixed
with 1 ml cell fractionation buffer and centrifuged at 800 × g for
20 min at room temperature. Subsequently, TRIzol® and
TRIzol LS reagents (Invitrogen; Thermo Fisher Scientific, Inc.)
were independently used to obtain total RNA from the nuclear pellet
and cell supernatant. Total RNA was reverse-transcribed into cDNA
using a cDNA reverse transcription kit (Roche Diagnostics).
Subsequently, qPCR was performed using the SYBR Premix Ex Taq kit
(Beyotime Institute of Biotechnology) and an FTC-3000P real-time
PCR system (Shanghai Fengling Biotechnology Co., Ltd.). The
thermocycling conditions were as follows: Initial denaturation at
95°C for 20 sec; followed by 40 cycles of denaturation at 95°C for
15 sec, annealing at 60°C for 30 sec and extension at 72°C for 25
sec. The primer sequences were as follows: circRNA DOCK1 forward,
5′-CCTAGACGCGGAGTTTCCTG-3′ and reverse, 5′-CCGCTCCTCTGGCATCATAG-3′;
SMAD2 forward, 5′-ATGTCGTCCATCTTGCCATTC-3′ and reverse,
5′-AACCGTCCTGTTTTCTTTAGCTT-3′; GAPDH forward,
5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAAGT-3′; miR-654-5p forward,
5′-AGTGGAAAGATGGTGGGCCG-3′ and reverse,
5′-GCTTCTAAAGGTGATGGTCAGCAG-3′; and U6 forward,
5′-CGCTTCACGAATTTGCGT-3′ and reverse, 5′-CTCGCTTCGCAGCACA-3′. U6
and GAPDH were used as internal references for circRNA DOCK1,
miR-654-5p and SMAD2, respectively. The expression levels were
analyzed using the 2−ΔΔCq method (21).
Cell Counting Kit-8 (CCK-8) assay
Hep3b cells of each group were seeded (100 µl cell
suspension/well; 2×103 cells) into 96-well plates.
Following culture for 24, 48 or 72 h, cells were analyzed using a
CCK-8 kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Absorbance was measured at a wavelength of
450 nm using a Thermomax microplate reader (Thermo Fisher
Scientific, Inc.).
Colony formation assay
Hep3b cells of each group were seeded (1 ml cell
suspension/well; 5×102 cells) into 6-well plates with
DMEM supplemented with 10% FBS. Following culture for 14 days,
cells were fixed with 4% formaldehyde at room temperature for 20
min and stained with 0.1% crystal violet solution at room
temperature for 20 min. The number of colonies (>50 cells of
each colony) was counted using a light microscope (magnification,
×100; Olympus Corporation).
Wound healing assay
Hep3b cells of each group were seeded into 6-well
plates (5×104 cells/well) and incubated in serum-free
DMEM at 37°C with 5% CO2. At 90% confluence, a 200-µl
sterile pipette tip was used to scratch the cell monolayer. Cells
were washed twice with PBS to remove cell debris. The wound was
observed at 0 and 24 h using a light microscope (magnification,
×100; Olympus Corporation). Five fields were used for
quantification.
Transwell assay
The Transwell inserts were precoated with Matrigel
at 37°C for 30 min (Becton-Dickinson and Company). Subsequently,
200 µl serum-free DMEM containing 1×105 Hep3b cells was
plated into the upper chamber. Then, 500 µl DMEM supplemented with
10% FBS was plated into the lower chamber. Following incubation for
24 h at 37°C with 5% CO2, cells were fixed with 4%
paraformaldehyde for 30 min at room temperature and stained with
hematoxylin for 30 min at room temperature. Invading cells were
observed using a light microscope (magnification, ×100; Olympus
Corporation). Five fields were used for quantification.
Western blotting
Total protein was extracted from cells
(8×105) using RIPA reagent (Beyotime Institute of
Biotechnology). Protein concentration was determined by a BCA
assay. Proteins (30 µg) were separated via 10% SDS-PAGE, and
separated proteins were subsequently transferred to PVDF membranes.
Following blocking with 5% skimmed milk at room temperature for 2
h, the membranes were incubated at 4°C for 12 h with primary
antibodies targeted against SMAD2 (cat. no. ab40855; 1:2,000;
Abcam) and GAPDH (cat. no. ab9485; 1:1,000; Abcam). Following
washing three times with TBS with 0.05% Tween-20, the membranes
were incubated with a HRP-conjugated goat anti-rabbit secondary
antibody (cat. no. ab6721; 1:2,000; Abcam) at 37°C for 1 h. Protein
bands were visualized with ECL reagent (Bio-Rad Laboratories, Inc.)
using a gel imager (Thermo Fisher Scientific, Inc.). The gray value
of the target protein/internal reference protein was calculated by
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Dual-luciferase reporter assay
The binding sites of circRNA DOCK1 to miR-654-5p and
miR-654-5p to SMAD2 were analyzed using the Encyclopedia of RNA
Interactomes (ENCORI) database (http://starbase.sysu.edu.cn/index.php). Hep3b cells (1
ml cell suspension/well; 5×102 cells) seeded into 6-well
plates were co-transfected with mimic-NC (50 nM) or miR-654-5p
mimic (50 nM) and DOCK1 wild-type (WT)/mutant (MT) (50 nM) or SMAD2
WT/MT (50 nM) with Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). The mutated 3′-UTR was generated
using a site directed mutagenesis kit (Agilent Technologies, Inc.).
At 48 h post-transfection, luciferase activities were measured
using a dual-luciferase assay kit (Beijing Solarbio Science &
Technology Co., Ltd.). Renilla luminescence was used as the
internal reference.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 20.0; IBM Corp). Data are presented as the mean ±
SD of at least three independent experiments. Comparisons among
multiple groups were analyzed using one-way ANOVA followed by the
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Interference with circRNA DOCK1
inhibits HCC cell proliferation, invasion and migration
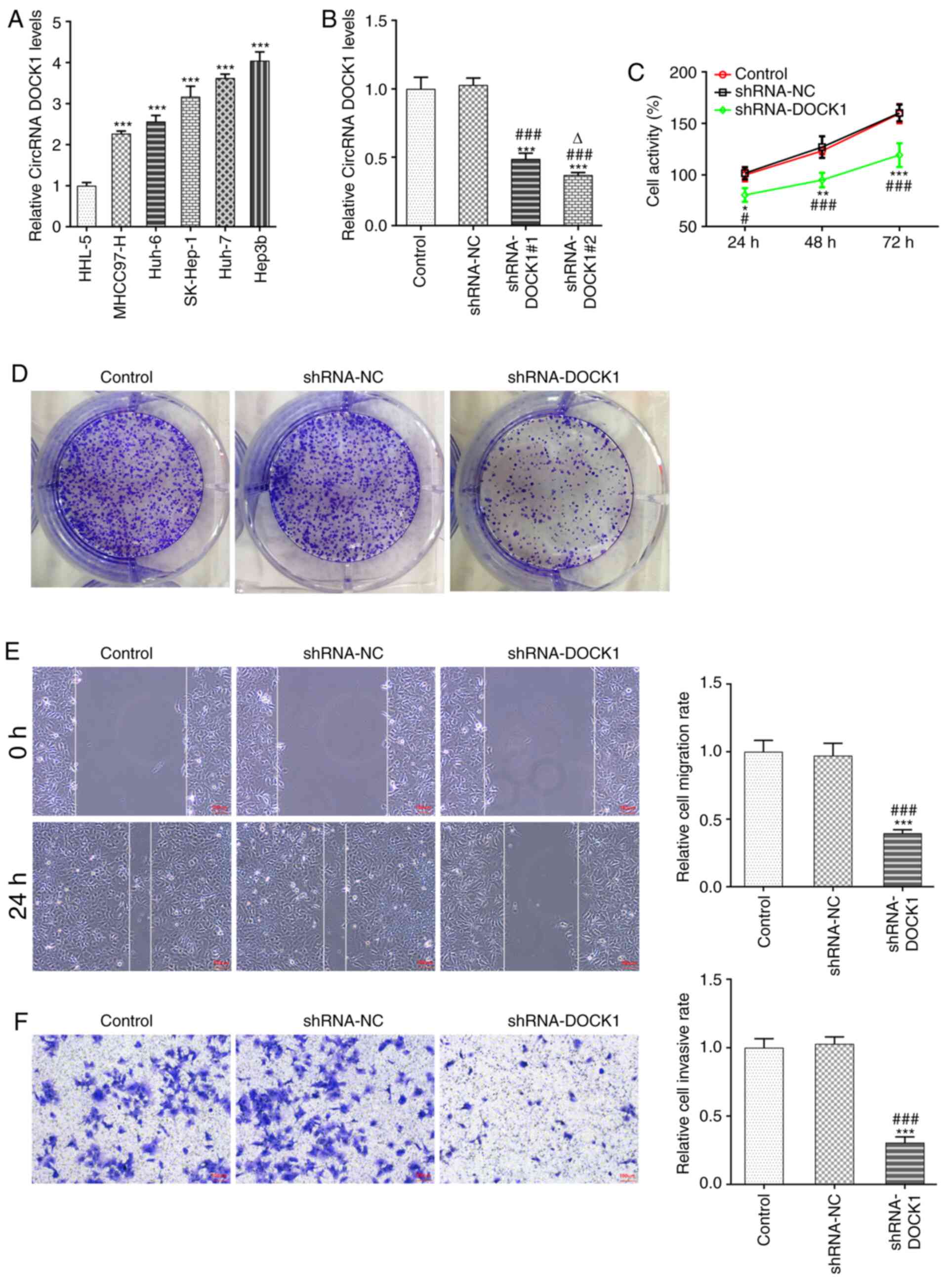
DOCK1 expression in HCC cells was higher compared
with that in HHL-5 cells, and DOCK1 expression was the highest in
Hep3b cells among the HCC cell lines; therefore, Hep3b cells were
selected for the subsequent experiments (Fig. 1A). Following transfection with
shRNA-DOCK1#1/2, DOCK1 expression was decreased. DOCK1 expression
in shRNA-DOCK1#2-transfected Hep3b cells was lower compared with in
shRNA-DOCK1#1 transfected Hep3b cells; therefore, shRNA-DOCK1#2 was
selected for the subsequent experiments (Fig. 1B). DOCK1 knockdown suppressed Hep3b
cell activity (Fig. 1C),
proliferation (Fig. 1D), migration
(Fig. 1E) and invasion (Fig. 1F).
circRNA DOCK1 serves as a sponge of
miR-654-5p to negatively regulate miR-654-5p expression
DOCK1 expression was higher in the cytoplasm
compared with the nucleus, which was confirmed by cell
fractionation and RT-qPCR (Fig.
2A).
Using the ENCORI database, it was predicted that
circRNA DOCK1 could bind to miR-654-5p (Fig. 2B). The dual-luciferase reporter
assay results indicated that the level of luc/R-Luc in Hep3b cells
transfected with DOCK1 WT and miR-654-5p mimic was decreased, which
indicated that DOCK1 bound to miR-654-5p (Fig. 2C). miR-654-5p expression in HCC
cells was lower compared with HHL-5 cells, and miR-654-5p
expression was the lowest in Hep3b cells among the HCC cell lines;
therefore, Hep3b cells were selected for subsequent experiments
(Fig. 2D). Following transfection
with miR-654-5p mimic or miR-654-5p inhibitor, miR-654-5p
expression was significantly increased in the miR-654-5p mimic
group, but decreased in the miR-654-5p inhibitor group (Fig. 2E). Following transfection with
shRNA-DOCK1, miR-654-5p expression was upregulated in Hep3b cells
(Fig. 2F).
miR-654-5p directly targets SMAD2
SMAD2 mRNA and protein expression levels in HCC
cells were higher compared with HHL-5 cells, and SMAD2 expression
was the highest in Hep3b cells among the HCC cell lines; therefore,
Hep3b cells were selected for subsequent experiments (Fig. 3A and B). Using the ENCORI database,
it was previously predicted that miR-654-5p could bind to SMAD2
(Fig. 3C). The level of luc/R-Luc
in Hep3b cells transfected with SMAD2 WT and miR-654-5p mimic was
decreased, which indicated that miR-654-5p bound to SMAD2 (Fig. 3D). Following transfection with
miR-654-5p mimic, SMAD2 mRNA and protein expression levels were
decreased (Fig. 3E and F).
Following transfection with shRNA-DOCK1, SMAD2 mRNA and protein
expression levels were decreased (Fig.
3G and H).
HCC cell proliferation, invasion and
migration are promoted by the circRNA DOCK1/miR-654-5p/SMAD2
axis
SMAD2 mRNA and protein expression levels in Hep3b
cells transfected with pcDNA3.1-SMAD2 were increased (Fig. 4A and B). Interference with DOCK1
decreased Hep3b cell activity (Fig.
4C), proliferation (Fig. 4D),
migration (Fig. 4E) and invasion
(Fig. 4F), which was partially
reversed by miR-654-5p knockdown and SMAD2 overexpression.
Discussion
HCC represents a serious public health concern
worldwide and it is associated with low survival rates (22). To improve the survival rate of
patients with HCC, it is important to elucidate the mechanism
underlying HCC occurrence and identify novel diagnostic markers and
therapeutic targets.
Recent studies demonstrated that circRNAs were
implicated in the occurrence and development of HCC, serving an
important role in numerous biological processes; therefore,
circRNAs may serve as potential diagnostic biomarkers and
therapeutic targets (23,24). A number of circRNAs have been
reported to be involved in cancer cell proliferation, invasion and
migration. For example, hsa_circ_0005986 was found to be decreased
in HCC tissues, and hsa_circ_0005986 reduced the expression levels
of target gene Notch1 by binding to miR-129-5p. In addition,
hsa_circ_0005986 downregulation promoted HCC cell proliferation via
cell cycle transformation (25).
Furthermore, circβ-catenin was demonstrated to be highly expressed
in liver cancer tissues, and knockdown significantly inhibited the
malignant phenotype in vivo and in vitro, which
activated the Wnt signaling pathway via the internal ribosomal
binding site to activate the translation of proteins and promote
cancer cell migration (26).
Finally, DOCK1 expression has been found to be increased in thyroid
cancer, bladder cancer and oral squamous cell carcinoma, and
promotes cancer development (11–13).
In the present study, it was found that DOCK1 was mainly expressed
in the cytoplasm, and the mechanism of competing endogenous RNA
takes place in the cytoplasm. In addition, DOCK1 expression was
increased in Hep3b cells and the knockdown of DOCK1 suppressed the
proliferation, migration and invasion of Hep3b cells, which we
speculate could be a potential treatment target of HCC. Therefore,
the role of DOCK1 overexpression was not investigated in the
present study.
miR-654-3p has been shown to be downregulated in
osteosarcoma (OS) tissues and cells, and miR-654-5p overexpression
suppressed OS cell proliferation, invasion and migration (27). miR-654-5p was found to be
downregulated in breast cancer cells and miR-654-5p overexpression
inhibited proliferation and invasion, and promoted apoptosis of
breast cancer cells (28). Salt
inducible kinase 2 knockdown suppressed the migration and invasion
of paclitaxel-resistant ovarian cancer cells, which could be
partially reversed by miR-654-5p downregulation (29). In the present study, the results
indicated that miR-654-5p directly targeted SMAD2. It was
previously reported that inhibition of SMAD2 inhibited the
regression of HCC (18–20). In the present study, miR-654-5p
expression was decreased and SMAD2 expression was increased in HCC
cells. miR-654-5p knockdown or SMAD2 overexpression reversed the
inhibitory effects of interference with DOCK1 on HCC cells.
In conclusion, the expression levels of DOCK1 and
SMAD2 were increased, and that of miR-654-5p was decreased in HCC
cells. Interference with circRNA DOCK1 inhibited HCC cell
proliferation, invasion and migration by upregulating miR-654-5p
and downregulating SMAD2. In addition, miR-654-5p knockdown or
SMAD2 overexpression promoted HCC cell proliferation, invasion and
migration. However, the present study has some limitations. The
research would be improved if more cell lines were included for the
mechanistic aspect of the study. Since HCC is heterogeneous tumor,
it is paramount to investigate multiple in vitro models
using different cell lines in order to bring about wholistic
changes in the discovery of therapeutic targets. In addition,
DOCK1, miR-654-5p and SMAD2 expression levels in HCC tissues and
their association with prognosis of patients with HCC, as well as
verification of the molecular mechanisms of these genes in HCC
in vivo animal models, should be investigated in the
future.
Acknowledgements
Not applicable.
Funding
This study was funded by The National Natural
Science Foundation of China (grant no. 81572427).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and JZ acquired the data, confirmed the
authenticity of all the raw data, and contributed to the analysis
and interpretation of data. YL and YW contributed to the design of
the study and drafted the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roayaie S, Jibara G, Tabrizian P, Park JW,
Yang J, Yan L, Schwartz M, Han G, Izzo F, Chen M, et al: The role
of hepatic resection in the treatment of hepatocellular cancer.
Hepatology. 62:440–451. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kulik L and El-Serag HB: Epidemiology and
management of hepatocellular carcinoma. Gastroenterology.
156:477–491.e1. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galle PR, Forner A, Llovet JM, Mazzaferro
V, Piscaglia F, Raoul JL, Schirmacher P and Vilgrain V; European
Association for the Study of the Liver. Electronic address, :
easloffice@easloffice.eu; European Association for the Study of the
Liver: EASL Clinical Practice Guidelines: Management of
hepatocellular carcinoma. J Hepatol. 69:182–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu AX, Kudo M, Assenat E, Cattan S, Kang
YK, Lim HY, Poon RT, Blanc JF, Vogel A, Chen CL, et al: Effect of
everolimus on survival in advanced hepatocellular carcinoma after
failure of sorafenib: The EVOLVE-1 randomized clinical trial. JAMA.
312:57–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lopez PM, Villanueva A and Llovet JM:
Systematic review: Evidence-based management of hepatocellular
carcinoma - an updated analysis of randomized controlled trials.
Aliment Pharmacol Ther. 23:1535–1547. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han TS, Hur K, Cho HS and Ban HS:
Epigenetic associations between lncRNA/circRNA and miRNA in
hepatocellular carcinoma. Cancers (Basel). 12:26222020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui W and Xue J: Circular RNA DOCK1
downregulates microRNA-124 to induce the growth of human thyroid
cancer cell lines. Biofactors. 46:591–599. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu P, Li X, Guo X, Chen J, Li C, Chen M,
Liu L, Zhang X and Zu X: Circular RNA DOCK1 promotes bladder
carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5
signalling pathway. Cell Prolif. 52:e126142019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Wei Y, Yan Y, Wang H, Yang J,
Zheng Z, Zha J, Bo P, Tang Y, Guo X, et al: CircDOCK1 suppresses
cell apoptosis via inhibition of miR 196a 5p by targeting BIRC3 in
OSCC. Oncol Rep. 39:951–966. 2018.PubMed/NCBI
|
|
14
|
Kong R: Circular RNA hsa_circ_0085131 is
involved in cisplati-resistance of non-small-cell lung cancer cells
by regulating autophagy. Cell Biol Int. 44:1945–1956. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang F, Wu X, Wei M, Guo H, Li H, Shao Z,
Wu Y and Pu J: miR-654-5p Targets HAX-1 to regulate the nalignancy
behaviors of colorectal cancer cells. BioMed Res Int.
2020:49147072020.PubMed/NCBI
|
|
16
|
Majem B, Parrilla A, Jiménez C,
Suárez-Cabrera L, Barber M, Marín A, Castellví J, Tamayo G,
Moreno-Bueno G, Ponce J, et al: MicroRNA-654-5p suppresses ovarian
cancer development impacting on MYC, WNT and AKT pathways.
Oncogene. 38:6035–6050. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang JX, He WL, Feng ZH, Chen DL, Gao Y,
He Y, Qin K, Zheng ZS, Chen C, Weng HW, et al: A positive feedback
loop consisting of C12orf59/NF-κB/CDH11 promotes gastric cancer
invasion and metastasis. J Exp Clin Cancer Res. 38:1642019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng S, Jia Q, Shen H, Xu X, Ling J, Jing
C and Zhang B: Treatment with the herbal formula Songyou Yin
inhibits epithelial-mesenchymal transition in hepatocellular
carcinoma through downregulation of TGF-β1 expression and
inhibition of the SMAD2/3 signaling pathway. Oncol Lett.
13:2309–2315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng X, Gai X, Han S, Moser CD, Hu C,
Shire AM, Floyd RA and Roberts LR: The human sulfatase 2 inhibitor
2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor
effect in hepatocellular carcinoma mediated via suppression of
TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer.
52:225–236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Liu G, Li Q, Wang F, Xie F, Zhai
R, Guo Y, Chen T, Zhang N, Ni W, et al: Mucin1 promotes the
migration and invasion of hepatocellular carcinoma cells via
JNK-mediated phosphorylation of Smad2 at the C-terminal and linker
regions. Oncotarget. 6:19264–19278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S,
Chen Z, Xiao B and Hu Y: Decreased expression of hsa_circ_0003570
in hepatocellular carcinoma and its clinical significance. J Clin
Lab Anal. 32:e222392018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y
and Guo J: Hsa_circ_0005986 inhibits carcinogenesis by acting as a
miR-129-5p sponge and is used as a novel biomarker for
hepatocellular carcinoma. Oncotarget. 8:43878–43888. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, et al: Translation of the
circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu XZ, Song H, Zhao Y and Zhang L:
MiR-654-5p regulated cell progression and tumor growth through
targeting SIRT6 in osteosarcoma. Eur Rev Med Pharmacol Sci.
24:3517–3525. 2020.PubMed/NCBI
|
|
28
|
Tan YY, Xu XY, Wang JF, Zhang CW and Zhang
SC: MiR-654-5p attenuates breast cancer progression by targeting
EPSTI1. Am J Cancer Res. 6:522–532. 2016.PubMed/NCBI
|
|
29
|
Li ZY, Wang XL, Dang Y, Zhu XZ, Zhang YH,
Cai BX and Zheng L: Long non-coding RNA UCA1 promotes the
progression of paclitaxel resistance in ovarian cancer by
regulating the miR-654-5p/SIK2 axis. Eur Rev Med Pharmacol Sci.
24:591–603. 2020.PubMed/NCBI
|