Introduction
Lower varicose veins are a common disorder that
mainly affect the great saphenous vein (1). Vascular smooth muscle cells (VSMCs)
are the main cellular components of the normal vein wall, and
phenotypic transition of VSMCs is a pathophysiological process that
occurs in vascular diseases. Previous studies have reported that
the transition of VSMCs from a contractile phenotype to a synthetic
phenotype may lead to vascular remodeling and subsequently result
in varicose veins (2,3).
MicroRNAs (miRNAs/miRs) exhibit different expression
levels in varicose veins compared with in normal veins (4). Notably, miR-199a-5p has been reported
to participate in several pathological processes, such as
osteoclast differentiation (5),
spinal cord injury (6) and tumor
proliferation (7). In addition,
overexpression of miR-199a-5p has been shown to promote pulmonary
artery hypertension by downregulating Smad3 in a previous study
(8). Moreover, VSMCs transfected
with miR-199a-5p mimics could inhibit nitric oxide levels and
increase Ca2+ levels (8). Huang et al (9) transfected human airway smooth muscle
cells with miR-199a-5p mimics and revealed that miR-199a-5p
upregulation may contribute to neutrophilic asthma pathogenesis by
modulating the inflammatory process. However, the role of
miR-199a-5p and VSMC phenotypic transition in varicose veins
remains unclear.
miR-199a-5p can suppress mRNA expression by directly
targeting the 3′-untranslated region (3′-UTR) (5,6,8,9).
Notably, forkhead box C2 (FOXC2) is a predicted target of
miR-199a-5p (10). FOXC2 is a key
transcription factor that participates in regulating adipose cell
metabolism, and serves an important role in blood vessel and
lymphatic vessel development (11).
A recent report revealed that FOXC2 may be associated with venous
valve dysfunction (12). In
addition, FOXC2 has been reported to be upregulated in venous cells
and to promote high expression of Notch pathway-related proteins
(Dll4 and Hey2) (13). Notably, the
Notch pathway has an essential role in the development of vascular
networks (14). These findings
indicated that miR-199a-5p/FOXC2 may be involved in varicose vein
pathogenesis.
The present study aimed to investigate the roles of
miR-199a-5p in varicose veins and in the phenotypic transition of
VSMCs. Bioinformatics analysis was performed to identify the
potential roles of miR-199a-5p/FOXC2 in varicose vein pathogenesis.
The miR-199a-5p/FOXC2 axis may provide a novel mechanistic insight
into the pathogenesis of varicose veins and may serve as a
promising diagnostic biomarker and therapeutic target for the
treatment of varicose veins.
Materials and methods
Tissue collection
The present study was conducted in accordance with
the Declaration of Helsinki and was approved by the Ethics
Committee of Qianwei Hospital of Jilin Province (Changchun, China;
approval no. QW202000224). The varicose vein tissues were obtained
from patients with varicose veins (n=10; average age, 63.2±8.9
years; six patients were female; four had been diagnosed with
hypertension; and one had been diagnosed with diabetes mellitus).
Normal great saphenous vein tissues were obtained from individuals
undergoing coronary artery bypass grafting surgery (n=10; average
age, 63.6±8.7 years; five patients were female; five had been
diagnosed with hypertension; and two had been diagnosed with
diabetes mellitus). The inclusion and exclusion criteria of
varicose vein was based on the criteria from the Updated 2020
Clinical, Etiologic, Anatomic and Pathophysiologic (CEAP)
classification guidelines (15).
All patients agreed to the use of their samples in scientific
research and provided written informed consent.
Bioinformatics analysis
TargetScan (version 7.1; www.targetscan.org) online tool (10) was used to predict the relevant
miRNAs that had binding sites with FOXC2 (Table SI). Conserved miRNAs were used for
further confirmation. miR-199a was selected as the target miRNA as
it has been more widely studied than miR-199b.
VSMC transfection
miR-199a-5p mimics and pcDNA3.1-FOXC2 vector were
used to induce the overexpression of miR-199a-5p and FOXC2,
respectively. miR-199a-5p inhibitor and small interfering (si)RNA
against FOXC2 (si-FOXC2) were used to knock down miR-199a-5p and
FOXC2, respectively. Negative control (NC) mimics, NC inhibitor,
pCDNA3.1 NC (empty plasmid) and NC siRNA (siNC) were used as the
control groups. All mimics, inhibitors, plasmids and siRNAs were
designed and synthesized by Shanghai GenePharma Co., Ltd.
miR-199a-5p mimics and inhibitor were tagged with FAM.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to transfect VSMCs (70–80% confluence)
with 250 pmol (0.5 µg) mimic or inhibitor for ~5 h at 37°C. Total
RNA and protein was harvested 48 h post-transfection. All
experiments were conducted 48 h post-transfection. Sequences were
as follows: miR-199a-5p mimics forward,
5′-CCCAGUGUUCAGACUACCUGUUC-3′ and reverse,
5′-ACAGGUAGUCUGAACACUGGGUU-3′; NC mimics forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-199a-5p inhibitor,
5′-GAACAGGUAGUCUGAACACUGGG-3′; NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAA-3′; si-FOXC2 forward,
5′-CUACCUGAGCGAGCAGGUAGTT-3′ and reverse,
5′-AUUCUGCUCGCUCAGGUAGTT-3′; and NC siRNA forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
RT-qPCR
RNA was extracted from transfected or normal VSMCs
and tissues using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). A NanoDrop ND-1000 (NanoDrop; Thermo
Fisher Scientific, Inc.) was used to detect the integrity and
concentration of the RNA samples. Total RNA was reverse transcribed
to cDNA using PrimeScript RT reagent kit with gDNA Eraser (Takara
Bio, Inc.) according to the manufacturer's instructions. qPCR was
performed using FastStart Universal SYBR Green Master (ROX) (Roche
Diagnostics) and an Applied Biosystems 7500 Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) to
confirm the relative expression levels of miR-199a-5p and FOXC2.
qPCR thermocycling conditions were as follows: Initial denaturation
at 95°C for 5 min; followed by 40 cycles at 95°C for 10 sec, 55°C
for 20 sec and 72°C for 20 sec, and a final extension step at 72°C
for 2 min. The relative expression levels of genes were calculated
using the 2−ΔΔCq method (16) after normalization to GAPDH (used for
mRNA normalization) or U6 (used for miRNA normalization). The
primer sequences were as follows: FOXC2 forward,
5′-CGGCCCAGCAGCAAACTTTCC-3′ and reverse,
5′-AGAGGCGGCGTGGATCTGTAG-3′; GAPDH forward,
5′-CGGACCAATACGACCAAATCCG′ and reverse, 5′-AGCCACATCGCTCAGACACC-3′;
miR-199a-5p forward, 5′-CCGGGATCCGCAAACTCAGCTTTAC-3′ and reverse,
5′-CGGAATTCGTGGCGACCGTGATACC-3′; U6 forward,
5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; SM22α
forward, 5′-AACAGCCTGTACCCTGATGG-3′ and reverse,
5′-CGGTAGTGCCCATCATTCTT-3′; Calponin forward,
5′-AGCTAAGAGAAGGGCGGAAC-3′ and reverse, 5′-CATCTGCAGGCTGACATTGA-3′;
SMA forward, 5′-GCGTGGCTATTCCTTCGTTA-3′ and reverse,
5′-ATGAAGGATGGCTGGAACAG-3′; and Myosin Heavy Chain 11 (MYH11)
forward, 5′-TGGAACTTCATCGACTTTGGG-3′ and reverse,
5′-ACAGCTTCTCCACGAAAGAC-3′.
Cell culture
The human VSMC cell line was purchased from the
Shanghai Institute of Biochemistry and Cell Biology and cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in a humidified incubator containing 5% CO2. The medium
was replaced every 3 days. All cells used in the present study were
between passages 5 and 7. VSMCs were serum starved (0.5% fetal
bovine serum) for 24 h and stimulated using platelet-derived growth
factor (PDGF)-BB (20 ng/ml; BioVision, Inc.) at 37°C for 24 h prior
to experimentation.
Western blot analysis
Proteins were extracted from cells and tissues using
RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd.)
and were quantified using a BCA assay (Thermo Fisher Scientific,
Inc.). Total cell proteins (30 µg/lane) were separated by SDS-PAGE
on 10% gels (cat. no. P1200; Beijing Solarbio Science &
Technology Co., Ltd.) and transferred to nitrocellulose membranes
(0.45 µm; EMD Millipore). Blots were blocked with 5% nonfat milk
with 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA) for 2 h at 37°C.
Subsequently, membranes were incubated overnight at 4°C with the
following primary antibodies: FOXC2 (1:500; cat. no. ab245510;
Abcam), smooth muscle 22α (SM22α; 1:500; cat. no. 10493-1-AP;
ProteinTech Group, Inc.), smooth muscle actin (SMA; 1:800; cat. no.
55135-1-AP; ProteinTech Group, Inc.), MYH11 (1:1,000; cat. no.
21404-1-AP; ProteinTech Group, Inc.), calponin (1:1,000; cat. no.
bs-0095R; BIOSS), proliferating cell nuclear antigen (PCNA;
1:3,000; cat. no. ab92552; Abcam) and β-actin (1:5,000; cat. no.
bs-0061R; BIOSS), followed by incubation with secondary antibodies
(1:5,000; cat. no. bs-0296G-HRP; BIOSS) at room temperature for 2.5
h. Protein bands were visualized with an ECL western blotting kit
(Cell Signaling Technology, Inc.) using cSeries Imager (Azure
Biosystems, Inc.). Protein expression levels were semi-quantified
using ImageJ software (version 4.62; National Institutes of
Health).
Dual luciferase reporter assay
293T cells were purchased from the Shanghai
Institute of Biochemistry and Cell Biology. The wild-type (WT) or
mutant (MUT) FOXC2 3′-UTRs were cloned into the pGL6-miR vector
(Beyotime Institute of Biotechnology). FOXC2 mutation was induced
using the Quick-Change Site-Directed Mutagenesis kit (Agilent
Technologies, Inc.). Subsequently, 2×104 293T cells per
well were seeded into 6-well plates for 24 h at 37°C before
transfection. A Dual Luciferase Reporter Gene Assay kit (Promega
Corporation) was used to assess luciferase activity after 293T
cells were co-transfected with either pGL6-FOXC2-WT (0.5 µg) or
pGL6-FOXC2-MUT (0.5 µg) and either miR-199a-5p mimics (0.5 µg) or
mimics NC (0.5 µg) using Lipofectamine 2000. A total of 24 h
post-transfection, luciferase activity was measured at 560 nm.
Luciferase activity was normalized to Renilla
luciferase.
Cell proliferation assay
The proliferation of VSMCs transfected with
miR-199a-5p mimics or inhibitor was determined using the Cell
Counting Kit-8 (CCK-8) assay (TransGen Biotech Co., Ltd.) according
to the manufacturer's instructions. Cells (3×104 per
well) transfected with miR-199a-5p mimics or inhibitor were seeded
into a 96-well plate for 24, 48 and 72 h. Subsequently, 10 µl CCK-8
Solution was added to the medium and incubated for 4 h at 37°C. The
absorbance was detected at 450 nm using an Epoch microplate
(Bio-Tek Instruments, Inc.).
Transwell migration assay
Cell migration was assessed by performing a
Transwell migration assay using Transwell chambers with 8-µm filter
inserts (24-well inserts). Transfected cells (5×104)
cultured in serum-free medium were seeded into the upper chamber of
Transwell inserts, whereas 500 µl DMEM supplemented with 20% FBS
was added to the lower chamber. After 36 h at 37°C, a cotton swab
was used to remove the cells remaining in the upper chamber. Cells
that had migrated to the lower chamber were fixed with 100%
methanol at 37°C and stained with 0.1% crystal violet. The number
of migrated cells was then counted under a laser scanning confocal
microscope (magnification, ×40; Leica Microsystems GmbH).
Wound healing assay
Cells (2.5×105 cells/well) were seeded
into a 6-well plate and transfected with miR-199a-5p mimics or
inhibitor for 48 h. Cells underwent serum starvation in medium
containing 0.5% FBS for 48 h, after which, a linear wound was
created in the center of the cell layer using a 200-µl pipette tip.
Cells were then incubated in serum-free medium for 48 h. The
floating cells were removed by washing the cells with PBS. The
distance of cell migration was determined by the mean value of the
width of the gap between the top, middle and bottom of the wound.
The wounds were observed in five fields of view using a light
microscope (magnification, ×100).
Statistical analysis
All statistical analyses were performed using SPSS
20.0 (IBM Corp.). Data are presented as the mean ± standard
deviation. The experiments were repeated at least three times.
Comparisons among groups were analyzed using one-way ANOVA followed
by Bonferroni's post hoc test, or a paired Student's t-test for
comparison of differences between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-199a-5p regulates VSMC phenotypic
transition
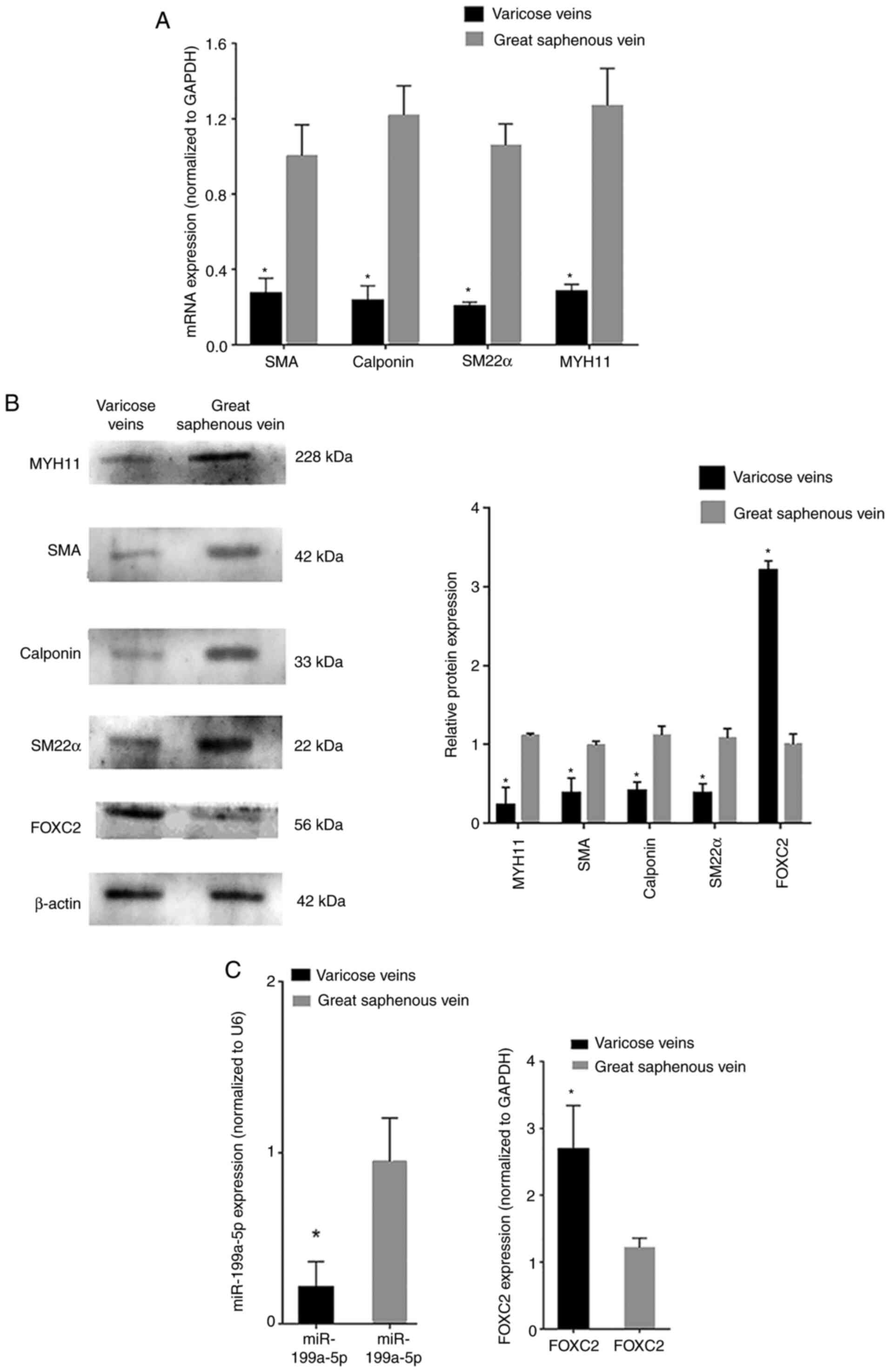
RT-qPCR analysis revealed that the mRNA expression
levels of phenotypic transition biomarkers, such as SM22α,
calponin, SMA and MYH11, were decreased in varicose vein tissues
compared with those in normal great saphenous vein tissues
(Fig. 1A). Western blot analysis
revealed that the protein expression levels of SM22α, calponin, SMA
and MYH11 were also decreased in varicose vein tissues compared
with those in normal great saphenous vein tissues (Fig. 1B). RT-qPCR analysis revealed that
miR-199a-5p was downregulated in varicose vein tissues, whereas
FOXC2 was upregulated in varicose vein tissues compared with that
in normal great saphenous vein tissues (Fig. 1C). In addition, western blot
analysis confirmed that the protein expression levels of FOXC2 were
increased in varicose vein tissues compared with those in normal
great saphenous vein tissues (Fig.
1B). These results indicated that miR-199a-5p may be involved
in VSMC proliferation during varicose vein pathogenesis and could
regulate VSMC phenotypic transition. The transition of VSMCs from a
contractile phenotype to a synthetic phenotype was inferred from
the expression levels of phenotypic transition-related
biomarkers.
Downregulation of miR-199a-5p in
proliferative VSMCs
To verify the finding that miR-199a-5p may
participate in the regulation of VSMC proliferation, the expression
levels of miR-199a-5p and FOXC2 were detected in proliferating
VSMCs at 12, 24, 36, 48 and 60 h after stimulation with PDGF-BB (20
ng/ml) (17). miR-199a-5p (Fig. 2A) and SM22α mRNA (Fig. 2B) expression levels were
downregulated during VSMC proliferation, whereas FOXC2 mRNA
expression was upregulated (Fig.
2C). In addition, western blot analysis confirmed that SM22α
was downregulated, but FOXC2 was upregulated in a time-dependent
manner (Fig. 2D). These results
indicated that miR-199a-5p was downregulated during VSMC
proliferation in a time-dependent manner.
miR-199a-5p regulates the expression
of phenotypic transition biomarkers involved in VSMC
differentiation
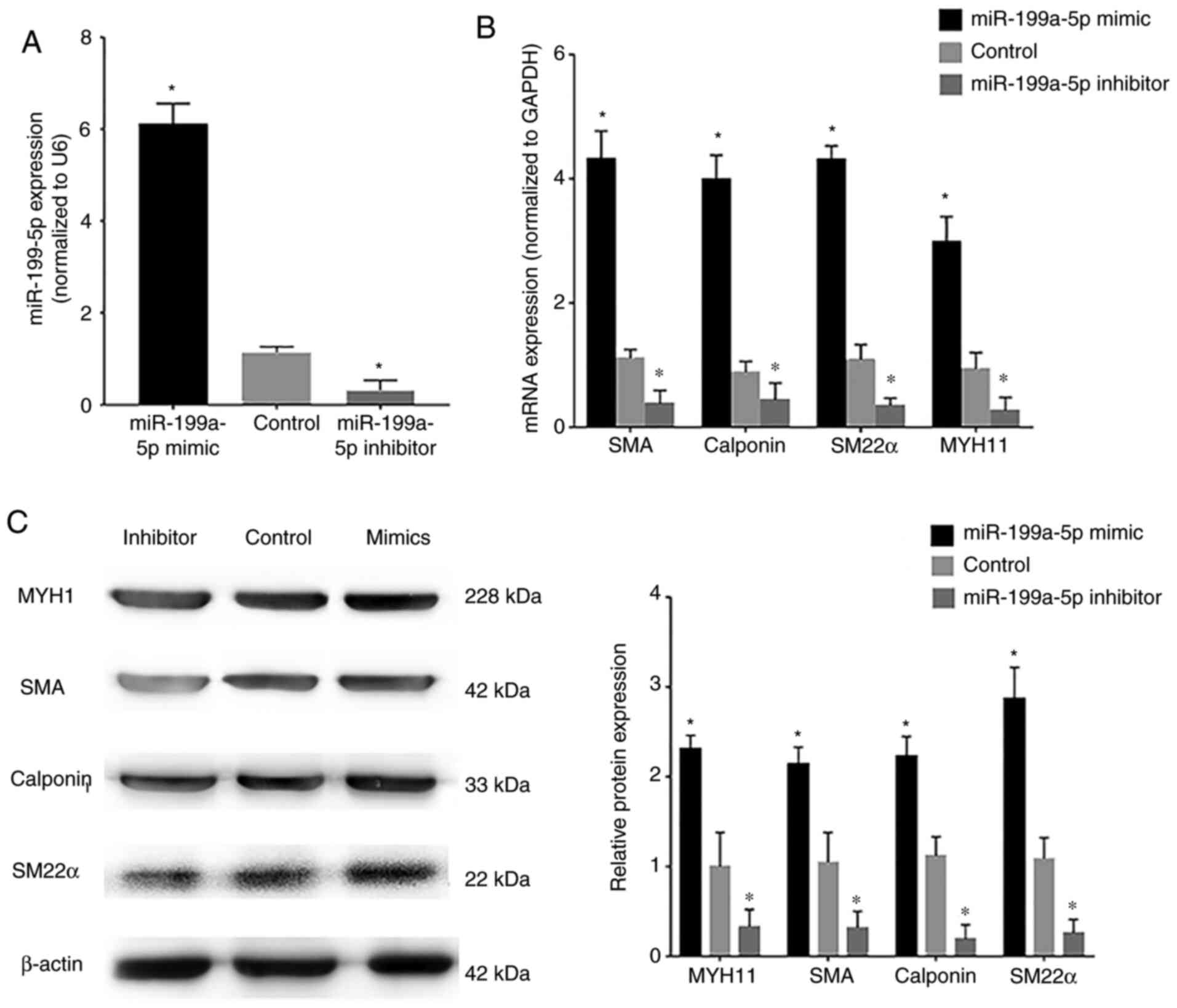
VSMCs were transfected with miR-199a-5p mimics or
miR-199a-5p inhibitor. Confirmation of transfection of VSMCs with
the FAM-labeled miR-199a-5p mimics and miR-199a-5p inhibitor is
shown in Fig. S1A and B,
respectively. The transfection efficiency of miR-199a-5p mimics and
inhibitor was assessed by RT-qPCR; transfection with miR-199a-5p
mimics induced an increase in miR-199a-5p expression in VSMCs,
whereas the miR-199a-5p inhibitor markedly reduced endogenous
miR-199a-5p expression levels compared with the control
(untransfected VSMCs) (Fig. 3A). In
addition, VSMCs were transfected with miR-199a-5p mimics, NC
mimics, miR-199a-5p inhibitor and NC inhibitor and FOXC2 protein
expression was detected by western blotting (Fig. S1C). Western blotting showed that
FOXC2 was upregulated in VSMCs transfected with inhibitor compared
with NC inhibitor, FOXC2 was downregulated in VSMCs transfected
with mimics compared with NC mimics. RT-qPCR and western blotting
revealed that the mRNA and protein expression levels, respectively,
of the phenotypic transition biomarkers SM22α, calponin, SMA and
MYH11, which are involved in VSMC differentiation, were
downregulated in cells transfected with the miR-199a-5p inhibitor
(Fig. 3B and C), whereas
miR-199a-5p overexpression was associated with upregulation of
these phenotypic transition biomarkers. These results suggested
that miR-199a-5p may regulate the expression of phenotypic
transition biomarkers involved in VSMC differentiation.
miR-199a-5p regulates VSMC
proliferation and migration
The effects of miR-199a-5p on VSMC migration were
determined by performing Transwell migration and wound healing
assays following miR-199a-5p mimics and inhibitor transfection. The
results revealed that miR-199a-5p knockdown significantly promoted
the migratory ability of VSMCs, whereas transfection with
miR-199a-5p mimics significantly inhibited the migratory ability of
VSMCs (Fig. 4A). A marked increase
in cell migration was also detected by wound healing assay after
transfection with the miR-199a-5p inhibitor at 48 h, whereas cells
transfected with the miR-199a-5p mimics exhibited reduced cell
migration ability at 48 h (Fig.
4B).
A CCK-8 proliferation assay was performed to detect
the proliferative ability of VSMCs. VSMCs were transfection with
the miR-199a-5p mimics or inhibitor, and cell proliferation was
measured at 24, 48 and 72 h. Untransfected cells were considered
the control group. VSMCs overexpressing miR-199a-5p exhibited a
decrease in proliferation compared with that in the control cells
(Fig. 4C), whereas VSMCs with
miR-199a-5p knockdown exhibited increased proliferation. Western
blot analysis revealed that the proliferation biomarker PCNA was
downregulated in miR-199a-5p mimics-transfected cells, but
upregulated in miR-199a-5p inhibitor-transfected cells (Fig. 4D). These results indicated that
miR-199a-5p may decrease VSMC proliferation and migration.
FOXC2 is a target gene of
miR-199a-5p
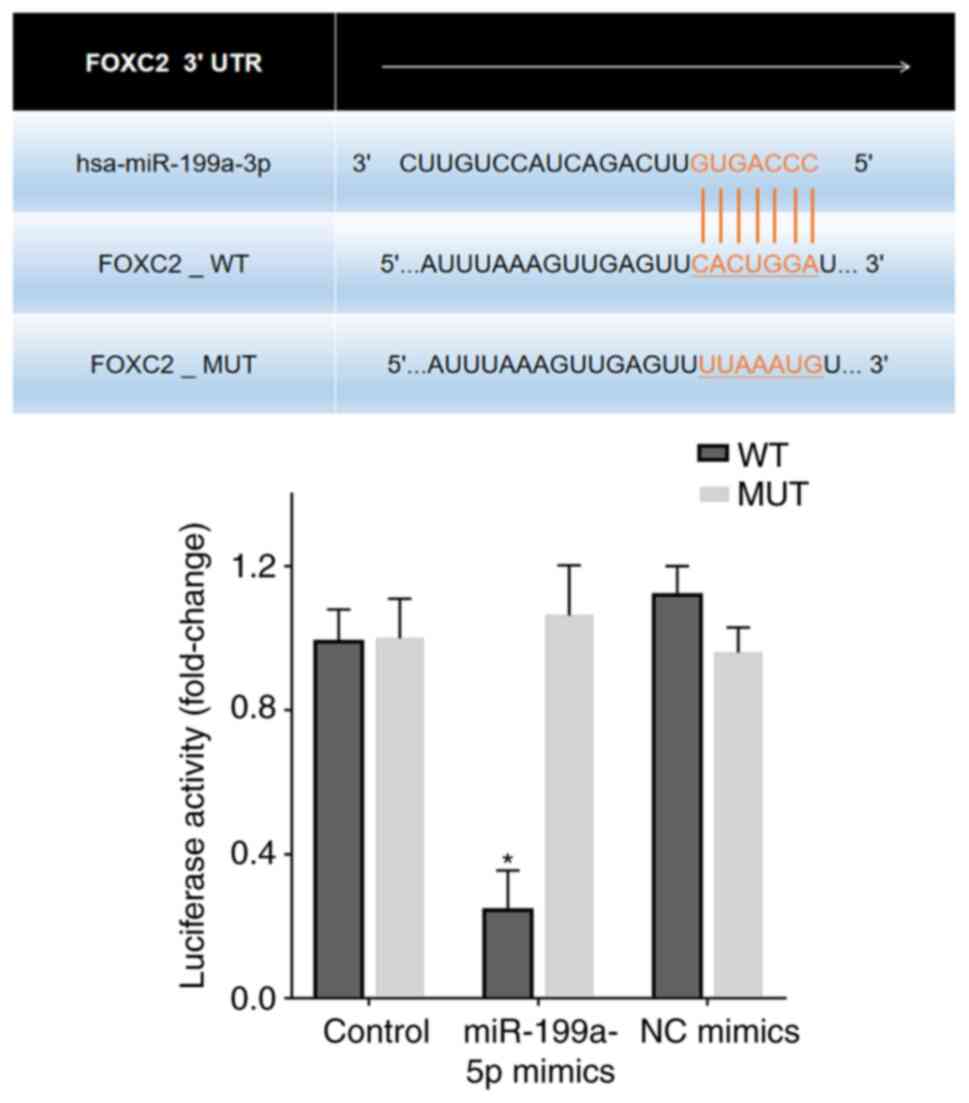
Bioinformatics analysis revealed putative
miR-199a-5p target sites in the FOXC2 3′-UTR region (Fig. 5). A dual luciferase reporter assay
was performed to confirm the effects of miR-199a-5p on FOXC2. 293T
cells transfected with miR-199a-5p mimics reduced luciferase
activity in cells expressing FOXC2-WT compared with that in
untransfected control cells or 293T cells transfected with NC
mimics and FOXC2-MUT/FOXC2-WT (Fig.
5).
The pcDNA3.1-FOXC2 overexpression vector and
si-FOXC2 were transfected into VSMCs for rescue experiments. VSMCs
were transfected with FOXC2 vector, pCDNA3.1 NC, si-FOXC2 and siNC,
and the transfection efficiency was confirmed by western blotting
(Fig. S2). si-FOXC2 decreased the
expression levels of FOXC2 compared with siNC, which increased the
expression levels of MYH11, SMA, Calponin and SM22. FOXC2 vector
increased the expression levels of FOXC2 compared with pcDNA3.1 NC,
which decreased the expression levels of MYH11, SMA, Calponin and
SM22. RT-qPCR was also performed to verify the transfection
efficiency of the pcDNA3.1-FOXC2 vector and FOXC2 siRNA;
untransfected cells were considered the control group.
pcDNA3.1-FOXC2 vector transfection increased the expression levels
of FOXC2 in VSMCs compared with those in the control group
(Fig. 6A). VSMCs transfected with
miR-199a-5p mimics + pcDNA3.1-FOXC2 vector exhibited higher
proliferation (Fig. 6B) and
migration (Fig. 6C) compared with
that in cells transfected with only the miR-199a-5p mimics.
Phenotypic transition biomarkers involved in VSMC differentiation,
such as SM22α, calponin, SMA and MYH11, were downregulated in VSMCs
transfected with miR-199a-5p mimics + pcDNA3.1-FOXC2 vector
compared with those in VSMCs transfected with the miR-199a-5p
mimics only (Fig. 6D and E).
 | Figure 6.FOXC2 rescue experiments. (A) RT-qPCR
confirmed that FOXC2 was overexpressed and knocked down
post-transfection with pcDNA3.1-FOXC2 vector or FOXC2 siRNA,
respectively. *P<0.05 vs. control (n=3). (B) CCK-8 confirmed
that VSMC proliferation was enhanced after transfection with
miR-199a-5p mimics + FOXC2 vector compared with miR-199a-5p mimics
alone. *P<0.05 (n=10). (C) Transwell migration assays revealed
that FOXC2 enhanced VSMC migration. Magnification, ×40. *P<0.05
(n=10). (D) Western blot analysis revealed that the expression
levels of VSMC differentiation biomarkers were decreased in cells
transfected with miR-199a-5p mimics + FOXC2 vector compared with
those in cells transfected with miR-199a-5p mimics only. *P<0.05
(n=3). (E) RT-qPCR revealed that FOXC3 decreased the expression
levels of VSMC differentiation biomarkers compared with those in
cells transfected with miR-199a-5p mimics only. *P<0.05 (n=10).
(F) RT-qPCR was used to detect the expression levels of phenotypic
transition biomarkers. *P<0.05 vs. control (n=10). (G) CCK-8
confirmed that proliferation of VSMCs was reduced in response to
FOXC2 silencing, but increased in response to FOXC2 overexpression.
*P<0.05 vs. control (n=3). (H) Wound healing assay revealed that
migration of VSMCs was reduced post-transfection with the FOXC2
siRNA, but increased following the overexpression of FOXC2 compared
with the control group. *P<0.05 vs. control (n=3). FOXC2,
forkhead box C2; miR-199a-5p, microRNA-199a-5p; MYH11, myosin heavy
chain 11; RT-qPCR, reverse transcription-quantitative PCR; siRNA,
small interfering RNA; SM22α, smooth muscle 22α; SMA, smooth muscle
actin; VSMC, vascular smooth muscle cell; CCK-8, Cell Counting
Kit-8. |
In addition, the role of FOXC2 in VSMC
proliferation, migration and phenotypic switch was assessed.
Expression levels of the differentiation biomarkers were increased
in VSMCs transfected with the FOXC2 siRNA compared with those in
the control group (Fig. 6F). In
addition, a marked decrease in cell proliferation was detected in
cells transfected with the FOXC2 siRNA (Fig. 6G). As shown in Fig. 6H, a marked decrease in the migration
of cells transfected with the FOXC2 siRNA was revealed at 48 h.
Discussion
Major biological functions of the vascular system
are performed in VSMCs. Notably, phenotypic switching of VSMCs has
been reported to serve a critical role in varicose vein
pathogenesis (4). VSMCs acquire
proliferative and migratory abilities when they transition to a
synthetic phenotype, resulting in vascular remodeling. In the
present study, contractile markers, such as SM22α, SMA, calponin
and MYH11, were downregulated in varicose vein tissues compared
with those in normal vein tissues, suggesting that the contractile
phenotype is a normal phenotype of VSMCs. To confirm that
miR-199a-5p specifically participated in regulating VSMC
proliferation, the expression levels of miR-199a-5p were detected
in VSMCs at different time points, which revealed that miR-199a-5p
was downregulated in VSMCs during cell proliferation.
In the present study, RT-qPCR confirmed that the
expression of miR-199a-5p was downregulated in varicose vein
tissues, which revealed that miR-199a-5p may promote VSMC
transition to the synthetic phenotype by reducing VSMC contractile
markers, such as SM22α, SMA, calponin and MYH11. The function of
miR-199a-5p has been reported in several other diseases. For
example, miR-199a-5p downregulation has been reported to promote
non-small cell lung cancer proliferation (18). In addition, miR-199a-5p upregulation
could inhibit the progression of papillary thyroid carcinoma by
reducing the migratory ability of cells (19). In addition, miR-199a-5p upregulation
promoted colorectal cancer cell proliferation and migration
(20). Therefore, the roles of
miR-199a-5p vary depending on the type of tissues involved in the
disease. Ahmadi et al (18)
showed that the downregulation of miR-199a-5p promoted lung cancer
cell proliferation and migration via targeting GRP78 within the
unfolded protein response pathway. Ma et al (19) reported that the downregulation of
miR-199a-5p promoted papillary thyroid carcinoma cell proliferation
and migration by targeting SNAI1. Thus, the present findings are
consistent with the findings of previous studies (18,19),
indicating that miR-199a-5p downregulation may promote VSMC
proliferation and migration. In the present study, VSMCs were
transfected with a miR-199a-5p inhibitor, after which, the
expression levels of the contractile markers SM22α, SMA, calponin
and MYH11 were downregulated, suggesting that VSMCs switched to a
synthetic phenotype. Conversely, it has previously been reported
that miR-199a-5p upregulation may promote cell proliferation and
metastasis in cervical carcinoma (21), and miR-199a-5p downregulation could
inhibit osteoblast differentiation (5). Therefore, the function of miR-199a-5p
may vary in different cell types.
Phenotypic switching of VSMCs serves as an initial
stage in varicose vein formation; however, the mechanism involved
in phenotypic switching during varicose vein formation remains
unclear. The role of miR-199a-5p also depends on its target genes.
After confirming that miR-199a-5p was downregulated in varicose
vein tissues, the present study aimed to identify the potential
target genes, as miRNAs regulate translation by binding to the
3′-UTRs of target genes. FOXC2 is a pathogenic gene that has been
reported to be associated with venous valve dysfunction (22). A number of studies have found that
FOXC2 plays an important role in a number of cancers. FOXC2 has
been reported to be overexpressed in breast (23), stomach (24), cervical (25) and ovarian (26) cancer. In addition, the transcription
factor FOXC2 has been shown to promote cell proliferation and also
induce the EMT via multiple pathways in malignancies. Moreover,
FOXC2 can also act as a crucial mediator in both angiogenesis and
lymphangiogenesis (27), thus it
could act as a potential transcriptional mediator involved in
increased pathological angiogenesis and neovascularization
(28). The present study confirmed
that FOXC2 was upregulated in varicose vein tissues and promoted
VSMC proliferation; this finding is consistent with that of a
previous report (29). After
identifying an opposite trend in miR-199a-5p and FOXC2 expression
levels in varicose vein tissues, it was hypothesized that
miR-199a-5p downregulation seemed to be associated with varicose
vein pathogenesis by binding FOXC2 These findings indicated that
miR-199a-5p downregulation may promote the development of varicose
veins by upregulating FOXC2.
In conclusion, miR-199a-5p was downregulated in
varicose vein tissues compared with that in normal vein tissues. In
addition, miR-199a-5p inhibited FOXC2 by targeting the 3′-UTR,
which regulated VSMC phenotypic switching. Therefore, the
miR-199a-5p/FOXC2 axis may provide a novel mechanistic insight into
the pathogenesis of varicose veins and may serve as a promising
diagnostic biomarker and therapeutic target for the treatment of
varicose veins.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was partly supported by the Development of
Science and Technology in Jilin Province Project, China (grant no.
20190303045SF).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC and ZC designed and supervised the study. YC and
WW performed the experiments. WW, XJ and LL analyzed and
interpreted the data. YC and ZC confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and was approved by the Ethics
Committee of Qianwei Hospital of Jilin Province (approval no.
QW202000224; Changchun, China). All patients agreed to the use of
their samples in scientific research and provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
FOXC2
|
forkhead box C2
|
|
miRNA
|
microRNA
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
VSMCs
|
vascular smooth muscle cells
|
References
|
1
|
Piazza G: Varicose veins. Circulation.
130:582–587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jacobs BN, Andraska EA, Obi AT and
Wakefield TW: Pathophysiology of varicose veins. J Vasc Surg Venous
Lymphat Disord. 5:460–467. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu Y, Bei Y, Li Y and Chu H: Phenotypic
and functional transformation in smooth muscle cells derived from
varicose veins. J Vasc Surg Venous Lymphat Disord. 5:723–733. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang X, Liu Z, Shen L, Jin Y, Xu G, Zhang
Z, Fang C, Guan W and Liu C: Augmentation of miR-202 in varicose
veins modulates phenotypic transition of vascular smooth muscle
cells by targeting proliferator-activated receptor-γ
coactivator-1α. J Cell Biochem. 120:10031–10042. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi XB, Jia B, Wang W, Xu GH, Guo JC, Li X
and Liu JN: Role of miR-199a-5p in osteoblast differentiation by
targeting TET2. Gene. 726:1441932020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao N, Fang B, Lv H, Jiang Y, Chen F, Wang
Z and Ma H: Upregulation of miR-199a-5p protects spinal cord
against ischemia/reperfusion-induced injury via downregulation of
ECE1 in rat. Cell Mol Neurobiol. 38:1293–1303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu H: Forkhead box transcription factors
in embryonic heart development and congenital heart disease. Life
Sci. 144:194–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Liu G, Zhang H and Wang J:
MiRNA-199a-5p influences pulmonary artery hypertension via
downregulating Smad3. Biochem Biophys Res Commun. 473:859–866.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Y, Zhang S, Fang X, Qin L, Fan Y,
Ding D, Liu X and Xie M: Plasma miR-199a-5p is increased in
neutrophilic phenotype asthma patients and negatively correlated
with pulmonary function. PLoS One. 13:e01935022018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mellor RH, Brice G, Stanton AW, French J,
Smith A, Jeffery S, Levick JR, Burnand KG and Mortimer PS;
Lymphoedema Research Consortium, : Mutations in FOXC2 are strongly
associated with primary valve failure in veins of the lower limb.
Circulation. 115:1912–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo Z, Luo C, Zhu T, Li L and Zhang W:
Elevated c-fos expression is correlated with phenotypic switching
of human vascular smooth muscle cells derived from lower limb
venous varicosities. J Vasc Surg Venous Lymphat Disord. 9:242–251.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Surendran S, Ramegowda KS, Suresh A, Binil
Raj SS, Lakkappa RK, Kamalapurkar G, Radhakrishnan N and C Kartha
C: Arterialization and anomalous vein wall remodeling in varicose
veins is associated with upregulated FoxC2-Dll4 pathway. Lab
Invest. 96:399–408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baeten JT and Lilly B: Notch signaling in
vascular smooth muscle cells. Adv Pharmacol. 78:351–382. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Porter JM and Moneta GL: Reporting
standards in venous disease: An update. International Consensus
Committee on Chronic Venous Disease. J Vasc Surg. 21:635–645. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Y, Yu S, Liu Y, Zhang J, Han L and Xu
Z: MicroRNA-124 controls human vascular smooth muscle cell
phenotypic switch via Sp1. Am J Physiol Heart Circ Physiol.
313:H641–H649. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ahmadi A, Khansarinejad B, Hosseinkhani S,
Ghanei M and Mowla SJ: miR-199a-5p and miR-495 target GRP78 within
UPR pathway of lung cancer. Gene. 620:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma S, Jia W and Ni S: miR-199a-5p inhibits
the progression of papillary thyroid carcinoma by targeting SNAI1.
Biochem Biophys Res Commun. 497:181–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu QD, Zhou QQ, Dong L, Huang Z, Wu F and
Deng X: MiR-199a-5p inhibits the growth and metastasis of
colorectal cancer cells by targeting ROCK1. Technol Cancer Res
Treat. 17:15330346187755092018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu D, Yang Y and Huang X: miR-199a-5p
promotes proliferation and metastasis and epithelial-mesenchymal
transition through targeting PIAS3 in cervical carcinoma. J Cell
Biochem. 120:13562–13572. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim CS and Davies AH: Pathogenesis of
primary varicose veins. Br J Surg. 96:1231–1242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pham TND, Perez White BE, Zhao H,
Mortazavi F and Tonetti DA: Protein kinase C α enhances migration
of breast cancer cells through FOXC2-mediated repression of
p120-catenin. BMC Cancer. 17:8322017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren YH, Liu KJ, Wang M, Yu YN, Yang K,
Chen Q, Yu B, Wang W, Li QW, Wang J, et al: De-SUMOylation of FOXC2
by SENP3 promotes the epithelial-mesenchymal transition in gastric
cancer cells. Oncotarget. 5:7093–7104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng CH, Quan Y, Li YY, Deng WG, Shao WJ
and Fu Y: Expression of transcription factor FOXC2 in cervical
cancer and effects of silencing on cervical cancer cell
proliferation. Asian Pac J Cancer Prev. 15:1589–1595. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu B, Han SM, Tang XY, Han L and Li CZ:
Overexpressed FOXC2 in ovarian cancer enhances the
epithelial-to-mesenchymal transition and invasion of ovarian cancer
cells. Oncol Rep. 31:2545–2554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu X and Liu NF: FOXC2 transcription
factor: A novel regulator of lymphangiogenesis. Lymphology.
44:35–41. 2011.PubMed/NCBI
|
|
28
|
Sano H, Leboeuf JP, Novitskiy SV, Seo S,
Zaja-Milatovic S, Dikov MM and Kume T: The Foxc2 transcription
factor regulates tumor angiogenesis. Biochem Biophys Res Commun.
392:201–206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang C, Li H and Guo X: FOXC2-AS1
regulates phenotypic transition, proliferation and migration of
human great saphenous vein smooth muscle cells. Biol Res.
52:592019. View Article : Google Scholar : PubMed/NCBI
|