Introduction
Endometrial cancer (EC) comprises a group of
epithelial malignant tumors that are found in the endometrium,
which often occur in perimenopausal and postmenopausal women
(1). EC is the 15th most common
type of malignant tumor worldwide, and it is the most common
gynecological malignant tumor in developed countries (2). Surgical resection and postoperative
adjuvant treatments for EC have been standardized and comprise
hysterectomy and bilateral salpingo-oophorectomy, followed by
paclitaxel/platinum conventional chemotherapy (3). However, women with relapsed or
advanced disease have a low response rate to conventional
treatments and extremely poor clinical outcomes (4,5).
Therefore, it remains necessary to develop further targeted
treatments for these patients based on the precise molecular
pathogenesis of EC.
Our previous study demonstrated that long non-coding
RNA (lncRNA) small nucleolar RNA host gene 12 (SNHG12) promoted
cell proliferation, invasion and migration by targeting
microRNA-4429 in EC (6). However,
the upstream mediators and involved underlying mechanisms were not
identified. Zic family member 2 (ZIC2) is a member of the ZIC
family of C2H2-type zinc finger proteins, which function as
transcription factors and regulate tissue specific gene expression
(7). A previous study reported that
the differential expression of ZIC2 in EC was associated with lymph
node metastasis (7). ZIC2 has also
been indicated to serve oncogenic roles in numerous types of human
cancer, including hepatocellular carcinoma (HCC) (8), cervical cancer (9), acute myeloid leukemia (10) and nasopharyngeal carcinoma (NPC)
(11). However, to the best of our
knowledge, the specific role of ZIC2 in EC has not been
determined.
The lncRNA SNHG12 was demonstrated to promote cell
proliferation and metastasis, and predicted a poor prognosis in NPC
by modulating the Notch signaling pathway (12). In addition, the knockdown of SNHG12
inhibited the activation of epithelial-mesenchymal transition and
Notch-1 signaling pathways in NPC (12). Dysregulated Notch signaling was
discovered to affect the tumorigenicity and proliferation of cells
in various types of cancer. For example, in EC, the expression
levels of Notch1 and Notch3 were revealed to be upregulated and the
elevated expression levels of Notch1 were associated with cancer
aggressiveness and poor prognosis in patients (13). Blocking the Notch signaling pathway
could also enhance the effect of the EGFR inhibitor by targeting
CD133+ EC cells (14).
Therefore, the inhibition of the Notch signaling pathway may
inhibit tumorigenesis and provide a novel therapeutic strategy for
EC (14).
Notably, analysis using the JASPAR database
predicted that ZIC2 could bind to the promoter of SNHG12.
Therefore, the present study aimed to investigate whether ZIC2
could bind to SNHG12 to regulate EC cell proliferation and
migration via activating Notch signaling.
Materials and methods
Cell lines and culture
The human EC cell lines, Ishikawa, KLE, RL95-2 and
AN3 CA, were obtained from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences and the cultured human normal
endometrial stromal cell (ESC) line (cat. no. BNCC340262;
http://www.bnbio.com/pro/p1/1/p_3323.html) was
obtained from BeNa Culture Collection; Beijing Beina Chunglian
Institute of Biotechnology. All cells were cultured in DMEM
(Sigma-Aldrich; Merck KGaA) supplemented with 15% FBS
(Sigma-Aldrich; Merck KGaA) and 1% penicillin/streptomycin, and
maintained in an atmosphere with 5% CO2 at 37°C.
Bioinformatics analysis
To analyze the overall level of ZIC2 differential
expression between normal people and cancer patients, the clinical
data in this study was obtained by searching the ENCORI database
(15) from The Cancer Genome Atlas
(TCGA) Genomics data (16), which
is an authoritative online genome analysis technology dominantly
based on large-scale sequencing, and at the same time, the
information that can identify the patient in the clinical data is
hidden.
Cell transfection
Short hairpin RNA (shRNA/sh) targeting ZIC2
(shRNA-ZIC2-1:
5′-CCGGCCGGAGTCTTTGAAGCTGAAACTCGAGTTTCAGCTTCAAAGACT-CCGGTTTTTG-3′;
and shRNA-ZIC2-2:
5′-CCGGCGGAAGCACATGAAGGTCC-ATCTCGAGATGGACCTTCATGTGCTTCCGTTTTTG-3′)
and scrambled negative control (NC) shRNA (shRNA-NC)
(5′-ACTTGCGCTTGCGAAAATCTATATAGC-3′) were designed and synthesized
by GenScript. The ZIC2 and SNHG12 overexpression plasmids were
established by inserting the full-length human ZIC2 and SNHG12 cDNA
sequences (Aksomics, Inc.) into the pcDNA3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.); empty vectors served as the NCs
(Oe-NC). All plasmids (50 nM) were transfected for 2 h at 37°C into
RL95-2 cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and the transfection efficiency was
evaluated at 48 h post-transfection using reverse
transcription-quantitative PCR (RT-qPCR).
Cell counting Kit-8 (CCK-8) assay
RL95-2 cells were seeded at a density of
2×103 cells/well into 96-well plates. Following
transfection with the indicated vectors for 24, 48 or 72 h, the
cells were incubated with 10 µl CCK-8 solution (MedChemExpress) for
2 h at 37°C. The absorbance of the cells was measured at a
wavelength of 450 nm using a microplate reader.
Colony formation assay
For the colony formation assays, the RL95-2 cell
suspension was resuspended in 1 ml medium and then plated into
24-well plates and incubated at 37°C for 2 weeks. Following
incubation, cells were stained for 20 min at room temperature with
2% crystal violet and colonies (>50 cells) were counted under a
light microscope (magnification, ×100).
Wound healing assay
RL95-2 cells were seeded into a six-well plate at a
density of 5×104 cells/well and cultured in an
atmosphere with 5% CO2 at 37°C. Upon cell confluence
reaching 70–80%, the cell monolayer was scratched with a 100-µl
pipette tip to create an artificial wound. The cells were then
cultured in serum-free DMEM for 24 h at 37°C. Following incubation,
cell migration was observed at 0 and 24 h using a light microscope
(magnification, ×100; Olympus Corporation). The cell migration rate
(%) was calculated using the following equation: [(Initial distance
at 0 h-final distance at 24 h)/initial distance] ×100.
Transwell assay
The Transwell assay was performed using 24-well
culture plates with 8-µm pore inserts (Falcon; Corning Life
Sciences). The lower chamber was filled with 600 µl DMEM
supplemented with 10% FBS. The upper chamber was filled with 200 µl
RL95-2 cell suspension (5×104 cells/well) suspended in
serum-free DMEM. Following 24 h of incubation, the invasive cells
were fixed with 4% methanol for 10 min at 37°C and then stained
with 0.1% crystal violet solution for 15 min at 37°C. The number of
invasive cells in five randomly selected fields of view were
counted using a counting chamber under a light microscope
(magnification, ×100).
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology). Total protein
was quantified using a BCA kit (Thermo Fisher Scientific, Inc.) and
40 µg protein/lane was separated via 8–12% SDS-PAGE. The separated
proteins were subsequently transferred onto PVDF membranes (Bio-Rad
Laboratories, Inc.) and blocked with 5% non-fat milk at 37°C for 2
h. The membranes were subsequently incubated with the following
primary antibodies overnight at 4°C: Anti-ZIC2 (1:5,000; cat. no.
ab150404), anti-Ki67 (1:5,000; cat. no. ab92742),
anti-proliferating cell nuclear antigen (PCNA; 1:1,000; cat. no.
ab29), anti-MMP2 (1:1,000; cat. no. ab92536), anti-MMP9 (1:1,000;
cat. no. ab137867), anti-activated Notch1 (1:500; cat. no. ab8925),
anti-hes family bHLH transcription factor 1 (HES-1; 1:500; cat. no.
ab108937) and anti-GAPDH (1:10,000; cat. no. ab181603; Abcam).
Following the primary antibody incubation, the membranes were
incubated with a goat anti-rabbit IgG HRP-conjugated secondary
antibody (1:10,000; cat. no. ab205718; Abcam) at room temperature
for 2 h. Protein bands were visualized using an
electrochemiluminescence kit (Invitrogen; Thermo Fisher Scientific,
Inc.) and imaged on a gel imaging system (Bio-Rad Laboratories,
Inc.). ImageJ software (version 1.52v; National Institutes of
Health) was used for densitometric analysis.
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using a
PrimeScript cDNA Synthesis kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. qPCR was subsequently
performed using a SYBR Green Master mix (Takara Biotechnology Co.,
Ltd.) on an ABI 7500 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following thermocycling
conditions were used for the qPCR: Initial denaturation for 10 min
at 95°C; followed by 40 cycles of denaturation at 95°C for 20 sec,
annealing at 60°C for 30 sec and polymerization at 72°C for 20 sec.
The following primer sequences were used for the qPCR: ZIC2
forward, 5′-AAGGACCCACACAGGGGAGAA-3′ and reverse,
5′-AACATGATCACAAGGTGCCCTC-3′; SNHG12 forward,
5′-TCTGGTGATCGAGGACTTCC-3′ and reverse,
5′-ACCTCCTCAGTATCACACACT-3′; and GAPDH forward,
5′-GAAAGCCTGCCGGTGACTAA-3′ and reverse, 5′-TTCCCGTTCTCAGCCTTGAC-3′.
The relative mRNA expression was calculated using the
2−∆∆Cq method (17). The
expression levels of ZIC2 and SNHG12 were normalized to the
expression levels of GAPDH.
Dual luciferase reporter assay
The JASPAR database (http://jaspar.genereg.net) was used to predict that
ZIC2 could bind to the promoter of SNHG12. For the dual-luciferase
reporter assay, wild-type (WT; 5′-GCAACAGCAGGATC-3′) or mutant
(MUT; 5′-ATGGTGATGAAGCT-3′) SNHG12 sequences were amplified and
cloned downstream of the luciferase reporter gene in pMIR-REPORT
luciferase vectors (Thermo Fisher Scientific, Inc.). Cells were
co-transfected with pcDNA3.1-ZIC2 or NC vectors and WT or MUT
SNHG12 pMIR-REPORT luciferase vectors. Following 48 h of
transfection, the relative luciferase activity was measured using a
Dual Luciferase Reporter assay system (Promega Corporation).
Luciferase activity was normalized to Renilla luciferase
activity. All experiments were performed in triplicate.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed according to a standard
protocol (18). Briefly,
5×107 RL95-2 cells were washed with cold PBS and then
fixed with 1% formaldehyde for 10 min at 37°C. Subsequently, cells
were subjected to a ChIP assay according to the manufacturer's
protocol of a High-Sensitivity kit (Abcam; cat. no. ab185913). The
following antibodies, Anti-ZIC2 (1:1,000; Abcam; cat. no. ab150404)
and anti-IgG (as the NC; 1:10,000; Abcam; cat. no. ab133470), were
used at 4°C overnight. The primer sequences used for the RT-qPCR
analysis of SNHG12 expression levels were: Forward,
5′-TCTGGTGATCGAGGACTTCC-3′ and reverse,
5′-ACCTCCTCAGTATCACACACT-3′.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (IBM Corp.). All experiments were performed at least three
times and data are presented as the mean ± SD. Statistical
differences between groups were determined using a one-way ANOVA
followed by a Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
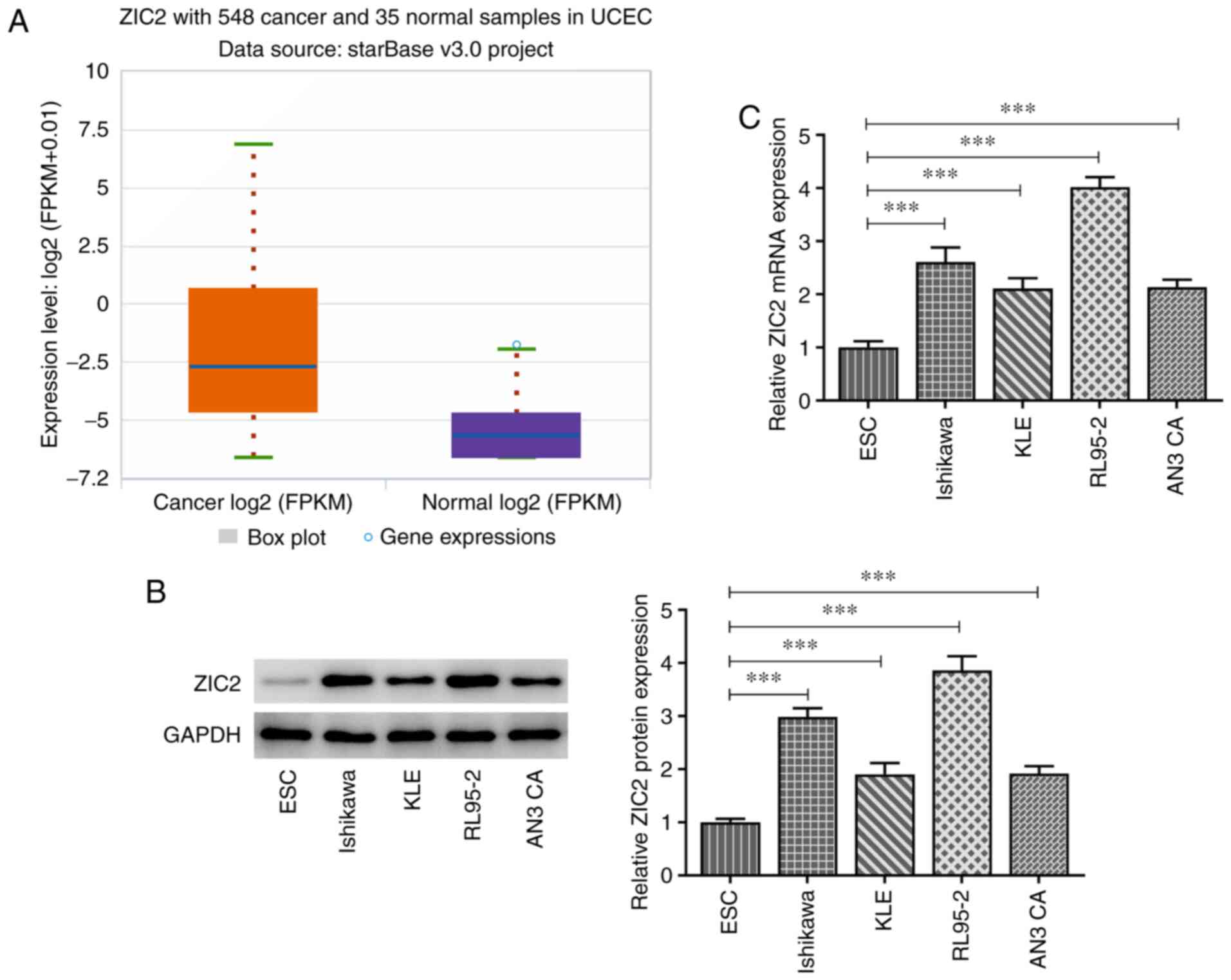
ZIC2 expression levels are upregulated
in patients with EC and EC cell lines
To determine whether ZIC2 was involved in EC, the
overall level of ZIC2 differential expression between healthy
individuals and cancer patients was analyzed by searching the
ENCORI database (15) from The
Cancer Genome Atlas (TCGA) Genomics data (16), and the analysis revealed that the
expression levels of ZIC2 were significantly upregulated in
patients with EC compared with healthy individuals (Fig. 1A). The protein and mRNA expression
levels of ZIC2 in the ESC normal endometrial stromal cell line and
EC cell lines, Ishikawa, KLE, RL95-2 and AN3 CA, were analyzed.
Similarly, the expression levels of ZIC2 were upregulated in EC
cell lines, especially in RL95-2 cells, compared with ESC cells
(Fig. 1B and C). Therefore, the
RL95-2 cell line was selected for use in subsequent experiments as
it expressed the highest level of ZIC2.
Knockdown of ZIC2 inhibits RL95-2 EC
cell proliferation, migration and invasion
The expression of ZIC2 was knocked down in RL95-2
cells using shRNA-ZIC2-1 and shRNA-ZIC2-2. As shRNA-ZIC2-1 exerted
a greater inhibitory effect on ZIC2 expression, it was selected for
use in subsequent studies (Fig. 2A and
B). The knockdown of ZIC2 significantly inhibited RL95-2 cell
proliferation, as evidenced by the decreased proliferation rate
(Fig. 2C), number of formed
colonies (Fig. 2D) and protein
expression levels of Ki67 and PCNA (Fig. 2E). The results of the wound healing
assay demonstrated that ZIC2 knockdown prevented RL95-2 cells from
migrating into the wounded area after 24 h, thus indicating that
ZIC2 knockdown was able to inhibit cell migration (Fig. 3A). Transwell assays were used to
determine the invasive ability of RL95-2 cells. As demonstrated in
Fig. 3B, cells transfected with
shRNA-ZIC2 had a significantly lower invasion rate compared with
cells in the shRNA-NC and untransfected control groups. In
addition, the protein expression levels of MMP2 and MMP9 were
downregulated following ZIC2 knockdown (Fig. 3C). These results indicated that ZIC2
knockdown may inhibit RL95-2 cell proliferation, migration and
invasion.
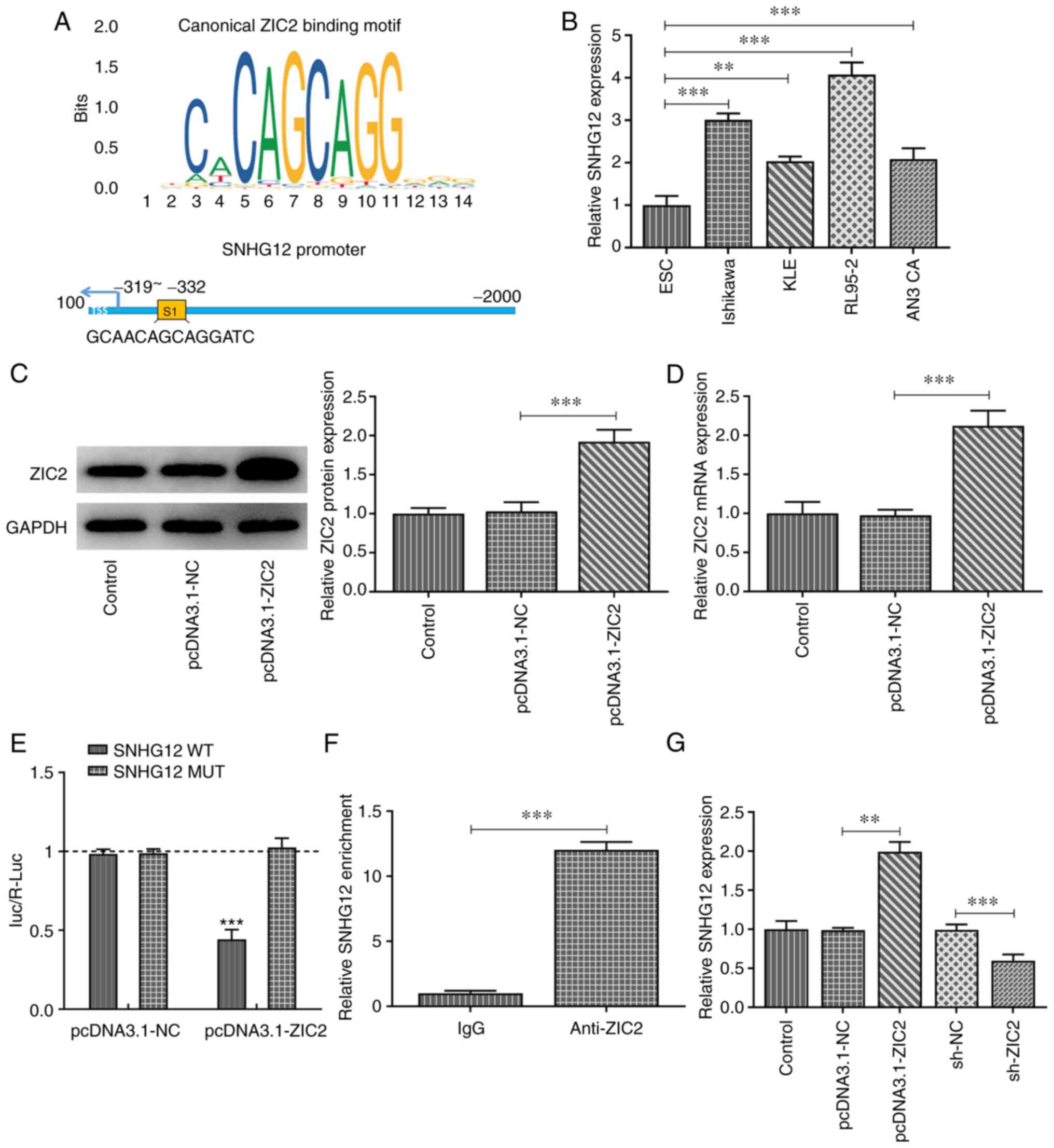
ZIC2 binds to SNHG12 and positively
regulates its expression
Using the JASPAR database, ZIC2 was predicted to
bind to the promoter of SNHG12 (Fig.
4A). Consistent with the results of a recent study (6), SNHG12 expression levels were revealed
to be upregulated in EC cell lines, most notably in RL95-2 cells
(Fig. 4B). ZIC2 was subsequently
overexpressed in RL95-2 cells (Fig. 4C
and D) and a dual luciferase reporter assay was performed. The
results revealed that ZIC2 overexpression significantly decreased
the relative luciferase activity of SNHG12-WT, but not SNHG12-MUT
vectors (Fig. 4E), indicating the
direct interaction between ZIC2 and SNHG12, which was further
validated using a ChIP assay (Fig.
4F). In addition, SNHG12 expression levels in RL95-2 cells were
upregulated by ZIC2 overexpression, whereas they were downregulated
by ZIC2 knockdown, indicating the positive regulatory effect of
ZIC2 on SNHG12 expression (Fig.
4G).
SNHG12 overexpression blocks the
effect of ZIC2 on RL95-2 EC cell proliferation, migration and
invasion while activating Notch signaling
SNHG12 was overexpressed in RL95-2 cells by
transfection with pcDNA 3.1-SNHG12 vectors (Fig. 5A), while ZIC2 expression was also
knocked down in RL95-2 cells. Rescue assays were subsequently
performed to evaluate whether ZIC2 regulated EC cellular processes
by targeting SNHG12. As revealed in Fig. 5B-D, cell proliferation was markedly
inhibited following ZIC2 silencing; however, SNHG12 overexpression
significantly rescued cell proliferation. Furthermore, ZIC2
knockdown-induced inhibition of RL95-2 cell migration and invasion
was partially reversed by SNHG12 overexpression, as evidenced by
the increased migration and invasion rates and upregulated
expression levels of MMP2 and MMP9 in the cells in the ZIC2
knockdown + SNHG12 overexpression group compared with the ZIC2
knockdown + Oe-NC group (Fig.
5E-J).
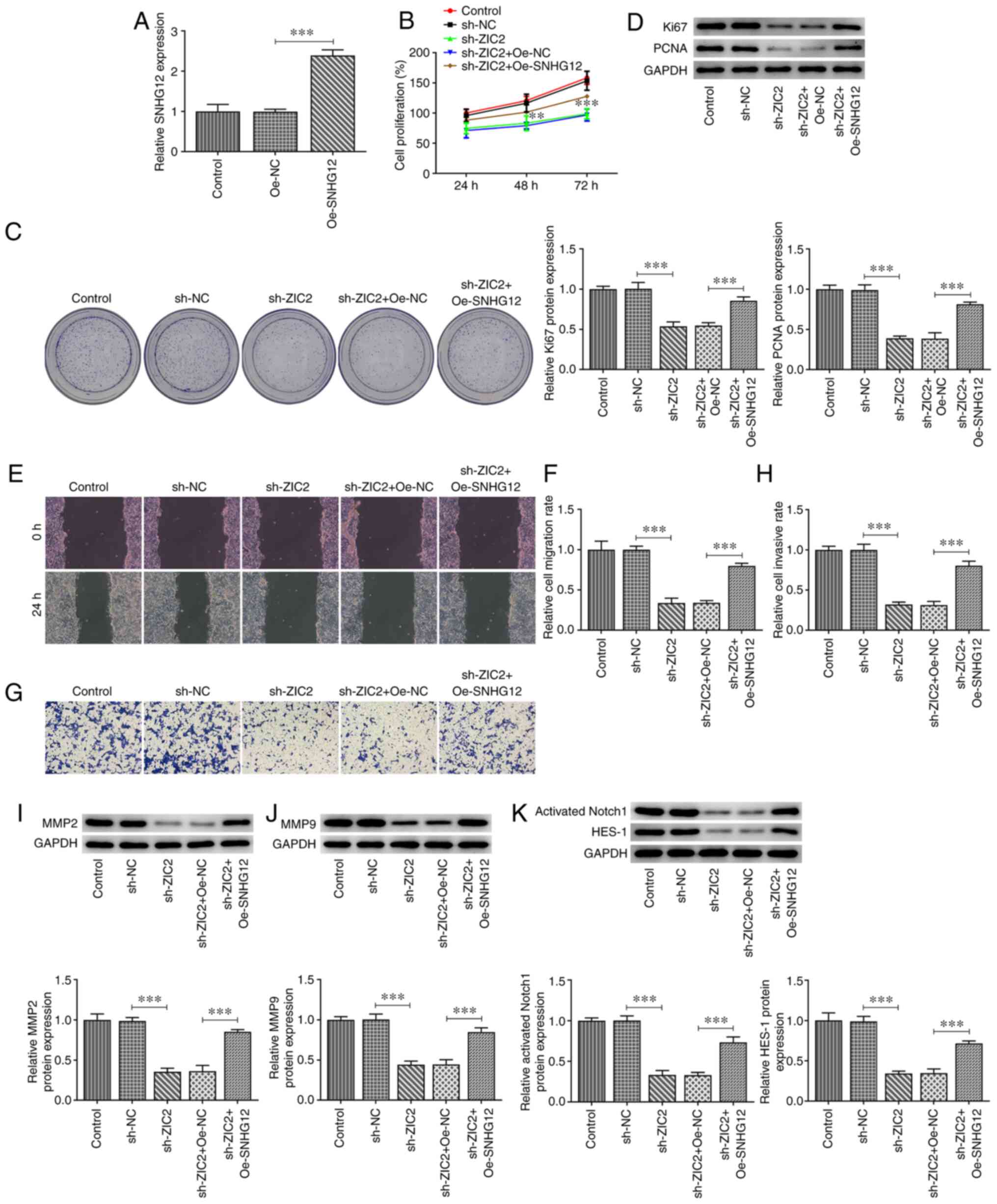 | Figure 5.SNHG12 overexpression partially
reverses the effect of ZIC2 knockdown in RL95-2 endometrial cancer
cells. (A) mRNA expression levels of SNHG12 in RL95-2 cells before
and after the overexpression of SNHG12. (B) Cell Counting Kit-8
assay was performed to determine the proliferation of RL95-2 cells
before and after transfection with the indicated vectors. (C)
Representative images for the colony formation assay of RL95-2
cells before and after transfection with the indicated vectors
(magnification, ×100). (D) Protein expression levels of Ki67 and
PCNA in RL95-2 cells. (E and F) Wound healing and (G and H)
Transwell assays were used to determine the migration and invasion,
respectively, of RL95-2 cells before and after transfection with
the indicated vectors (magnification, ×100). Expression levels of
(I) MMP2, (J) MMP9 and (K) Notch signaling pathway-related proteins
in RL95-2 cells before and after transfection with the indicated
vectors were analyzed using western blotting. **P<0.01 and
***P<0.001. SNHG12, small nucleolar RNA host gene 12; ZIC2, Zic
family member 2; PCNA, proliferating cell nuclear antigen; Oe,
overexpression; NC, negative control; sh, short hairpin. |
Finally, the expression levels of proteins
associated with Notch signaling were analyzed in RL95-2 cells
following ZIC2 knockdown and SNHG12 overexpression to determine
whether Notch signaling was involved in the effects of the
ZIC2/SNHG12 signaling axis in EC. As revealed in Fig. 5K, the protein expression levels of
activated Notch1 and HES-1 were significantly downregulated
following ZIC2 knockdown, while they were partially rescued by
SNHG12 overexpression, indicating that ZIC2 may activate Notch
signaling by upregulating the expression levels of SNHG12 in RL95-2
cells.
Discussion
Although the 5-year survival rate following early
diagnosis and treatment of EC can be as high as 90%, the 5-year
survival rate for patients with stage III and IV disease remains
low (4,5). Understanding the precise molecular
mechanisms of proliferation and metastasis is important for
developing improved therapeutic strategies for patients with EC.
Therefore, it is pivotal to determine the key molecular mechanisms
involved in EC cell proliferation and metastasis. In our previous
study, SNHG12 was identified as a biomarker that could be used for
the early diagnosis and targeted therapy of EC (6). The results of the present study
demonstrated that the expression levels of ZIC2 were upregulated in
patients with EC and EC cell lines, and ZIC2 bound to SNHG12 to
positively regulate its expression. Further results revealed that
the knockdown of ZIC2 inhibited EC cell proliferation, migration
and invasion by regulating SNHG12 expression. Consequently, the
ZIC2/SNHG12 signaling axis may serve as a target for novel
therapeutic strategies for EC.
The ZIC protein was discovered to be essential for
developing vertebrate embryos and numerous human cancer types
(19). ZIC2 was only found in
normal brain and testicular tissues, but was highly expressed in
tumors (19). The expression levels
of ZIC2 were also revealed to be upregulated and play key roles in
multiple types of cancer. For example, ZIC2 knockdown inhibited the
proliferation, migration and invasion of breast cancer cells
(20). In another previous study,
ZIC2 expression was gradually upregulated from normal to cancer to
metastatic tissues, and promoted tumor growth and metastasis in HCC
(8). ZIC expression was also found
to be upregulated in NPC, and knocking down ZIC2 expression was
indicated to inhibit NPC lymphangiogenesis and lymph node
metastasis (21). The findings of
the present study revealed that the expression levels of ZIC2 were
upregulated in patients with EC and EC cell lines. In addition, the
upregulated expression of ZIC2 is associated with a poor overall
survival rate, indicating that ZIC2 may play an oncogenic role in
EC (22). ZIC2 expression was
subsequently knocked down in EC cells and the results demonstrated
that the proliferation rate, colony formation ability, protein
expression levels of Ki67, PCNA, MMP2 and MMP9, and the migration
and invasion rates of EC cells, were all significantly suppressed
following ZIC2 knockdown. According to previous studies, the
protein expression of MMP2 and MMP9 along with wound healing and
Transwell assays were revealed to be sufficient to indicate the
migration and invasion of tumor cells (23,24).
Consequently, these results indicated that the knockdown of ZIC2
may inhibit EC cell proliferation, migration and invasion.
ZIC2 was predicted to bind to the promoter of
SNHG12, which was demonstrated to promote EC progression in our
previous study (6). ZIC2 has been
reported to be dominantly located in the nucleus (25). In the present study, dual luciferase
reporter and ChIP assays were performed to validate their
interaction. ZIC2 was observed to positively regulate SNHG12
expression. It was therefore hypothesized that ZIC2 may bind to
SNHG12 to upregulate SNHG12 expression, thereby exerting its
stimulatory effect on EC cell proliferation, migration and
invasion. SNHG12 was subsequently overexpressed in EC cells with
knocked down ZIC2 expression to determine whether the effect of
ZIC2 knockdown on EC cells could be modified. Consistent with the
present study hypothesis, the effects of ZIC2 knockdown on EC cells
were notably reversed by the overexpression of SNHG12. These data
indicated that ZIC2 may exert its effects on EC via targeting
SNHG12. However, SNHG12 overexpression did not completely reverse
the effects of ZIC2 knockdown, suggesting that the oncogenic
effects of ZIC2 in EC may also be mediated via other downstream
mechanisms.
The present results also revealed that the knockdown
of ZIC2 inhibited the activation of Notch signaling, whereas SNHG12
overexpression reversed this effect. SNHG12 has been revealed to
modulate the Notch signaling pathway in cancer cells (26,27).
In addition, inhibition of the Notch signaling pathway was reported
to inhibit the progression of EC (13,14).
The present findings identified Notch signaling as the downstream
pathway via which SNHG12 regulated EC cell proliferation, migration
and invasion. However, future experiments using human samples and
animal models need to be performed to validate our findings. The
correlation between expression levels of ZIC2 and SNHG12 in EC
using clinical human samples will also be analyzed.
In conclusion, the findings of the present study
indicated that ZIC2 may act as an upstream mediator of SNHG12 in EC
cells. The knockdown of ZIC2 inhibited EC cell proliferation,
migration and invasion via downregulating SNHG12 expression to
inhibit Notch signaling. These findings may contribute to the
further understanding of the role and mechanism of SNHG12 in EC and
indicated that components of the ZIC2/SNHG12 signaling axis may
serve as potential diagnostic and prognostic biomarkers, or targets
for novel therapeutic strategies for EC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PC and GL acquired the data. MW, HB and BZ
contributed to the study design and analysis of the data. MW
drafted the manuscript. HB and BZ revised it critically for
important intellectual content. HB, PC and GL are responsible for
confirming the authenticity if the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sorosky JI: Endometrial cancer. Obstet
Gynecol. 120:383–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brooks RA, Fleming GF, Lastra RR, Lee NK,
Moroney JW, Son CH, Tatebe K and Veneris JL: Current
recommendations and recent progress in endometrial cancer. CA
Cancer J Clin. 69:258–279. 2019.PubMed/NCBI
|
|
3
|
Inoue F, Sone K, Toyohara Y, Takahashi Y,
Kukita A, Hara A, Taguchi A, Tanikawa M, Tsuruga T and Osuga Y:
Targeting epigenetic regulators for endometrial cancer therapy: Its
molecular biology and potential clinical applications. Int J Mol
Sci. 22:23052021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ueda SM, Kapp DS, Cheung MK, Shin JY,
Osann K, Husain A, Teng NN, Berek JS and Chan JK: Trends in
demographic and clinical characteristics in women diagnosed with
corpus cancer and their potential impact on the increasing number
of deaths. Am J Obstet Gynecol. 198:218.e1–6. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cai P, Wu M, Zhang B, Wu S, Wei H and Wei
L: Long non-coding RNA SNHG12 regulates cell proliferation,
invasion and migration in endometrial cancer by targeting miR-4429.
Mol Med Rep. 22:2842–2850. 2020.PubMed/NCBI
|
|
7
|
Bidus MA, Risinger JI, Chandramouli GV,
Dainty LA, Litzi TJ, Berchuck A, Barrett JC and Maxwell GL:
Prediction of lymph node metastasis in patients with endometrioid
endometrial cancer using expression microarray. Clin Cancer Res.
12:83–88. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu SX, Zhang CZ, Luo RZ, Wang CH, Liu LL,
Fu J, Zhang L, Wang H, Xie D and Yun JP: Zic2 promotes tumor growth
and metastasis via PAK4 in hepatocellular carcinoma. Cancer Lett.
402:71–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang YF, Yang HY, Shi XQ and Wang Y:
Upregulation of microRNA-129-5p inhibits cell invasion, migration
and tumor angiogenesis by inhibiting ZIC2 via downregulation of the
Hedgehog signaling pathway in cervical cancer. Cancer Biol Ther.
19:1162–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Yang S, Zeng J and Chen M:
miR-1271-5p inhibits cell proliferation and induces apoptosis in
acute myeloid leukemia by targeting ZIC2. Mol Med Rep. 19:508–514.
2019.PubMed/NCBI
|
|
11
|
Shen ZH, Zhao KM and Du T: HOXA10 promotes
nasopharyngeal carcinoma cell proliferation and invasion via
inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci.
21:945–952. 2017.PubMed/NCBI
|
|
12
|
Liu ZB, Tang C, Jin X, Liu SH and Pi W:
Increased expression of lncRNA SNHG12 predicts a poor prognosis of
nasopharyngeal carcinoma and regulates cell proliferation and
metastasis by modulating Notch signal pathway. Cancer Biomark.
23:603–613. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jonusiene V and Sasnauskiene A: Notch and
endometrial cancer. Adv Exp Med Biol. 1287:47–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shang C, Lang B and Meng LR: Blocking
NOTCH pathway can enhance the effect of EGFR inhibitor through
targeting CD133+ endometrial cancer cells. Cancer Biol Ther.
19:113–119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen C, Shao R, Li B, Zhai Y, Wang T, Li
X, Miao L, Huang J, Liu R, Liu E, et al: Neoisoliquiritin exerts
tumor suppressive effects on prostate cancer by repressing androgen
receptor activity. Phytomedicine. 85:1535142021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu P, Wang Y, He L, Huang G, Du Y, Zhang
G, Yan X, Xia P, Ye B, Wang S, et al: ZIC2-dependent OCT4
activation drives self-renewal of human liver cancer stem cells. J
Clin Invest. 125:3795–3808. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang P, Yang F, Luo Q, Yan D and Sun S:
miR-1284 inhibits the growth and invasion of breast cancer cells by
targeting ZIC2. Oncol Res. 27:253–260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu D, Han GH, Zhao X, Liu X, Xue K, Wang D
and Xu CB: MicroRNA-129-5p suppresses nasopharyngeal carcinoma
lymphangiogenesis and lymph node metastasis by targeting ZIC2. Cell
Oncol (Dordr). 43:249–261. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv Z, Qi L, Hu X, Mo M, Jiang H, Fan B and
Li Y: Zic family member 2 (ZIC2): A potential diagnostic and
prognostic biomarker for pan-cancer. Front Mol Biosci.
8:6310672021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu F, Li Q, Wang Z and Cao X: Sinomenine
inhibits proliferation, migration, invasion and promotes apoptosis
of prostate cancer cells by regulation of miR-23a. Biomed
Pharmacother. 112:1085922019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wan G, Liu Y, Zhu J, Guo L, Li C, Yang Y,
Gu X, Deng LL and Lu C: SLFN5 suppresses cancer cell migration and
invasion by inhibiting MT1-MMP expression via AKT/GSK-3β/β-catenin
pathway. Cell Signal. 59:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marchini S, Poynor E, Barakat RR, Clivio
L, Cinquini M, Fruscio R, Porcu L, Bussani C, D'Incalci M, Erba E,
et al: The zinc finger gene ZIC2 has features of an oncogene and
its overexpression correlates strongly with the clinical course of
epithelial ovarian cancer. Clin Cancer Res. 18:4313–4324. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou S, Yu L, Xiong M and Dai G: LncRNA
SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by
upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res
Commun. 495:1822–1832. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J,
Cheng W and Zhu L: FTO demethylates m6A modifications in HOXB13
mRNA and promotes endometrial cancer metastasis by activating the
WNT signalling pathway. RNA Biol. Nov 5–2020.(Epub ahead of print).
View Article : Google Scholar
|