Introduction
Lung cancer is one of the most prevalent types of
cancer in China, and its incidence and morbidity remain high due to
various independent factors, such as smoking, atmospheric
pollution, cooking oil fume and chronic neonatal lung disease
(1,2). Lung cancer can be histologically
classified into small cell lung carcinoma and non-small cell lung
carcinoma (NSCLC), and ~85% of patients with lung cancer are
diagnosed with NSCLC (3). NSCLC can
be further categorized into squamous-cell carcinoma, lung
adenocarcinoma (ADC) and large-cell carcinoma. Among these
subtypes, lung ADC is the most common type of NSCLC (4). Despite advances being made in the
treatment methods available for NSCLC, the 5-year survival rate of
patients has not improved and remains at <15% (5).
Circular RNAs (circRNAs/circ) are newly identified
non-coding RNAs (ncRNAs) that are present in the cytoplasm of
eukaryotic organisms (6). circRNAs
have been reported as potential molecular diagnosis markers in
multiple types of cancer, including gastric cancer, oral squamous
cell carcinoma and breast cancer (7,8). A
previous study reported that circRNA plasmacytoma variant
translocation 1 (circ-PVT1) played an oncogenic role in gastric
cancer by upregulating zinc finger E-box binding homeobox 1
expression via sponging microRNA (miRNA/miR)-124-3p (7). In addition, circ-PVT1 was found to
increase the proliferation of epithelial ovarian cancer cells, and
suppress apoptosis, via sponging miR-149, which promoted the
progression of ovarian cancer (9).
However, to the best of our knowledge, the specific role of
circ-PVT1 in lung ADC has not yet been reported to date. As
circRNAs can sponge miRNAs to regulate gene expression (10), the present study hypothesized that
circ-PVT1 may be involved in the development of lung ADC by
sponging certain miRNAs.
The association between circ-PVT1 and miR-429 was
predicted using ENCORI in the present study. A previous study
showed that the expression levels of miR-429 are downregulated in
the serum of patients with NSCLC, and the decreased expression
levels are associated with poor overall survival rate (11). Thus, miR-429 was suggested to be an
independent risk factor for the prognosis of these patients. A
previous study reported that the upregulated expression of long
non-coding RNA metastasis-associated lung adenocarcinoma transcript
1 in human ADC cells and tissues was associated with tumor size,
tumor node metastasis (TNM) stage and lymph node metastasis, and
its expression levels were negatively correlated with miR-429
expression levels (12). However,
other previous studies have suggested a role for miR-249 in
promoting the development of lung cancer. For example, upregulated
miR-429 expression was found to increase the sensitivity of
epithelial ovarian cancer cells to cisplatin (DDP) treatment
(13). Similarly, in another study,
the overexpression of miR-429 induced DDP sensitivity in ovarian
cancer cells (14). In fact, the
chemoresistance of ovarian cancer organoids and cells has been
found to be enhanced by controlling cellular senescence and glucose
metabolism in ovarian cancer organoids and cells via the
Aurora-A/SOX8/ forkhead box k1 (FOXK1) signaling axis (15). Furthermore, FOXK1 has also been
identified to act as an oncogene in lung cancer (16).
The present study aimed to determine the role of
circ-PVT1 in lung ADC and to investigate whether its effects in
lung ADC were associated with the regulation of miR-429 and FOXK1.
The current findings may provide novel insights into potential
targets for the treatment of lung ADC.
Materials and methods
Cell culture and treatment
Human normal lung epithelial cells (BEAS-2B) and
human lung ADC cells (HCC827, H1299, PC-9 and A549) were obtained
from Shanghai Fuheng Biological Technology Co., Ltd. The
DDP-resistant A549 cell line (A549/DDP) was obtained from JRDUN
Biotechnology (Shanghai) Co., Ltd. All cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Thermo Fisher Scientific, Inc.). A549/DDP
and A549 cells were cultured in medium supplemented with DDP
(Sigma-Aldrich; Merck KGaA) at different concentrations (0, 1.25,
2.5, 5, 10, 20 and 40 µM) at 37°C for 24 h. Three cell-culture
passages were studied and three replicates were performed in each
passage.
Cell transfection
Short hairpin (sh)RNA-negative control (NC),
shRNA-circ-PVT1#1/2, overexpression (Oe)-NC, Oe-circ-PVT1,
pcDNA3.1-NC and pcDNA3.1-FOXK1 were constructed by Shanghai
GeneChem Co., Ltd. The NC used for the overexpression of FOXK1 was
an empty pcDNA3.1 plasmid. The sequences for the miR-429 mimic,
inhibitor and NC transfections were as follows: miR-429 mimic
forward, 5′-UAAUACUGUCUGGUAAAACCGU-3′ and reverse,
5′-GGUUUUACCAGACAGUAUUAUU-3′; NC mimic forward,
5′-UUCUCCGAACGUGUCACGUUT−3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT−3′; miR-429 inhibitor,
5′-GCTGATTTAAAGGCTTAG−3′; and NC inhibitor,
5′-CAAATGTAGGTAGAGGA-3′.
miR-429 mimic, inhibitor and NC were obtained from
Guangzhou RiboBio Biotechnology Co., Ltd. A549 cells
(3×105) were seeded in 6-well plates and transfected
with the aforementioned plasmids or oligonucleotides (500 µM) using
Lipofectamine® 2000 reagent (cat. no. 11668019;
Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. The time interval between transfection
and subsequent experimentation was 48 h.
Reverse transcription-quantitative PCR
(RTqPCR)
Total RNA was extracted from A549 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (2 µg) was reverse-transcribed into cDNA using a
Prime Script™ RT Master Mix reverse transcription kit (cat. no.
RR036B; Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. qPCR was subsequently performed using
TaqMan™ 2X Universal PCR Master mix without AmpErase™ UNG (Applied
Biosystems; Thermo Fisher Scientific, Inc.) on an ABI 7500
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The following thermocycling conditions were used for qPCR:
Initial denaturation at 95°C for 10 min; followed by 40 cycles at
95°C for 15 sec and 60°C for 1 min. The following primers (Sangon
Biotech Co., Ltd.) were used for qPCR: circ-PVT1 forward,
5′-ATCGGTGCCTCAGCGTTCGG-3′ and reverse, 5′-CTGTCCTCGCCGTCACACCG-3′;
miR-429 forward, 5′-ACACTCCAGCTGGGTAATACTGTCTGGTAA−3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; FOXK1 forward, 5′-ACGTTTGGTGACCAGAGGGA-3′
and reverse, 5′-CGACAGAATTCAAGCCGCAC-3′; GAPDH forward,
5′-GAGTCAACGGATTTGGTCGTATTG−3′ and reverse,
5′-CCTGGAAGATGGTGATGGGATT-3′; and U6 forward,
5′-AUAAAUCCCUUUACACCUCTT−3′ and reverse,
5′-AAUAAAUCCCUUUACACCUCTT-3′. GAPDH and U6 were used as the
internal controls for mRNA and miRNA, respectively. The relative
mRNA expression levels were calculated using SDS version 2.0.1
software (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Quantification of the relative mRNA expression levels were
performed using the 2−ΔΔCq method (17).
Western blotting
Total protein was extracted from A549 cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology)
supplemented with protease and phosphatase inhibitors. Protein was
quantified using the BCA method and 30 µg protein/lane was loaded
and separated via 12% SDS-PAGE. The separated proteins were
subsequently transferred onto nitrocellulose membranes and blocked
with 5% non-fat milk at room temperature for 1 h. The membranes
were then incubated with primary antibodies against matrix
metalloproteinase (MMP)2 (cat. no. ab92536; 1:1,000; Abcam), MMP9
(cat. no. ab76003; 1:1,000; Abcam), FOXK1 (cat. no. ab18196;
1:1,000; Abcam) and GAPDH (cat. no. ab9485; 1:2,500; Abcam) at 4°C
overnight. Following the primary antibody incubation, the membranes
were washed with TBS with Tween-20 (0.05%) twice and incubated with
HRP-conjugated goat anti-rabbit IgG (cat. no. ab6721; 1:2,000;
Abcam) secondary antibody at room temperature for 1 h. Protein
bands were visualized using Immobilon™ Western Chemiluminescent HRP
substrate (EMD Millipore). ImageJ software (version 1.8.0; National
Institutes of Health) was used for densitometric analysis.
Transwell assay
The 96-well Transwell chambers were precoated with
Matrigel (BD Biosciences) overnight at 37°C. A549 cells
(0.5×106 cells/ml) were suspended in serum-free DMEM and
plated into the upper chamber. The lower chamber of the 96-well
Transwell plate (cat. no. 3380; Corning, Inc.) was filled with DMEM
supplemented with 10% FBS (18).
Following 24 h of incubation at 37°C, the invasive cells were fixed
with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 10 min at
room temperature and stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. Stained
cells were visualized using an inverted microscope (Olympus
Corporation) in five randomly selected fields of view
(magnification, ×100).
Wound healing assay
A549 cells (3×104 cells/well) were plated
into a 96-well plate and cultured until they reached 100%
confluency. A 200-µl sterile pipette tip was subsequently used to
make a single vertical scratch in the cell monolayer. The cells
were then washed with PBS three times to remove non-adherent cells
and cultured in serum-free DMEM at 37°C for 24 h. Cells were
visualized and imaged under a light microscope (magnification,
×100).
Colony formation assay
A549 cells (2×103 cells/well) were seeded
into 96-well plates and cultured in a humidified incubator at room
temperature. Following 2 weeks of incubation, the cells were fixed
with 4% paraformaldehyde for 30 min at room temperature and stained
with 0.5% gentian violet for 1 h at room temperature. Stained cells
were observed and imaged with a camera (Canon, Inc.).
MTT assay
A549 cells (3×103 cells/well) were seeded
into a 96-well culture plate containing DMEM supplemented with 10%
FBS and exposed to different doses of DDP (1.25, 2.5, 5, 10, 20 and
40 µM) for 48 h. Following incubation, 10 µl MTT (10 mg/ml) was
added/well and incubated for 4 h. The culture medium was
subsequently removed, and 100 µl DMSO (Beyotime Institute of
Biotechnology) was added to the wells to dissolve the purple
formazan crystals. Following incubation for 30 min, the optical
density (OD) of each well was measured using a microplate reader at
a wavelength of 570 nm.
Caspase-3 activity assay
Caspase-3 activity was measured in A549 cells using
a colorimetric assay kit (cat. no. C1115; Beyotime Institute of
Technology), according to the manufacturer's protocol.
Dual-luciferase reporter assay
The binding sites between miR-429 and circ-PVT1, and
between FOXK1 and miR-429 were predicted using ENCORI (http://starbase.sysu.edu.cn/index.php).
The wild-type (WT) or mutant (MUT) 3′-untranslated regions (UTRs)
of circ-PVT1 and FOXK1 were constructed and cloned downstream of
the pmirGLO firefly luciferase reporter vector (Promega
Corporation), which were co-transfected with miR-429 mimic or
mimic-NC into A549 cells using Lipofectamine 2000 reagent.
Mutations in the 3′-UTRs were generated using a QuikChange Multi
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.). The
relative luciferase activity was measured using a Dual Luciferase
Reporter Assay kit (Promega Corporation) at 48 h post-transfection.
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.) and GraphPad Prism 7 software (GraphPad
Software, Inc.). All experiments were performed in triplicate and
data are expressed as the mean ± SD. Statistical differences
between two groups were determined using an unpaired student's
t-test, whereas statistical differences among multiple groups were
performed using one-way ANOVA followed by Tukey's post hoc test.
The correlation between miR-429 expression and circ-PVT1 expression
in human normal lung epithelial cells and human lung ADC cells was
determined using Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Knockdown of circ-PVT1 inhibits the
proliferation, invasion and migration of A549 cells
To validate the role of circ-PVT1 in the progression
of lung ADC, the mRNA expression levels of circ-PVT1 in human
normal lung epithelial cells and lung ADC cells were analyzed using
RT-qPCR. The expression levels of circ-PVT1 were significantly
upregulated in lung ADC cells compared with human normal lung
epithelial cells (Fig. 1A). The
expression levels of circ-PVT1 were the highest in A549 cells;
therefore, A549 cells were selected for use in subsequent
experiments. The expression of circ-PVT1 in A549 cells was knocked
down by transfection with shRNA-circ-PVT1. The expression levels of
circ-PVT1 were downregulated to a greater extent in cells
transfected with shRNA-circ-PVT1#2 compared with shRNA-circ-PVT1#1;
therefore, shRNA-circ-PVT1#2 was used for the following
transfections (Fig. 1B). Compared
with the control and shRNA-NC groups, cell viability (Fig. 1C), proliferation (Fig. 1D and E), invasion and migration
(Fig. 2A-D) were all decreased in
the shRNA-circ-PVT1 group. Moreover, the expression levels of MMP2
and MMP9 were significantly downregulated in A549 cells transfected
with shRNA-circ-PVT1 (Fig. 2E and
F). These results suggested that the knockdown of circ-PVT1 may
inhibit the proliferation, invasion and migration of A549
cells.
Expression levels of circ-PVT1 are
upregulated in A549/DDP cells and are involved in DDP-induced
apoptosis
The association between circ-PVT1 and DDP resistance
was investigated in A549 and A549/DDP cells. The expression levels
of circ-PVT1 were upregulated in A549/DDP cells compared with A549
cells, suggesting the involvement of circ-PVT1 in DDP resistance
(Fig. 3A). To determine the effects
of circ-PVT1 on DDP-resistant lung ADC cells, A549/DDP cells were
transfected with shRNA-circ-PVT1 and A549 cells were transfected
with Oe-circ-PVT1 plasmids (Fig. 3B and
E). The cells were then treated with different concentrations
of DDP. The results of the MTT assay revealed that the transfection
of cells with shRNA-circ-PVT1 significantly reduced the viability
of A549/DDP cells treated with increasing doses of DDP compared
with control and shRNA-NC-transfected cells (Fig. 3C). In addition, the transfection of
cells with the Oe-circ-PVT1 plasmid increased the viability of A549
cells treated with increasing doses of DDP (Fig. 3F). Notably, treatment with 5 and 10
µM DDP markedly affected cell viability at the early stage.
Caspase-3 activity was also increased following DDP treatment in
A549/DDP cells transfected with shRNA-circ-PVT1 (Fig. 3D), while caspase-3 activity was
decreased following DDP treatment in A549 cells transfected with
the Oe-circ-PVT1 plasmid (Fig. 3G).
These findings indicated that the expression levels of circ-PVT1
may be upregulated in A549/DDP cells and involved in DDP-induced
apoptosis.
circ-PVT1 negatively regulates the
expression levels of miR-429
The binding sites between miR-429 and circ-PVT1 are
shown in Fig. 4A. The relative
luciferase activity was reduced in the miR-429 mimic + circ-PVT1 WT
group compared with the mimic-NC + circ-PVT1 WT group, while it
remained unchanged between the miR-429 mimic + circ-PVT1 MUT group
and mimic-NC + circ-PVT1 groups (Fig.
4B). Notably, miR-429 expression was decreased in lung ADC
cells compared with human normal lung epithelial cells (Fig. 4C). miR-429 expression was negatively
correlated with circ-PVT1 expression in human normal lung
epithelial cells and human lung ADC cells using a Pearson's
correlation analysis (Fig. 4D). The
expression levels of miR-429 were significantly upregulated in
cells transfected with the shRNA-circ-PVT1 compared with the
control groups (Fig. 4E). The
expression levels of miR-429 were upregulated in the miR-429 mimic
group compared with the mimic-NC group, while they were decreased
in the miR-429 inhibitor group compared with the NC inhibitor group
(Fig. 4F). These results suggested
that circ-PVT1 may negatively regulate the expression of
miR-429.
 | Figure 4.circ-PVT1 negatively regulates the
expression of miR-429. (A) It was predicted that miR-429 may target
the 3′UTR of circ-PVT1. (B) The relationship between circ-PVT1 and
miR-429 was measured via a dual-luciferase reporter assay.
***P<0.001 vs. miR-429 mimic + circ-PVT1 MUT group. (C) The mRNA
levels of miR-429 in human normal lung epithelial cells (BEAS-2B)
and human lung adenocarcinoma cells (HCC827, H1299, PC-9, A549)
were measured via reverse transcription-quantitative PCR.
***P<0.001 vs. BEAS-2B group. (D) The correlation between
circ-PVT1 and miR-429 was determined using Pearson's correlation
coefficient in human normal lung epithelial cells and human lung
ADC cells. (E) The mRNA expression of miR-429 after knockdown of
circ-PVT1 was determined. ***P<0.001 vs. Control group;
###P<0.001 vs. shRNA-NC group. (F) Proof of
transfection for miR-429 mimic and inhibitor transfections.
***P<0.001 vs. Control group; ###P<0.001 vs.
mimic-NC group; $P<0.05 vs. NC inhibitor group.
circ-PVT1, circular RNA plasmacytoma variant translocation 1; miR,
microRNA; shRNA, short hairpin RNA; NC, negative control; Oe,
overexpression; WT, wild-type; MUT, mutant. |
FOXK1 is a target gene of miR-429
The binding sites between FOXK1 and miR-429 were
predicted and are presented in Fig.
5A. The results of the dual-luciferase reporter assay validated
the hypothesis that miR-429 may regulate the expression of FOXK1
(Fig. 5B). The expression levels of
FOXK1 were subsequently determined using RT-qPCR and western
blotting in BEAS-2B cells and multiple lung ADC cell lines. The
expression levels of FOXK1 were upregulated in lung ADC cells, and
the expression levels of FOXK1 were the highest in A549 cells
(Fig. 5C and D). As shown in
Fig. 5E and F, transfection with
the miR-429 mimic downregulated the expression levels of FOXK1 in
A549 cells. Following the knockdown of circ-PVT1 in A549 cells,
FOXK1 expression levels were also downregulated (Fig. 5G and H).
circ-PVT1/miR-429/FOXK1 signaling axis
promotes the proliferation, invasion and migration of A549 cells,
which may occur through DDP-induced apoptosis
Finally, whether circ-PVT1 could regulate the
expression of FOXK1 via sponging miR-429 to affect the cellular
behaviors of A549 cells was investigated. The pcDNA3.1-FOXK1 vector
was constructed and transfected into cells; the expression levels
of FOXK1 were upregulated in cells transfected with the
pcDNA3.1-FOXK1 vector compared with the control or empty vector
groups (Fig. 6A and B).
Transfection of cells with shRNA-circ-PVT1 decreased the cell
viability, proliferation, invasion and migration, which was
subsequently reversed by transfection with either the miR-429
inhibitor or pcDNA3.1-FOXK1 plasmid (Fig. 6C-I). The expression levels of MMP2
and MMP9 were also downregulated by shRNA-circ-PVT1, while
transfection with the miR-429 inhibitor or pcDNA3.1-FOXK1 could
upregulate the expression levels (Fig.
6J and K). In addition, the shRNA-circ-PVT1-induced reduction
in cell viability and increased caspase-3 activity in A549/DDP
cells were both partially abolished by transfection with the
miR-429 inhibitor (Fig. 7A and B).
The Oe-circ-PVT1-induced increase in cell viability and decreased
caspase-3 activity in A549 cells were also partially abolished by
transfection with the miR-429 inhibitor or pcDNA3.1-FOXK1 (Fig. 7C and D). These results suggested
that the circ-PVT1/miR-429/FOXK1 signaling axis may promote the
proliferation, invasion and migration of A549 cells, which may
occur through DDP-induced apoptosis.
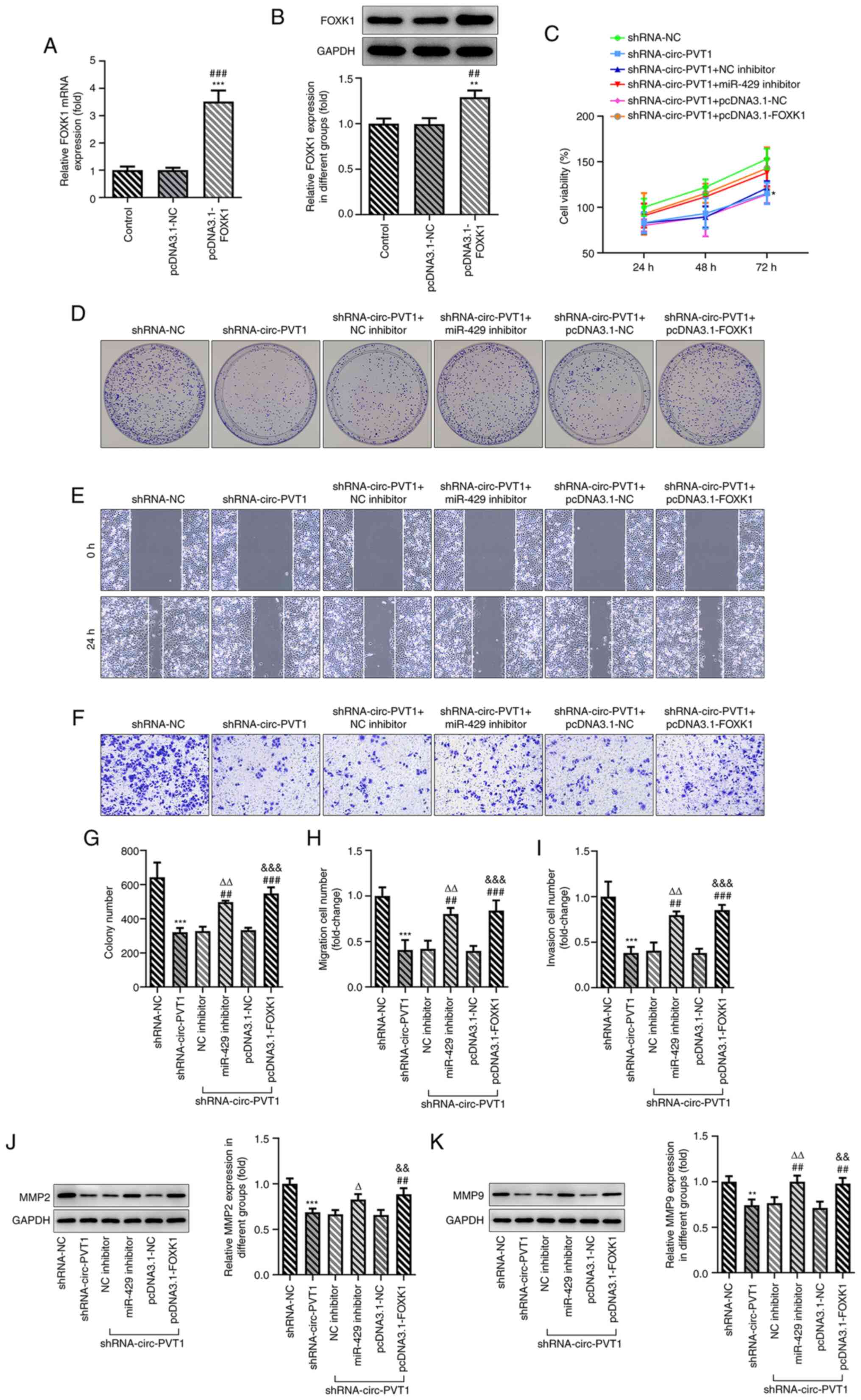 | Figure 6.circ-PVT1/miR-429/FOXK1 axis promotes
the proliferation, invasion and migration of A549 cells and
involves cisplatin-induced apoptosis. (A and B) The expression of
FOXK1 following transfection with pcDNA3.1-FOXK1 was determined via
reverse transcription-quantitative PCR and western blotting.
**P<0.01 and ***P<0.001 vs. Control group;
##P<0.01 and ###P<0.001 vs. pcDNA3.1-NC
group. (C) The cell viability of A549 cells was measured after the
cells were transfected with the corresponding plasmids. *P<0.05
vs. shRNA-NC group. (D-I) The proliferation, migration and invasion
of A549 cells were detected by colony formation, wound healing and
Transwell assays, respectively. ***P<0.001 vs. shRNA-NC group;
##P<0.01 and ###P<0.001 vs.
shRNA-circ-PVT1 group; ∆∆P<0.01 vs. shRNA-circ-PVT1 +
NC inhibitor group; &&&P<0.001 vs.
shRNA-circ-PVT1 + pcDNA3.1-NC group. (J and K) The expression
levels of MMP2 and MMP9 were measured after the cells were
transfected with the corresponding plasmids. **P<0.01 and
***P<0.001 vs. shRNA-NC group; ##P<0.01 vs.
shRNA-circ-PVT1 group; ∆P<0.05 and
∆∆P<0.01 vs. shRNA-circ-PVT1 + NC inhibitor group;
&&P<0.01 vs. shRNA-circ-PVT1 + pcDNA3.1-NC
group. FOXK1, forkhead box k1; miR, microRNA; NC, negative control;
circ-PVT1, circular RNA plasmacytoma variant translocation 1;
shRNA, short hairpin RNA; MMP, matrix metalloproteinase. |
Discussion
circRNAs are ncRNAs that have high stability and
tissue-specific expression patterns (19). However, to the best of our
knowledge, the functions of the majority of circRNAs are poorly
understood. Increasingly mature high-throughput sequencing
technologies have provided evidence to suggest that circRNAs may be
associated with various diseases, including cancer (20). To date, circRNAs have been reported
to participate in the development of lung cancer. For example,
circRNA itchy E3 ubiquitin protein ligase was observed to inhibit
the proliferation of lung cancer via inactivating Wnt/β-catenin
signaling (21). circ-homeodomain
interacting protein kinase 3, as an oncogene, has been found to
modulate autophagy and has been suggested to be a potential
prognostic marker in lung cancer (22). In addition, circ-mannosidase α class
2B member 2 has been reported to sponge miR-1275 and promote the
development of lung cancer (23).
To date, the role of circ-PVT1 has been extensively studied in
multiple types of cancer; however, to the best of our knowledge,
its role in lung cancer has not been discussed.
The aberrant expression of miRNAs has been reported
to be involved in multiple processes of cancer cells, including
proliferation, apoptosis and differentiation (24). Accumulating evidence has
demonstrated the significant role of miRNAs in the progression of
NSCLC. For example, the miR-200 family was shown to be implicated
in lung cancer development and drug sensitivity (24). Emerging evidence has also supported
the inhibitory role of miR-429, a member of the miR-200 family, on
the expression of tumor suppressor genes, such as PTEN and TIMP
metallopeptidase inhibitor 2, to enhance the proliferation,
migration and invasion of cells (25). Another study also reported a
significant role for miR-429 in promoting the proliferation of
NSCLC cells (25). Thus, the role
of miR-429 in lung ADC cells was further investigated in the
present study, and the results revealed that lung ADC cell lines
exhibited downregulated expression levels of miR-429.
circRNAs have been widely recognized to function by
interacting with miRNAs (26,27).
Certain circRNAs that interact with miRNA have been observed to be
overexpressed in lung cancer. For example, hsa_circ_100395 affected
the proliferation, migration and invasion of lung cancer cells by
targeting the miR-1228/transcription factor 21 axis; this axis has
been suggested to account for the lymph node metastasis, high TNM
stage and low survival rate of patients with lung cancer (28). circ-PVT1 has been confirmed to
interact with miR-124-3p and inversely regulate its expression to
affect drug sensitivity in paclitaxel-resistant gastric cancer
cells (7). The results of the
present study revealed that circ-PVT1 could interact with miR-429
to regulate the functions of A549 cells.
The circRNA/miRNA/mRNA network plays a role in the
mechanism through which circ-PVT1 affects the progression of lung
ADC. The present results identified and validated the negative
regulatory association between miR-429 and FOXK1 using a Pearson's
correlation analysis and dual-luciferase reporter assays. FOXK1 has
been reported to play a significant oncogenic role in various types
of cancer, including esophageal, ovarian and colorectal cancer
(29–31). In addition, FOXK1 expression levels
have been found to be negatively associated with the overall
survival of patients with esophageal cancer (29). The results of the present study
revealed that the expression levels of FOXK1 were upregulated in
lung ADC cells, while transfection with either the pcDNA3.1-FOXK1
plasmid or miR-429 inhibitor could abolish the inhibitory effects
of circ-PVT1 knockdown on cell proliferation, migration and
invasion. These results indicated that the knockdown of circ-PVT1
may inhibit the progression of lung ADC via the miR-429/FOXK1
signaling axis. Moreover, the current results demonstrated that the
knockdown of circ-PVT1 enhanced the effect of DDP on the viability
of A549/DDP cells, suggesting the promoting role of circ-PVT1 on
the resistance of A549 cells to DDP. These findings are similar to
the findings of a previous study, which reported that circ-PVT1,
which was high expressed in doxorubicin- and DDP-resistant
osteosarcoma cells, promoted the chemotherapy resistance of
osteosarcoma cells (32).
In conclusion, the findings of the present study
indicated that the knockdown of circ-PVT1 may inhibit the
progression of lung ADC and enhance lung ADC cell sensitivity to
DDP via the miR-429/FOXK1 signaling axis, which may provide
potential novel targets for the treatment of lung ADC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the General Program
of National Natural Science Foundation of China (grant no.
81671905).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LC and DG conceived the study and performed the
experiments. LC, XZ and XD analyzed and interpreted the data, and
confirm the authenticity of all the raw data. LC wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu J, Zheng H, Yuan S, Zhou B, Zhao W, Pan
Y and Qi D: Overexpression of ANLN in lung adenocarcinoma is
associated with metastasis. Thorac Cancer. 10:1702–1709. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsutsui T, Yamaki H, Kumagai T, Omori C,
Kobayashi H, Kakizaki Y and Miyashita Y: Small cell lung cancer
with thyroid gland oligometastasis: A case report. Thorac Cancer.
12:387–390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death-based treatment of lung adenocarcinoma. Cell Death
Dis. 9:1172018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eberle A, Jansen L, Castro F, Krilaviciute
A, Luttmann S, Emrich K, Holleczek B, Nennecke A, Katalinic A and
Brenner H; GEKID Survival Working Group, : Lung cancer survival in
Germany: A population-based analysis of 132,612 lung cancer
patients. Lung Cancer. 90:528–533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu YY, Zhang LY and Du WZ: Circular RNA
circ-PVT1 contributes to paclitaxel resistance of gastric cancer
cells through the regulation of ZEB1 expression by sponging
miR-124-3p. Biosci Rep. 39:BSR201930452019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Momen-Heravi F and Bala S: Emerging role
of non-coding RNA in oral cancer. Cell Signal. 42:134–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun X, Luo L and Gao Y: Circular RNA PVT1
enhances cell proliferation but inhibits apoptosis through sponging
microRNA-149 in epithelial ovarian cancer. J Obstet Gynaecol Res.
46:625–635. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93, and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PLoS One. 9:e877802014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao H, Zhu Q and Zhou J: Long non-coding
RNA MALAT1 interaction with miR-429 regulates the proliferation and
EMT of lung adenocarcinoma cells through RhoA. Int J Clin Exp
Pathol. 12:419–430. 2019.PubMed/NCBI
|
|
13
|
Zou J, Liu L, Wang Q, Yin F, Yang Z, Zhang
W and Li L: Downregulation of miR-429 contributes to the
development of drug resistance in epithelial ovarian cancer by
targeting ZEB1. Am J Transl Res. 9:1357–1368. 2017.PubMed/NCBI
|
|
14
|
Wang L, Mezencev R, Svajdler M, Benigno BB
and McDonald JF: Ectopic over-expression of miR-429 induces
mesenchymal-to-epithelial transition (MET) and increased drug
sensitivity in metastasizing ovarian cancer cells. Gynecol Oncol.
134:96–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun H, Wang H and Wang X, Aoki Y and Wang
X, Yang Y, Cheng X, Wang Z and Wang X: Aurora-A/SOX8/FOXK1
signaling axis promotes chemoresistance via suppression of cell
senescence and induction of glucose metabolism in ovarian cancer
organoids and cells. Theranostics. 10:6928–6945. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Long Z and Wang Y: miR-195-5p suppresses
lung cancer cell proliferation, migration, and invasion via FOXK1.
Technol Cancer Res Treat. 19:15330338209225872020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Liang Y, Mao Q, Xia W, Chen B,
Shen H, Xu L, Jiang F and Dong G: Circular RNA circCRIM1 inhibits
invasion and metastasis in lung adenocarcinoma through the microRNA
(miR)-182/miR-93-leukemia inhibitory factor receptor pathway.
Cancer Sci. 110:2960–2972. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maass PG, Glazar P, Memczak S, Dittmar G,
Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC and
Rajewsky N: A map of human circular RNAs in clinically relevant
tissues. J Mol Med (Berl). 95:1179–1189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y,
Fan C, Li X, Li G, Li Y, et al: Circular RNAs (circRNAs) in cancer.
Cancer Lett. 425:134–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-Catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Mao R, Su W, Yang X, Geng Q, Guo
C, Wang Z, Wang J, Kresty LA, Beer DG, et al: Circular RNA
circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha
signaling in STK11 mutant lung cancer. Autophagy. 16:659–671. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma X, Yang X, Bao W, Li S, Liang S, Sun Y,
Zhao Y, Wang J and Zhao C: Circular RNA circMAN2B2 facilitates lung
cancer cell proliferation and invasion via miR-1275/FOXK1 axis.
Biochem Biophys Res Commun. 498:1009–1015. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu C, Hu W, Li LL, Wang YX, Zhou Q, Zhang
F, Song-Yang YY, Zhu W, Sun CC and Li DJ: Roles of miR-200 family
members in lung cancer: More than tumor suppressors. Future Oncol.
14:2875–2886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao P, Liu W and Zhou H: miR-429 promotes
the proliferation of non-small cell lung cancer cells via targeting
DLC-1. Oncol Lett. 12:2163–2168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen D, Ma W, Ke Z and Xie F: CircRNA
hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung
cancer progression. Cell Cycle. 17:2080–2090. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qu D, Yan B, Xin R and Ma T: A novel
circular RNA hsa_circ_0020123 exerts oncogenic properties through
suppression of miR-144 in non-small cell lung cancer. Am J Cancer
Res. 8:1387–1402. 2018.PubMed/NCBI
|
|
29
|
Chen D, Wang K, Li X, Jiang M, Ni L, Xu B,
Chu Y, Wang W, Wang H, Kang H, et al: FOXK1 plays an oncogenic role
in the development of esophageal cancer. Biochem Biophys Res
Commun. 494:88–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Gong M, Zhao Y, Zhao X and Li Q:
FOXK1 facilitates cell proliferation through regulating the
expression of p21, and promotes metastasis in ovarian cancer.
Oncotarget. 8:70441–70451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu M, Wang J, Tang W, Zhan X, Li Y, Peng
Y, Huang X, Bai Y, Zhao J, Li A, et al: FOXK1 interaction with FHL2
promotes proliferation, invasion and metastasis in colorectal
cancer. Oncogenesis. 5:e2712016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|





















