Introduction
Colorectal cancer (CRC) is a common gastrointestinal
cancer accounting for ~6.1% of all cancer cases (1). For patients with distant metastasis
and recurrent disease, the prognosis is poor (2). It is very important to clarify the
mechanism underlying CRC progression and identify novel targets for
the treatment of CRC.
It is reported that CRC is a heterogeneous disease,
and its occurrence and development is a multi-step process,
accompanied by complicated changes at the molecular level (3). Long non-coding RNAs (lncRNAs) are
incapable of encoding proteins and participate in regulating
various biological processes (4–8). For
instance, lnc-GNAT1-1 is downregulated in CRC, and it serves a
tumor-suppressive role by regulating Raf kinase inhibitor
protein/NF-κB/Snail signaling (6).
Moreover, lncRNA SNHG15 is highly expressed in CRC tissues, which
is associated with lymph node and liver metastases (7,8).
Notably, LINC00963 serves an important role in regulating the
proliferation and migration of CRC cells (9). However, the expression pattern,
biological functions and underlying mechanism of LINC00963 in CRC
have not been fully elucidated.
MicroRNAs (miRNAs/miRs) are non-coding small RNAs of
19–23 nucleotides in length, and are another class of regulatory
factors in CRC progression (10).
In eukaryotes, miRNAs modulate the expressions of >30% of genes
(11,12). Numerous miRNAs are dysregulated in
CRC, such as miR-533, miR-205, miR-203a-3p, miR-127-3p and
miR-18a-5p, and these miRNAs promote or repress the malignant
biological behaviors of CRC cells (13–16).
miR-1281 is reported to function as a tumor suppressor in multiple
cancer types (17,18), and notably, miR-1281 expression is
significantly downregulated in serum exosomes of patients with CRC
(19); however, the function and
mechanism of miR-1281 in CRC warrant further investigation.
The present study aimed to investigate the
expression pattern, biological functions and mechanism of LINC00963
in CRC progression. It was revealed that LINC00963 was highly
expressed in CRC, and it promoted the malignant phenotypes of CRC
cells by repressing miR-1281 expression and upregulating tripartite
motif-containing 65 (TRIM65) expression.
Materials and methods
Tissue collection
Tissue samples collected from patients with CRC
between 19 and 76 years old, including 27 females and 26 males,
from Qinghai Provincial People's Hospital (Qinghai, China) between
May 2016 and June 2019. After histological diagnosis, only the
patients who did not receive radiotherapy or chemotherapy prior to
surgery were enrolled. Pathological diagnoses of CRC were
determined by three pathologists according to the eighth edition of
the Union for International Cancer Control and the American Joint
Committee on Cancer tumor node metastasis classification (20,21).
CRC tissues and normal tissues (3.0 cm away from the tumor margin)
were collected during the surgery and, after resection, the samples
were rapidly frozen in liquid nitrogen, and then stored at −80°C.
The collection and use of human tissue samples were approved by the
Ethics Review Board of Qinghai Provincial People's Hospital
(approval no. KYLL180913), and written informed consent was
provided by all patients.
Cell lines and transfection
Human CRC cell lines (HCT116, SW480 and LoVo cells)
were purchased from the American Type Culture Collection, HT29
cells and the normal colon epithelial cell line NCM460 were
purchased from China Center for Type Culture Collection. The cells
were maintained in Dulbecco's modified Eagle's medium (DMEM)
containing 10% fetal bovine serum (FBS; both Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2. The
sequence of LINC00963 was cloned into pcDNA3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.) to construct overexpression
plasmids (pcDNA3.1-LINC00963, LINC00963), with empty vectors used
as the controls (pcDNA3.1-vector, NC). LINC00963 small interfering
(si)RNAs (si-LINC00963#1, 5′-TTGTACAGTTGGGTAAATCGAGG-3′; and
si-LINC00963#2, 5′-CCAGACACTGAACTGCCTT-3′), miR-1281 mimics
(5′-UCGCCUCCUCCUCUCCC-3′), miR-1281 inhibitor
(5′-AGCGGAGGAGGAGAGGG-3′), and corresponding negative controls
(NCs), including si-NC (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-NC
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′) and miR-NC inhibitor
(5′-GTGTAACACGTCTATACGCCA-3′), were obtained from Invitrogen
(Thermo Fisher Scientific, Inc.). The transient transfection was
conducted with final concentrations of 50 nM of siRNAs, miRNA
mimics and inhibitor, and 2 µg of overexpression plasmid by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Lipofectamine 2000 was added with oligonucleotide or plasmid, mixed
gently and incubated for 20 min at room temperature. Subsequently,
the mixture was added to cell suspension (5×106
cells/ml), and then incubated at 37°C and 5% CO2 for 6
h. Finally, the medium was replaced with DMEM supplemented with 10%
FBS. Cells were incubated at 37°C for 48 h, and were harvested for
reverse transcription-quantitative (RT-q)PCR, Cell Counting Kit-8
(CCK-8), colony formation, Transwell and western blot assays.
RT-qPCR
Total RNA of tissues and cells was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and then reverse transcribed to obtain cDNA with a
Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics)
according to the manufacturer's instructions. qPCR was performed
with SYBR-Green I Master (Roche Diagnostics) on the
LightCycler® 480 System (Roche Diagnostics). The RT-qPCR
cycling conditions were as follows: Initial denaturation at 95°C
for 10 min, followed by 40 cycles of denaturation at 95°C for 15
sec and annealing/elongation at 60°C for 60 sec. The relative gene
expression level of LINC00963 and TRIM65 was normalized to GAPDH,
and miR-1281 expression was normalized to U6, with the fold change
calculated by the 2−ΔΔCq method (22). The primers were designed by BGI
Genomics.
CCK-8 assay
Briefly, 100 µl of CRC cell suspension (containing
~2×103 cells) was added to each well of a 96-well plate.
Then, 10 µl of CCK-8 solution (Dojindo Molecular Technologies,
Inc.) was added to each well at 0, 24, 48 and 72 h. After the cells
were incubated at 37°C for 2 h, the value of optical density of
each well at 450 nm was measured by a microplate reader.
Colony formation assay
At 48 h after transfection, CRC cells were
dispersed, seeded into 6-well plates (1,000 cells/well) and
cultured for 2 weeks. Then, the medium was discarded, and the
colonies were washed using phosphate buffered saline (PBS), fixed
with 4% paraformaldehyde for 10 min at room temperature and stained
with 0.1% crystal violet for 15 min at room temperature. After the
colonies were washed using PBS again and dried, the colonies were
observed with a light microscope (magnification, ×200; Olympus
Corporation) and photographed with a camera (magnification, ×2.5;
Nikon Corporation), and the number of cell colonies (containing
>50 cells) was counted manually.
Cell invasion and migration
assays
To assess the migration and invasion of CRC cells,
4×104 transfected cells in 200 µl of serum-free medium
were transferred into the upper compartment of each Transwell
chamber (pore size, 8 µm, Corning Life Sciences), and 500 µl of
DMEM with 10% FBS was loaded into the lower compartment. After the
cells were cultured for 24 h, the migrated or invaded cells were
washed in PBS twice and fixed with 4% paraformaldehyde for 25 min
at room temperature and stained with 0.1% crystal violet for 10 min
at room temperature, successively. After that, the images were
captured under a light microscope (Olympus Corporation) at ×200
magnification, and cell counting was performed manually in three
randomly selected visual fields. For the invasion assay, the bottom
of the Transwell chambers was coated with a layer of Matrigel (1:8;
Corning Life Sciences) at 37°C for 30 min; the other procedures are
the same as those for the migration assay.
Luciferase reporter assay
LncBase Predicted v.2 database (threshold, 0.8;
http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted)
and TargetScanHuman database (version 7.2; http://www.targetscan.org/vert_72/) were used to
predict the potential binding sites between LINC00963 and miR-1281,
and miR-1281 and TRIM65 3′-untranslated region (3′UTR) (23,24).
For validating the predicted binding sites between LINC00963 and
miR-1281, and miR-1281 and TRIM65 3′UTR, pmiR-RB-REPORT™ (Guangzhou
RiboBio Co., Ltd.) reporter plasmids containing the wild-type (WT)
or mutant type (MUT) LINC00963 and TRIM65 3′UTR sequences with
predicted binding sites for miR-1281 were constructed. Then, 293T
cells (4×104 cells; China Center for Type Culture
Collection) were cultured in DMEM with 10% FBS and were
co-transfected with the reporter plasmids (80 ng) and miR-1281
mimics or control miRNA (50 nM) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Next, the cell culture was continued
for 48 h, before the cells in each group were collected and the
luciferase activities were determined with a dual-luciferase
reporter assay kit (Promega Corporation) according to the
instructions provided by the manufacturer. The relative luciferase
activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
assay
The RIP assay was performed with a Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (Millipore Inc.)
according to the manufacturer's instructions. In brief, RIP lysis
buffer was utilized to lyse the CRC cells, and 200 µl of cell
lysates were immunoprecipitated with anti-argonaute 2 (Ago2; cat.
no. ab32381; 1:50) or negative control anti-immunoglobulin G (IgG;
cat. no. ab190475; 1:100) antibodies (Abcam) at 4°C overnight.
Next, proteinase K and DNase (Beyotime Institute of Biotechnology)
were used to remove the proteins and DNA in the mixture, and then
the RNA was immunoprecipitated. Following that, the total RNA was
extracted using a RNeasy MinElute Cleanup kit (Qiagen China Co.,
Ltd.). Subsequently, RT-qPCR was performed to detect the enrichment
of LINC00963 as aforementioned.
RNA pull-down assay
Biotinylated miR-1281 or NC probes were conjugated
with streptavidin beads (Beyotime Institute of Biotechnology). The
biotinylated RNA was transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. RNA
pull-down assay was performed using a Magnetic RNA-Protein
Pull-Down kit (cat. no. 20164; Thermo Fisher Scientific, Inc.). The
cells transfected with probes were incubated with lysis buffer on
ice, and after 10 min, the mixtures were centrifuged at 13,000 × g
for 10 min at 4°C and cell debris was discarded. Subsequently, the
supernatants (10 mg) were incubated with magnetic beads for 2 h at
4°C, and then RNA was purified using an RNeasy Mini kit (Qiagen
GmbH) according to the manufacturer's instructions. Finally, the
abundance of LINC00963 was analyzed using RT-qPCR.
Western blot analysis
LoVo and HTC116 cells were lysed in RIPA lysis
buffer (Sigma-Aldrich; Merck KGaA). Then lysates were centrifuged
at 12,000 × g for 20 min at 4°C, and the total proteins were
extracted. Protein concentrations were detected by a BCA protein
assay kit (Thermo Fisher Scientific, Inc.). The protein sample (60
µg/lane) in each group was separated via 8% SDS-PAGE and then
transferred onto polyvinylidene fluoride membranes (Beyotime
Institute of Biotechnology). After that, the membranes were blocked
with 3% bovine serum albumin and then incubated overnight at 4°C
with primary antibodies against TRIM65 (Aviva Systems Biology; cat.
no. OAAB08057; 1:500), Ki67 (Abcam; cat. no. ab15580; 1:500),
matrix metalloproteinase 2 (Abcam; cat. no. ab92536; 1:500), matrix
metalloproteinase 9 (Abcam; cat. no. ab76003; 1:500) and GAPDH
(Abcam; cat. no. ab8245; 1:3,000). GAPDH was the internal
reference. Next, Tris-buffered saline with 0.05% Tween 20 (TBST)
was utilized for rinsing the membranes 3 times for 15 min each
time. Next, horseradish peroxidase-labeled goat anti-rabbit and
anti-mouse IgG (H + L) (Beyotime Institute of Biotechnology; cat.
nos. A0208 and A0216, respectively; 1:2,000) were used to incubate
with the membranes at room temperature for 30 min. After the
membranes were washed with TBST again, the protein bands were
developed using an enhanced chemiluminescence (ECL) kit (Beyotime
Institute of Biotechnology) and detected by ChemiDoc™ Touch Imaging
System (cat. no. 1708370; Bio-Rad Laboratories, Inc.). The
intensity of the bands was analyzed by ImageJ software version
1.8.0 (National Institutes of Health).
Statistical analysis
All experiments were performed in triplicate.
Experimental data were presented as the mean ± SD. Statistical
analysis was performed with SPSS 17.0 (SPSS, Inc.). Whether the
data are normally distributed or not was examined by the
Kolmogorov-Smirnov test. Student's unpaired t-test or one-way ANOVA
(with Tukey's post-hoc test) was employed to make comparisons.
Pearson's correlation analysis was used to analyze the correlation
between the expressions of LINC00963 and miR-1281 in CRC tissues.
The association of LINC00963 expression levels with
clinicopathological characteristics was analyzed using
χ2 test. P<0.05 was considered to indicate a
statistically significant difference.
Results
LINC00963 expression is upregulated in
CRC
Firstly, RT-qPCR was used to detect LINC00963
expressions in cancer tissues and adjacent tissues of 53 patients
with CRC. As shown, LINC00963 expression in CRC was significantly
upregulated, compared with that in normal tissues (Fig. 1A). Additionally, in CRC cell lines,
LINC00963 expression was higher compared with in normal colonic
epithelial cell line (Fig. 1B).
Statistical analyses revealed that high expression of LINC00963 was
significantly associated with larger tumor size (P=0.039) and lymph
node metastasis (P=0.020) of the patients (Table I).
 | Table I.Characteristics of LINC00963
expression in patients with colorectal cancer. |
Table I.
Characteristics of LINC00963
expression in patients with colorectal cancer.
|
|
| Expression of
LINC000963 |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total (n=53) | High (n=27) | Low (n=26) | P-value |
|---|
| Sex |
|
|
| 0.335 |
|
Male | 26 | 15 | 11 |
|
|
Female | 27 | 12 | 15 |
|
| Age, years |
|
|
| 0.907 |
|
<60 | 22 | 11 | 11 |
|
|
≥60 | 31 | 16 | 15 |
|
| Location |
|
|
| 0.132 |
|
Colon | 30 | 18 | 12 |
|
|
Rectum | 23 | 9 | 14 |
|
| Tumor size, cm |
|
|
| 0.039a |
|
<5 | 23 | 8 | 15 |
|
| ≥5 | 30 | 19 | 11 |
|
| Lymph node
metastasis |
|
|
| 0.020a |
| N0 | 26 | 9 | 17 |
|
|
N1-N2 | 27 | 18 | 9 |
|
|
Differentiation |
|
|
| 0.171 |
|
Well | 18 | 12 | 6 |
|
|
Moderate | 11 | 6 | 5 |
|
|
Poor | 24 | 9 | 15 |
|
| TNM stage |
|
|
| 0.131 |
|
I–II | 23 | 9 | 14 |
|
|
III–IV | 30 | 18 | 12 |
|
| Distant
metastasis |
|
|
| 0.464 |
| No | 21 | 12 | 9 |
|
|
Yes | 32 | 15 | 17 |
|
LINC00963 promotes proliferation,
migration and invasion of CRC cells
Given that among the CRC cell lines, LINC00963 had
the lowest expression in LoVo cells and the highest expression in
HCT116 cells, LoVo and HCT116 cells were selected as cell models
for further experiments. LINC00963 overexpression plasmids were
transfected into LoVo cells to construct a cell model of LINC00963
overexpression, and LINC00963 siRNAs were transfected into HCT116
cells to construct LINC00963 knockdown models. RT-qPCR confirmed
that the transfection was successful (Fig. 2A). The CCK-8 assay indicated that
LINC00963 overexpression significantly promoted the proliferation
of LoVo cells at 24, 48 and 72 h, while knocking down LINC00963 had
the opposite effects on HCT116 cells at 48 and 72 h (Fig. 2B). The colony formation assay showed
that LINC00963 overexpression markedly increased the colony-forming
ability of LoVo cells, but this colony-forming ability was
suppressed in HCT116 cells following LINC00963 knockdown (Fig. 2C). The Transwell assays suggested
that the migration and invasion of LoVo cells with LINC00963
overexpression were significantly increased, while the migration
and invasion of HCT116 cells with LINC00963 knockdown were
significantly decreased (Fig. 2D).
Additionally, western blot analysis revealed that LINC00963
overexpression promoted the expression levels of Ki67, MMP2 and
MMP9, while knockdown of LINC00963 caused the opposite effect
(Fig. 2E). These results suggested
that LINC00963 promoted the malignant phenotypes of CRC cells.
 | Figure 2.LINC00963 promotes the proliferation,
migration and invasion of colorectal cancer cells. (A) LINC00963
overexpression plasmids and siRNAs were transfected into LoVo and
HCT116 cells, respectively, and the relative expression of
LINC00963 in LoVo and HCT116 cells after the transfection was
detected by reverse transcription-quantitative PCR. (B) Cell
Counting Kit-8 assay was used to detect the proliferation of LoVo
cells with LINC00963 overexpression and HCT116 cells with LINC00963
knockdown. (C) Colony formation assay was used to detect the
colony-forming ability of LoVo cells with LINC00963 overexpression
and HCT116 cells with LINC00963 knockdown. (D) Transwell assay was
used to detect the migration and invasion of LoVo cells with
LINC00963 overexpression and HCT116 cells with LINC00963 knockdown.
Scale bar, 200 µm. (E) Western blot assay was used to detect the
expression of proliferation- and metastasis-related proteins (Ki67,
MMP2 and MMP9) in LoVo cells with LINC00963 overexpression and
HCT116 cells with LINC00963 knockdown. All experiments were
performed in triplicate. *P<0.05, **P<0.01, ***P<0.001 vs.
NC or si-NC. MMP, matrix metalloproteinase; NC, negative control;
si, small interfering. |
LINC00963 may function as a molecular
sponge for miR-1281
Using the LncBase database, it was revealed that
there was a potential binding site between LINC00963 (Gene_ID:
ENSG00000204054) and miR-1281 (Fig.
3A and Table SI). To verify
whether LINC00963 targets miR-1281, LINC00963-WT and LINC00963-MUT
luciferase reporter plasmids were co-transfected into LoVo and
HCT116 cells with miR-1281 mimics or miR-NC. The results revealed
that miR-1281 mimics significantly decreased the luciferase
activity of the LINC00963-WT reporter plasmid but had no impact on
that of LINC00963-MUT (Fig. 3B).
Additionally, RNA pull-down and RIP experiments demonstrated that
LINC00963 directly interacted with miR-1281 (Fig. 3C and D). Furthermore, RT-qPCR
revealed that miR-1281 expression in LoVo cells with LINC00963
overexpression was significantly inhibited, and in HCT116 cells
with LINC00963 knockdown, the expression of miR-1281 was markedly
increased (Fig. 3E). Besides, it
was found that miR-1281 expression in CRC was significantly
decreased compared with in in adjacent tissues (Fig. 3F), and Pearson's correlation
analysis showed that LINC00963 expression was negatively correlated
with miR-1281 expression in CRC tissue (Fig. 3G). Collectively, these results
implied that LINC00963 adsorbed miR-1281 and negatively regulated
its expression in CRC.
miR-1281 represses the malignant
biological behaviors of CRC cells
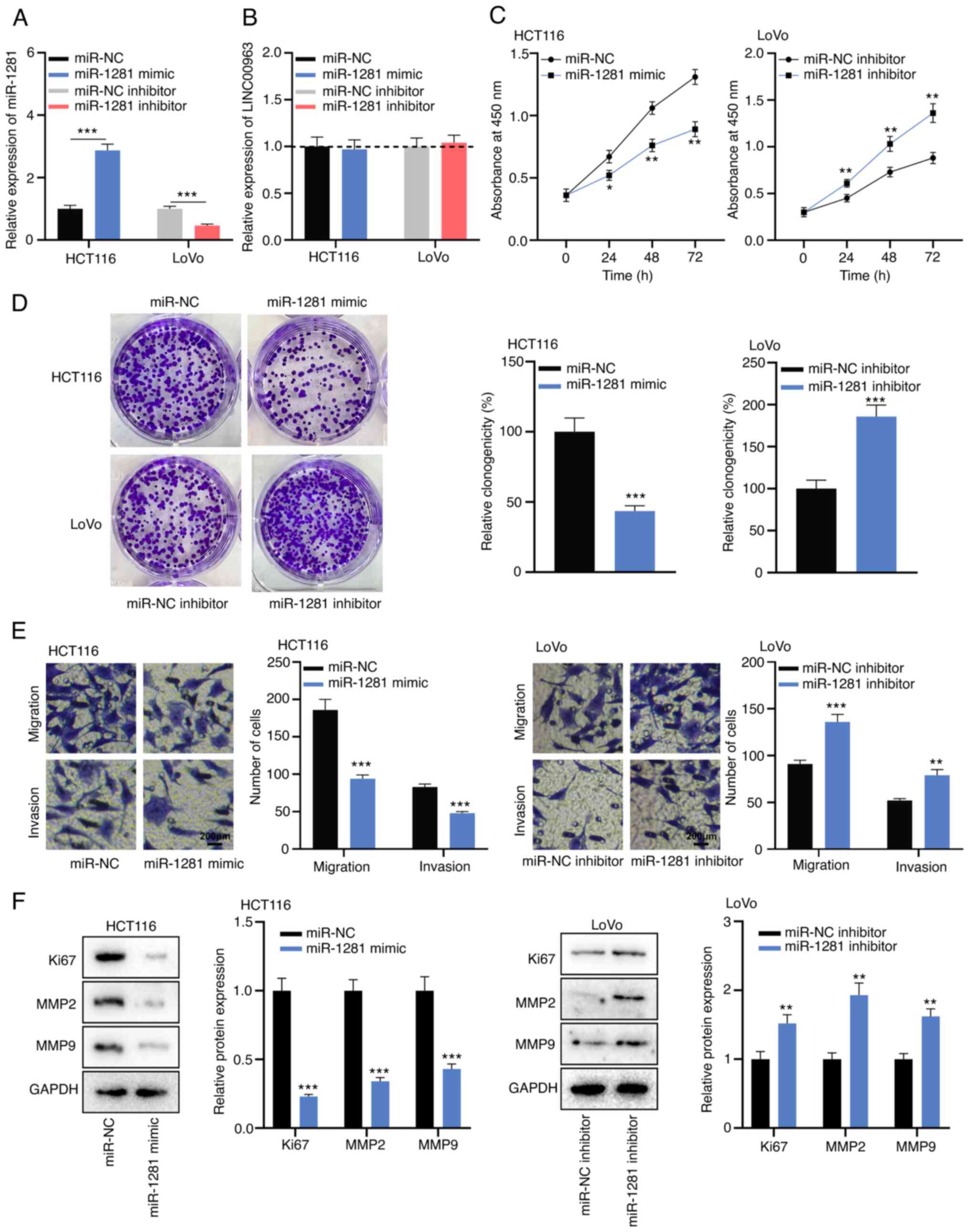
To elucidate the biological function of miR-1281,
miR-1281 mimics and inhibitors were transfected into HCT116 and
LoVo cells, and it was confirmed that the transfection was
successful via RT-qPCR (Fig. 4A).
RT-qPCR also revealed that in HCT116 cells with miR-1281
overexpression and LoVo cells with miR-1281 inhibition, there was
no significant change in LINC00963 expression (Fig. 4B). CCK-8, colony formation and
Transwell assays revealed that overexpression of miR-1281 inhibited
the proliferation, migration and invasion of HCT116 cells, while in
LoVo cells, knockdown of miR-1281 had the opposite effects
(Fig. 4C-E). Furthermore, western
blot analysis verified that the miR-1281 mimic inhibited the
expression levels of Ki67, MMP2 and MMP9, while the opposite
effects were observed after transfection of miR-1281 inhibitor
(Fig. 4F). These results suggested
that miR-1281 inhibited the malignant biological behaviors of CRC
cells.
TRIM65 is a target of miR-1281
TargetScan predicted that there was a complementary
binding site between miR-1281 and the 3′UTR of TRIM65 (Fig. 5A). To verify whether miR-1281 could
bind to the 3′UTR of TRIM65, TRIM65-WT and TRIM65-MUT reporter
plasmids were co-transfected into 293T cells with miR-1281 mimics
or miR-NC. The results confirmed that miR-1281 mimics significantly
reduced the luciferase activity of TRIM65-WT, while the luciferase
activity of TRIM65-MUT was not altered (Fig. 5B). RT-qPCR and western blot analysis
demonstrated that overexpression of miR-1281 significantly reduced
TRIM65 expression at both the mRNA and protein level in HCT116 and
LoVo cells, while LINC00963 overexpression had opposite effects;
moreover, overexpression of miR-1281 partially inhibited the
increase of TRIM65 expression caused by LINC00963 overexpression
(Fig. 5C and D).
LINC00963 regulates CRC cell
proliferation and metastasis via the miR-1281/TRIM65 axis
To clarify whether LINC00963 exerted its biological
function through miR-1281, miR-1281 mimics were transfected into
LoVo cells with LINC00963 overexpression, and miR-1281 inhibitors
were transfected into HCT116 cells with LINC00963 knockdown; the
transfection efficacy was verified via RT-qPCR (Fig. 6A). Functional experiments indicated
that miR-1281 mimics could partially abrogate the effects of
LINC00963 overexpression on the viability and metastasis of LoVo
cells; moreover, miR-1281 inhibition partially reversed the
inhibitory effect of knocking down LINC00963 on the proliferation
and metastasis of HCT116 cells (Fig.
6B-E). These results supported the hypotheses that LINC00963
regulates the malignant biological behaviors of CRC cells by
regulating miR-1281/TRIM65 axis.
Discussion
The importance of lncRNAs in cancer biology has been
increasingly recognized (25).
Although the roles of certain lncRNAs in CRC have been reported, a
large number of CRC-related lncRNAs have not been identified.
LINC00963, also known as MetaLnc9, is reported to be abnormally
expressed in various tumor types, and is involved in regulating the
proliferation, migration and invasion of tumor cells (26–29).
For example, it promotes the proliferation and invasion abilities
of osteosarcoma cells by regulating the miR-204-3p/fibronectin 1
(FN1) axis (26). Its expression is
upregulated in liver cancer, which promotes the proliferation of
cancer cells by activating the PI3K/AKT pathway (27). In head and neck carcinomas,
LINC00963 is highly expressed, and knockdown of LINC00963 reduces
the ability of self-renewal, invasion and colony formation in
cancer cells by regulating ABCB5 (28). LINC00963 also features prominently
in the transition of prostate cancer from androgen dependence to
androgen independence (29). In the
present study, it was observed that LINC00963 expression was
upregulated in CRC tissues and cells, which is consistent with a
previous study (9); moreover, the
high expression of LINC00963 was associated with larger tumor size
and lymph node metastasis in patients. Functionally, LINC00963
potentiated the proliferation, colony formation, migration and
invasion of CRC cells, implying that LINC00963 represents a tumor
promoter in CRC, and may be a promising novel therapeutic
target.
miRNAs, which are also non-coding RNAs, widely exist
in human organs and tissues, and partake in the vast majority of
biological processes (30).
Reportedly, miR-1281 has tumor-suppressive properties in certain
types of cancer; specifically, in osteosarcoma, miR-1281 expression
is modulated by p53, and it is upregulated under endoplasmic
reticulum stress, which promotes the apoptosis of cancer cells by
negatively regulating ubiquitin-specific peptidase 39 (17). In gastric and breast cancers,
miR-1281 also suppresses the malignant biological behaviors of
cancer cells (18,31). In muscle-invasive bladder cancer,
the expression of miR-1281 is also reduced (32). In the current study, it was observed
that miR-1281 is a target of LINC00963, miR-1281 expression was
downregulated in CRC tissues and cells, and the expression of
miR-1281 was negatively correlated with LINC00963 expression in CRC
tissues. Also, miR-1281 overexpression inhibited the proliferation,
colony formation, migration and invasion of CRC cells, suggesting
that miR-1281 is also a suppressor in CRC.
LncRNAs can function as competitive endogenous
(ce)RNAs to modulate miRNA expression levels (33). According to previous reports,
LINC00963 expression is upregulated in melanoma, and it promotes
melanoma progression by sponging miR-608 and upregulating nucleus
accumbens-associated protein 1 expression (34). In osteosarcoma, LINC00963
accelerates the growth and invasion of cancer cells by inhibiting
miR-204-3p expression and inducing the expression of FN1 (26). In prostate cancer, LINC00963
promotes the expression of NOP2 by sponging the tumor suppressor
miR-542-3p and facilitates the metastasis of prostate cancer
(35). In breast cancer, LINC00963
promotes tumorigenesis and radioresistance by adsorbing miR-324-3p
and inducing activated CDC42 kinase 1 expression (36). Additionally, LINC00963 can promote
the progression of breast cancer by targeting miR-625 and
upregulating high mobility group AT-hook 1 expression (37). In the present study, LINC00963 was
identified as a molecular sponge for miR-1281, and TRIM65 was
validated as one of the downstream targets of miR-1281. As a
ubiquitin ligase, TRIM65 is reported to exert oncogenic functions
in cancer biology. For example, TRIM65 negatively regulates the
tumor suppressor p53 via ubiquitination in lung cancer cells, and
silencing of TRIM65 inhibits the malignant phenotypes of lung
cancer cells (38,39). In hepatocellular carcinoma, TRIM65
activates β-catenin signaling via the ubiquitylation of Axin1
(40). Notably, TRIM65 expression
is upregulated in CRC, and TRIM65 targets Rho GTPase activating
protein 35 to cause protein degradation, as well as promoting the
proliferation, migration and invasion of CRC cells (41). A recent study reported that in
glioma, TRIM65 is a target gene of miR-1281 (42). In the present study, LINC00963 was
demonstrated to positively regulate TRIM65 expression by repressing
miR-1281 expression. miR-1281 overexpression reversed the effects
of LINC00963 overexpression on the phenotypes of CRC cells, which
may be achieved by regulating the expression of TRIM65. Therefore,
the present results proposed a ceRNA network consisting of
LINC00963, miR-1281 and TRIM65, which was involved in CRC
progression.
In conclusion, it was demonstrated that LINC00963
expression was upregulated and miR-1281 expression was
downregulated in CRC. Also, knockdown of LINC00963 or
overexpression of miR-1281 inhibited the proliferation, migration
and invasion of CRC cells, while overexpressing LINC00963 or
inhibiting miR-1281 induced the opposite effects. Additionally,
LINC00963 can upregulate TRIM65 expression by sponging miR-1281.
The present results offer useful indications for the diagnosis and
treatment of CRC. However, in subsequent work, in vivo
experiments are needed to further verify the current results.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GL and HL conceived and designed the study. HL and
DZ performed the experiments and analyzed the data. GL and HL
drafted and revised the manuscript. GL, DZ and HL confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The collection and use of human tissue samples were
approved by the Ethics Review Board of Qinghai Provincial People's
Hospital (approval no. KYLL180913), and written informed consent
was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jiang X, Li Q, Zhang S, Song C and Zheng
P: Long noncoding RNA GIHCG induces cancer progression and
chemoresistance and indicates poor prognosis in colorectal cancer.
Onco Targets Ther. 12:1059–1070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu L and Xie D, Xie H, Huang W, Zhang J,
Jin W, Jiang W and Xie D: ARHGAP10 inhibits the proliferation and
metastasis of CRC cells via blocking the activity of RhoA/AKT
signaling pathway. Onco Targets Ther. 12:11507–11516. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nguyen LH, Goel A and Chung DC: Pathways
of colorectal carcinogenesis. Gastroenterology. 158:291–302. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng W, He D, Shan B, Wang J, Shi W, Zhao
W, Peng Z, Luo Q, Duan M, Li B, et al: LINC81507 act as a competing
endogenous RNA of miR-199b-5p to facilitate NSCLC proliferation and
metastasis via regulating the CAV1/STAT3 pathway. Cell Death Dis.
10:5332019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abedini P, Fattahi A, Agah S, Talebi A,
Beygi AH, Amini SM, Mirzaei A and Akbari A: Expression analysis of
circulating plasma long noncoding RNAs in colorectal cancer: The
relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks.
J Cell Physiol. 234:22028–22033. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye C, Shen Z, Wang B, Li Y, Li T, Yang Y,
Jiang K, Ye Y and Wang S: A novel long non-coding RNA lnc-GNAT1-1
is low expressed in colorectal cancer and acts as a tumor
suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin
Cancer Res. 35:1872016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang L, Lin H, Kang L, Huang P, Huang J,
Cai J, Xian Z, Zhu P, Huang M, Wang L, et al: Aberrant expression
of long noncoding RNA SNHG15 correlates with liver metastasis and
poor survival in colorectal cancer. J Cell Physiol. 234:7032–7039.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun X, Bai Y, Yang C, Hu S, Hou Z and Wang
G: Long noncoding RNA SNHG15 enhances the development of colorectal
carcinoma via functioning as a ceRNA through
miR-141/SIRT1/Wnt/β-catenin axis. Artif Cells Nanomed Biotechnol.
47:2536–2544. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng K and Zhang TK: LncRNA LINC00963
promotes proliferation and migration through the miR-124-3p/FZD4
pathway in colorectal cancer. Eur Rev Med Pharmacol Sci.
24:7634–7644. 2020.PubMed/NCBI
|
|
10
|
Balacescu O, Sur D, Cainap C, Visan S,
Cruceriu D, Manzat-Saplacan R, Muresan MS, Balacescu L, Lisencu C
and Irimie A: The impact of miRNA in colorectal cancer progression
and its liver metastases. Int J Mol Sci. 19:37112018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y,
Tao Y, Shi W and Liu J: Lnc-ATB contributes to gastric cancer
growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem Biophys
Res Commun. 484:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hannafon BN, Cai A, Calloway CL, Xu YF,
Zhang R, Fung KM and Ding WQ: miR-23b and miR-27b are oncogenic
microRNAs in breast cancer: Evidence from a CRISPR/Cas9 deletion
study. BMC Cancer. 19:6422019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan L, You WQ, Sheng NQ, Gong JF, Hu LD,
Tan GW, Chen HQ and Wang ZG: A CREB1/miR-433 reciprocal feedback
loop modulates proliferation and metastasis in colorectal cancer.
Aging (Albany NY). 10:3774–3793. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li P, Xue WJ, Feng Y and Mao QS:
MicroRNA-205 functions as a tumor suppressor in colorectal cancer
by targeting cAMP responsive element binding protein 1 (CREB1). Am
J Transl Res. 7:2053–2059. 2015.PubMed/NCBI
|
|
15
|
Chen L, Gao H, Liang J, Qiao J, Duan J,
Shi H, Zhen T, Li H, Zhang F, Zhu Z and Han A: miR-203a-3p promotes
colorectal cancer proliferation and migration by targeting PDE4D.
Am J Cancer Res. 8:2387–2401. 2018.PubMed/NCBI
|
|
16
|
Zhang H, Zhu M, Shan X, Zhou X, Wang T,
Zhang J, Tao J, Cheng W, Chen G, Li J, et al: A panel of
seven-miRNA signature in plasma as potential biomarker for
colorectal cancer diagnosis. Gene. 687:246–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang J, Ma B, Li X, Jin W, Han C, Wang L
and Wang H: miR-1281, a p53-responsive microRNA, impairs the
survival of human osteosarcoma cells upon ER stress via targeting
USP39. Am J Cancer Res. 8:1764–1774. 2018.PubMed/NCBI
|
|
18
|
Liu G, Jiang Z, Qiao M and Wang F:
Lnc-GIHCG promotes cell proliferation and migration in gastric
cancer through miR-1281 adsorption. Mol Genet Genomic Med.
7:e7112019. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan S, Han B, Gao S, Wang X, Wang Z, Wang
F, Zhang J, Xu D and Sun B: Exosome-encapsulated microRNAs as
circulating biomarkers for colorectal cancer. Oncotarget.
8:60149–60158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bertero L, Massa F, Metovic J, Zanetti R,
Castellano I, Ricardi U, Papotti M and Cassoni P: Eighth edition of
the UICC classification of malignant tumours: An overview of the
changes in the pathological TNM classification criteria-what has
changed and why? Virchows Arch. 472:519–531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44(D1): D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Yin L, Li H, Liu LH and Xiao T:
The LncRNA LINC00963 facilitates osteosarcoma proliferation and
invasion by suppressing miR-204-3p/FN1 axis. Cancer Biol Ther.
20:1141–1148. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu JH, Tian XY, An QM, Guan XY and Hao CY:
LINC00963 promotes hepatocellular carcinoma progression by
activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci.
22:1645–1652. 2018.PubMed/NCBI
|
|
28
|
Lee SP, Hsieh PL, Fang CY, Chu PM, Liao
YW, Yu CH, Yu CC and Tsai LL: LINC00963 promotes cancer stemness,
metastasis, and drug resistance in head and neck carcinomas via
ABCB5 regulation. Cancers (Basel). 12:10732020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Han S, Jin G, Zhou X, Li M, Ying
X, Wang L, Wu H and Zhu Q: Linc00963: A novel, long non-coding RNA
involved in the transition of prostate cancer from
androgen-dependence to androgen-independence. Int J Oncol.
44:2041–2049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ganju A, Khan S, Hafeez BB, Behrman SW,
Yallapu MM, Chauhan SC and Jaggi M: miRNA nanotherapeutics for
cancer. Drug Discov Today. 22:424–432. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan LY, Shi KY, Xu D, Ren LP, Yang P,
Zhang L, Wang F and Shao GL: LncRNA GIHCG regulates microRNA-1281
and promotes malignant progression of breast cancer. Eur Rev Med
Pharmacol Sci. 23:10842–10850. 2019.PubMed/NCBI
|
|
32
|
Pignot G, Cizeron-Clairac G, Vacher S,
Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B,
Amsellem-Ouazana D and Bieche I: microRNA expression profile in a
large series of bladder tumors: Identification of a 3-miRNA
signature associated with aggressiveness of muscle-invasive bladder
cancer. Int J Cancer. 132:2479–2491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Wang G, Gao Y, Zhao C, Li X, Zhang
F, Jiang C and Wu B: Lnc-SNHG1 activates the TGFBR2/SMAD3 and
RAB11A/Wnt/β-catenin pathway by sponging MiR-302/372/373/520 in
invasive pituitary tumors. Cell Physiol Biochem. 48:1291–1303.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiao H, Jiang S, Wang H, Li Y and Zhang W:
Upregulation of LINC00963 facilitates melanoma progression through
miR-608/NACC1 pathway and predicts poor prognosis. Biochem Biophys
Res Commun. 504:34–39. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun F, Wu K, Yao Z, Mu X, Zheng Z, Sun M,
Wang Y, Liu Z and Zhu Y: Long noncoding RNA LINC00963 induces NOP2
expression by sponging tumor suppressor miR-542-3p to promote
metastasis in prostate cancer. Aging (Albany NY). 12:11500–11516.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang N, Zeng X, Sun C, Guo H, Wang T, Wei
L, Zhang Y, Zhao J and Ma X: LncRNA LINC00963 promotes
tumorigenesis and radioresistance in breast cancer by sponging
miR-324-3p and inducing ACK1 expression. Mol Ther Nucleic Acids.
18:871–881. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu Z, Wang W, Wang Y, Wang X, Sun S, Yao
Y, Zhang Y and Ren Z: Long noncoding RNA LINC00963 promotes breast
cancer progression by functioning as a molecular sponge for
microRNA-625 and thereby upregulating HMGA1. Cell Cycle.
19:610–624. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Ma C, Zhou T, Liu Y, Sun L and Yu Z:
TRIM65 negatively regulates p53 through ubiquitination. Biochem
Biophys Res Commun. 473:278–282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang XL, Shi WP, Shi HC, Lu SC, Wang K,
Sun C, He JS, Jin WG, Lv XX, Zou H and Shu YS: Knockdown of TRIM65
inhibits lung cancer cell proliferation, migration and invasion: A
therapeutic target in human lung cancer. Oncotarget. 7:81527–81540.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang YF, Zhang MF, Tian QH and Zhang CZ:
TRIM65 triggers β-catenin signaling via ubiquitylation of Axin1 to
promote hepatocellular carcinoma. J Cell Sci. 130:3108–3115.
2017.PubMed/NCBI
|
|
41
|
Chen D, Li Y, Zhang X, Wu H, Wang Q, Cai
J, Cui Y, Liu H, Lan P, Wang J, et al: Ubiquitin ligase TRIM65
promotes colorectal cancer metastasis by targeting ARHGAP35 for
protein degradation. Oncogene. 38:6429–6444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu G, Liu N, Wang H, Wang Y and Guo Z:
LncRNA LINC01857 promotes growth, migration, and invasion of glioma
by modulating miR-1281/TRIM65 axis. J Cell Physiol.
234:22009–22016. 2019. View Article : Google Scholar : PubMed/NCBI
|