Introduction
Bladder cancer is diagnosed in more than 430,000
patients worldwide every year, making it the ninth most common
malignancy (1). Ninety percent of
bladder cancers are transitional cell carcinomas, and the other 10%
are secondary deposits of squamous cell carcinoma, adenocarcinoma,
sarcoma, small cell carcinoma and other cancers of the body
(2). Bladder cancer has long been a
threat to human health due to its high morbidity and mortality
rates (3). In addition, genetic
mutations and a variety of external risk factors such as exposure
to carcinogens, smoking, chlorination of drinking water and
cyclophosphamide can lead to bladder cancer (4). Unfortunately, the etiology and
pathophysiology of bladder cancer are not fully understood.
The Rab protein family, a large number of small Rab
GTPases, mediates secretion, endoplasmic reticulum membrane
transport and the biogenesis of autophagosomes, and it is an
essential component of the vesicle transport mechanism (5). Overexpression of Rab GTPases is
related to cancer progression, and there are many mechanisms by
which Rab protein dysfunction has been linked to cancer development
(6). Elevated expression of
oncogenic Rab1, along with Rab1a proteins, promotes the growth of
tumors, often resulting in a poor prognosis (7). Overexpression of Rab23 has been linked
to gastric cancer (8), and Rab23
overexpression has been shown to facilitate malignant cell growth
and invasion in bladder cancer via the NF-κB pathway (9). Rab25 contributes to the invasiveness
of cancer cells by regulating integrin trafficking (10). Upregulation of Rab27b promotes the
malignant biological behavior, including F-actin recombination,
G1/S phase cell cycle transformation, cell proliferation, and
invasion enhancement of estrogen receptor-positive breast cancer
cells (11). Rab2a GTPase promotes
breast cancer stem cells and tumor progression via Erk signaling
activation (12). Abnormal
overexpression of Rab5a may stimulate the proliferation of ovarian
cancer cells through the APPL1-related epidermal growth factor
signal transduction pathway (13).
Rab6c, a newly identified Rab6 subfamily member, has attracted
recent attention because its aberrant expression might confer a
selective advantage to drug-resistant breast cancer cells (14). However, the role of Rab6c in bladder
cancer remains unknown.
miRNAs are small non-coding RNAs that regulate gene
expression by binding to the 3′ untranslated (3′UTR) of mRNA,
resulting in mRNA degradation or protein translation inhibition
(15). There are over 1,000 miRNAs
in the human genome, each potentially regulating hundreds of mRNAs.
Many miRNAs have been identified to exert important roles in
various cellular biological processes (16). The miR-218 located on chromosome
4p15.31, is associated with tumor growth, invasion as well as
metastasis (17). Accumulating
evidence has shown that the expression of miR-218 is abnormally low
in gastric cancer, cervical cancer, head and neck squamous cell
carcinoma, and breast cancer (18,19). A
recent bioinformatics analysis has suggested that miR-218 may be a
candidate tumor suppressor gene for bladder cancer, potentially
inhibiting the proliferation, migration, and invasion of bladder
cancer cells (20). Additionally,
several targets of miR-218 in bladder cancer have been reported
including LASP1 (21), BMI1
(22), and Glut1 (23). Given that miRNA targeting
transcripts is guided by complementary partial sequences, each
miRNA may regulate hundreds of genes (16). miRNAs have been the focus of bladder
cancer research in recent years (24). Therefore, the present study aimed to
investigate the role of miR-218 in bladder cancer and its
targets.
Materials and methods
Bioinformatics analysis of public
datasets
The Gene Expression Omnibus (GEO) and The Cancer
Genome Atlas (TCGA) databases provide an invaluable resource of
publicly available gene expression data that can be integrated and
analyzed to derive new hypothesis and knowledge. In this study, the
difference in Rab6c expression between tumor and normal of bladder
cancer samples were identified in the Gene Expression Omnibus (GEO)
database (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3167)
(25) and the database included 41
tumor tissue samples from bladder cancer patients and 9 normal
tissue samples. The data was unpaired and had no gender
distribution information. In addition, miR-218 expression in
bladder cancer samples was analyzed from Cancer Genome Atlas (TCGA)
database (portal.gdc.cancer.gov/) (26) and the database included 412 tumor
tissue samples from bladder cancer patients and 19 normal tissue
samples. The data was paired but there was no gender distribution
information. In addition, TargetScan human 7.1 (targetscan.org/vert_72/) was used to analyze the
target genes of miR-218 in bladder cancer.
Collection of clinical tissue
specimens from patients
Tumor tissue and matched adjacent normal tissue
samples were collected from 6 patients with bladder cancer (aged
55–67 years) undergoing surgery at the General Hospital of Shenyang
Military from 2008/1/31 to 2014/3/31 (Shenyang, China). The
inclusion criteria were male patients diagnosed with bladder cancer
by histology or cytology. The exclusion criteria were those who had
received systemic anti-cancer treatment for metastatic or
persistent/recurrent disease, or had a disease involving the
bladder during screening. Notably, <3 cm is adjacent normal
tissue, 3–5 cm is near cancer tissue, and greater than 5 cm is
distant cancer tissue (27).
Permission for the collection and application of patient samples
was granted by the Ethics Committee of the General Hospital of
Shenyang Military. In addition, each patient signed a written
informed consent before operation. All obtained clinical tissue
specimens were immediately frozen in liquid nitrogen and stored at
−80°C.
Cell culture and transfection
Bladder cancer cell lines (SV-HUC-1, T24 and EJ)
were purchased from the American Type Culture Collection and
cultured for 24 h at 37°C in a humidified atmosphere containing 5%
CO2 using DMEM (HyClone; Cytiva), containing 10% FBS
(HyClone; Cytiva) and antibiotics (100 U/ml penicillin and 100
mg/ml streptomycin). All T24 and EJ cells were authenticated by
short tandem repeat and confirmed negative for mycoplasma
contamination prior to the experiments. The cells were transferred
to the second generation for lentivirus transfection. Lentivirus
vectors plasmid were constructed by GenePharma. Following the
instructions of Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.), recombinant miR-218 lentivirus particles
(1×108 TU/ml) constructed with 5 µg GV309, Rab6c
lentivirus particles (1×108 TU/ml) constructed with 7.5
µg plent-EF1a-FH-CMV-GP were used to transfected cultured cells
(5×105) using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in DMEM with 10% FBS at a multiplicity of
infection of 10 for 20 min. The lentiviral vector with green
fluorescent protein (GFP) and resistance tag (Puromycin, 400 ng/ml)
was used to select the cells. 72 h after transfection, the
expression of GFP of the cells were observed under fluorescence
microscope (×200 magnification; Olympus Corporation) and cultured
for 24 h at 37°C in a humidified atmosphere containing 5%
CO2 for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol reagent (Invitrogen, Germany) was used to
obtain RNA from bladder cancer tissues and cells. According to the
manufacturer's instructions, a TaqMan MicroRNA reverse
transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used for reverse transcription of total RNA to obtain
cDNA to analyze miRNA expression. Subsequently, template cDNA was
used for qPCR analysis with PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and miR-218 or GAPDH primers, in
which GAPDH was used as an internal reference. mRNA expression
analysis of Rab6c was performed in the same manner. The
thermocycling conditions were as follows: Initial denaturation
95°C, 15 min; 40 of cycles of 55°C for 15 min and final extension
at 85°C for 2 min. The primer sequences used in this study were as
follows: Rab6c sense, 5′-AGGAGATCTGCCGCCGCGATCGC-3′ and antisense,
5′-CGAGCGGCCGCGTACGCGTCCTC-3′; miR-218 sense,
5′-CGAGTGCATTTGTGCTTGATCTA-3′ and antisense,
5′-TGGTGTCGTGGAGTCG-3′; U6 sense, 5′-CTCGCTTCGGCAGCACA-3′ and
antisense, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH sense,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and antisense,
5′-GCCATCACGCCACAGTTTC-3′. The relative mRNA expression of miR-218
and Rab6c was quantified with cycle threshold values and normalized
using the 2−∆∆Cq method (28). U6 was the internal control of
miR-218 expression, and GAPDH was the internal control of Rab6c
expression. The expression of miR-218 and Rab6c was relative to the
fold change of the corresponding negative controls, which was
defined as 1.0.
Western blotting
Total protein was extracted from cultured cells or
tissues using RIPA lysis buffer (Beyotime). Subsequently, protein
content was determined by the bicinchoninic acid method (BCA,
Pierce; Thermo Fisher Scientific, Inc.) and 30 µg of protein from
each group was separated by 10% SDS-PAGE and transferred to
nitrocellulose membranes (Amersham Bioscience). After the membranes
were blocked using TBST solution containing 5% skimmed milk and 0.5
ml/l Tween-20 at 4°C for 1 h, incubated with primary antibody
against Rab6c (1:500; Invitrogen; Thermo Fisher Scientific, Inc.;
PA5-39409) and GAPDH (1:1,000; Santa Cruz Biotechnology, sc-47724)
at room temperature for 2 h, they were incubated with horseradish
peroxidase-labeled secondary antibody (Santa Cruz Biotechnology)
with 1:5,000 dilution. The protein signal bands were visualized
using an enhanced chemiluminescence detection reagent (ECL; Thermo
Scientific, Inc.) and analyzed by ImageJ software 1.8.0.112
(National Institutes of Health).
CCK-8 assay
The T24 and EJ bladder cancer cell lines were seeded
at 2×103 cells/well and cultured in 6-well plates for 5
days. The manufacturer's instructions of the CCK-8 kit (Dojindo)
were strictly followed to perform the cell counting experiment
using a microplate reader (BioTek) to measure the optical density
(OD) at 450 nm.
Colony formation detection
The colony formation assay was performed as
previously described (29).
Briefly, transfected T24 and EJ bladder cancer cells were cultured
in 6-well plates at a density of 1,000 cells/well for 10 days.
After the cell colonies were treated with methanol for 15 min, they
were stained with 0.1% crystal violet for 10 min at room
temperature. After the number of cells in a single clone was
greater than 50, and the size between 0.3–1.0 mm, we started
counting and taking pictures under an optical microscope (Olympus).
The percentage of colony formation was calculated by setting the
control group to 100%.
Cell Transwell invasion assay
Briefly, 2×104 transfected T24 and EJ
cells were seeded in the upper Transwell invasion chambers
(24-well, 8-mm pore; Corning), which were coated with Matrigel (BD
Biosciences) at 37°C. The lower chamber was filled with medium
containing 10% FBS. After 48 h, the unmigrated cells were removed,
and the cells that migrated to the bottom were fixed with 70%
ethanol and stained with 0.1% crystal violet for 20 min at room
temperature. Next, the stained cells were photographed under
fluorescent microscope (200× magnification, Olympus Corporation)
and counted by ImageJ software 1.8.0.112 (National Institutes of
Health).
Luciferase reporter assay
The sequences of h-Rab6c-3′UTR-wild-type (WT) or
h-Rab6c-3′UTR-mutant (Mut) were synthesized, connected into the
pSI-Check2 vector (Hanbio) and extracted plasmids. The 293T cells
(Chinese Academy of Sciences, Shanghai, China) were cultured in
96-well plates for 24 h at 37°C until the cell density reached
5×104 cells/ml and co-transfected with the corresponding
plasmids (0.16 µg) with Lipofectamine 2000 (0.3 µl, 0.8 mg/ml;
Invitrogen;; Thermo Fisher Scientific, Inc.) at 37°C. After 6 h of
transfection, the cells were exchanged for fresh DMEM medium, and
cultured for 48 h at 37°C for subsequent Renilla luciferase
detection. Follow the instructions of the Dual Luciferase Reporter
Assay Kit (Promega Corporation), 100 µl Passive Lysis Buffer was
added to the 96-well plate and centrifuged at 1,200 × g at 4°C for
10 min, and then 100 µl Luciferase Assay Reagent II and 100 µl cell
lysate were added in sequence and mixed by pipetting 2–3 times. 100
µl STOP & GLO® reagent (Luciferase Assay Reagent;
Promega Corporation) was added and mixed 2–3 times to record the
Renilla luciferase value, which was the reporter gene
luminescence value.
Statistical analysis
The presented results were representative of
experiments repeated at least three times and all data as the mean
± standard deviation (SD). Statistical analysis was conducted with
SPSS 21.0 (IBM, Inc.) and GraphPad 8.0 (GraphPad Software). All
tests were analyzed using paired t-test and one-way ANOVA
followed by Bonferroni's post hoc test analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
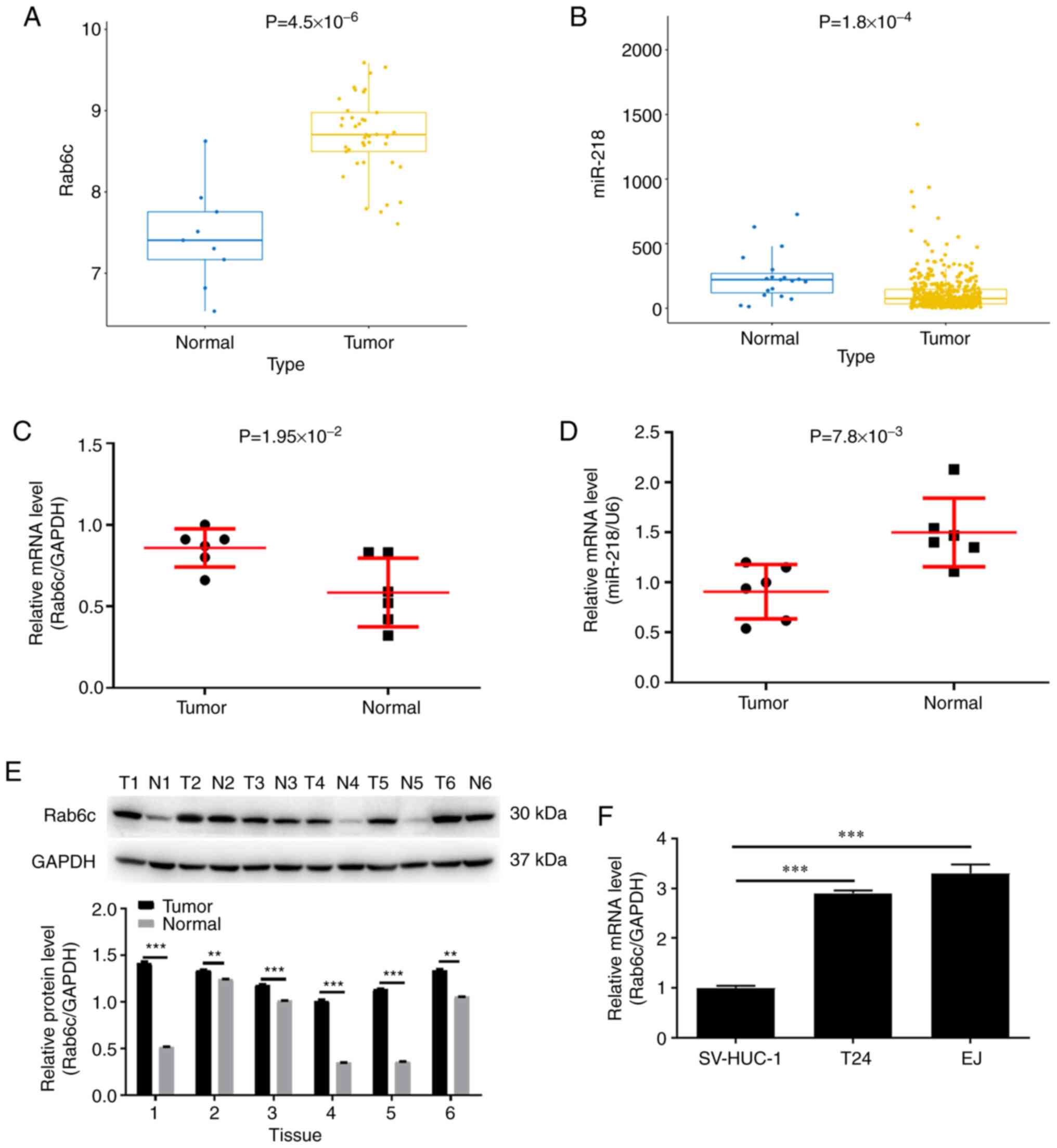
Rab6c is upregulated and miR-218 is
lowly expressed in bladder cancer
Previous studies have reported that miR-218 was
associated with the development of a variety of cancers, including
bladder cancer (23,30). In addition, Rab6c is a member of the
Rab family and is involved in drug resistance in MCF7 cells
(31). The relationship between
Rab6c and miR-218 in bladder cancer cell lines was examined in the
current study. First, the difference in Rab6c expression between
tumor and normal of bladder cancer samples was identified in the
GEO database. The results indicated that the expression level of
Rab6c in tumor tissues was significantly higher compared with that
in normal tissues (Fig. 1A).
Conversely, TCGA database results demonstrated that miR-218
expression was significantly lower in bladder cancer tissues
compared with that in normal tissues (Fig. 1B).
Subsequently, the expression levels of Rab6c and
miR-218 were detected in clinical samples of bladder cancer. As
expected, Rab6c mRNA expression was higher in tumor tissues
compared with in normal tissues (Fig.
1C). At the same time, it was identified that miR-218 was
expressed at low levels in bladder cancer (Fig. 1D). Consistently, Rab6c protein
expression was abnormally elevated in tumor tissue compared with
normal tissues (Fig. 1E). Due to
the individual differences and pathological grade stages of these 6
patients with bladder cancer, the expression level of Rab6c varied
among different patients. In fact, in patients with advanced
bladder cancer (cases 2, 3, 6), Rab6c was expressed in normal
tissues adjacent to the cancer, but it is still lower than that in
tumor tissues. In addition, Rab6c expression in T24 and EJ cells
was relatively higher compared with that in SV-HUC-1 cells
(Fig. 1F). In general, Rab6c was
upregulated in bladder cancer, while miR-218 was expressed at low
levels.
Rab6c and miR-218 are mutually
regulated in bladder cancer
Rab6c and miR-218 overexpressing cells were
constructed to examine their effects in bladder cancer cells. As
presented in Fig. 2A, miR-218
overexpression was established in T24 and EJ cells. It was found
that Rab6c protein expression was decreased in
miR-218-overexpressing T24 and EJ cells (Fig. 2B). Moreover, the expression level of
miR-218 was downregulated in Rab6c-overexpressing T24 and EJ cells
(Fig. 2C).
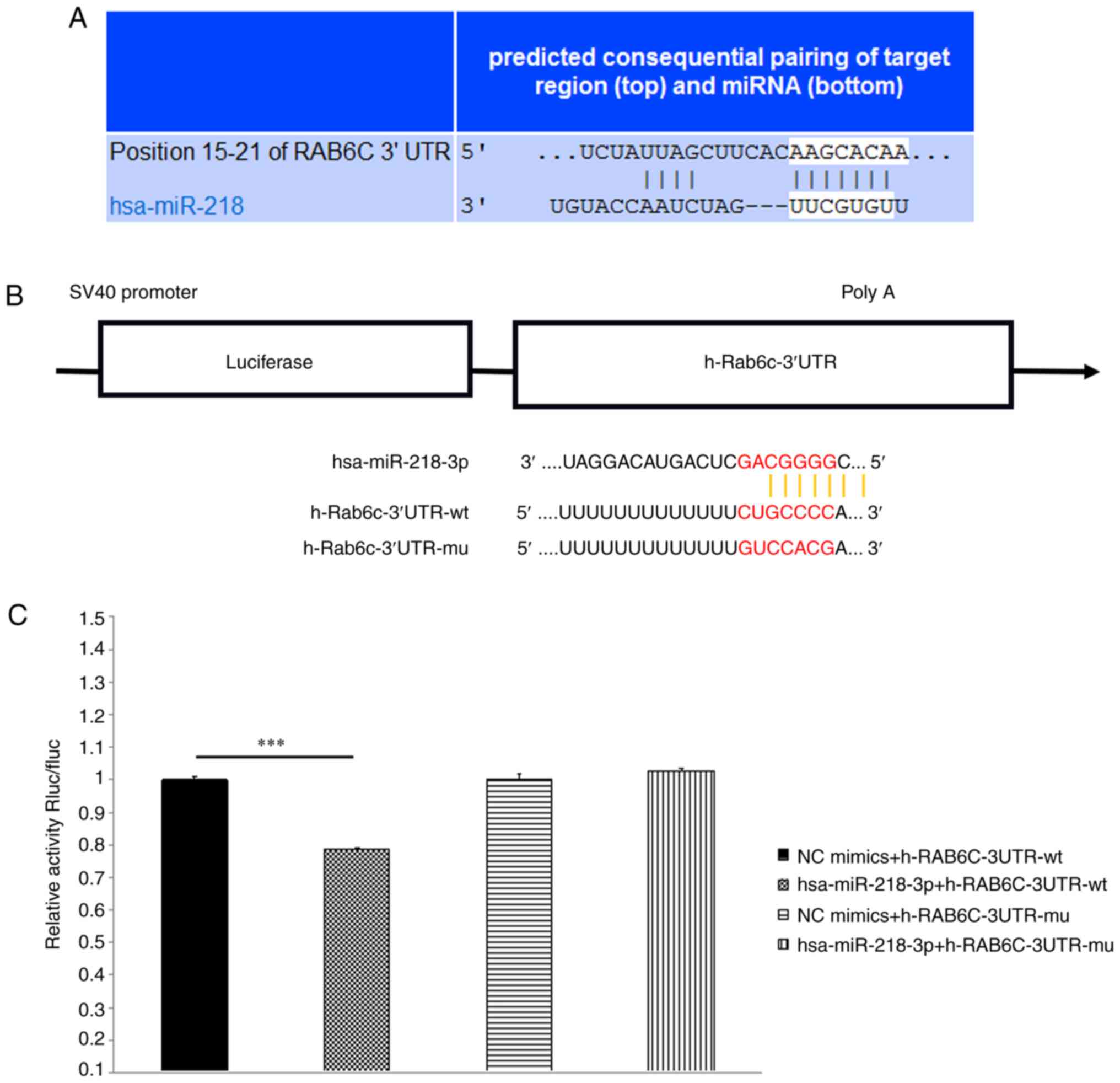
According to the conserved miR-218 binding site
(-AAGCACAA-) in the 3′UTR of Rab6c mRNA, as indicated by
TargetScan, a publicly available algorithm, Rab6c was preliminarily
identified as a promising target for miR-218 (Fig. 3A). Compared with the NC group,
hsa-miR-218-3p significantly downregulated the luciferase activity
in h-Rab6c-3′UTR-wt group, indicating an interaction between RAB6C
and miR-218. However, hsa-miR-218-3p failed to downregulate
luciferase activity in h-Rab6c-3′UTR-mu group (Fig. 3B and C). These results suggested
that there was a negative regulatory effect between Rab6c and
miR-218 in bladder cancer.
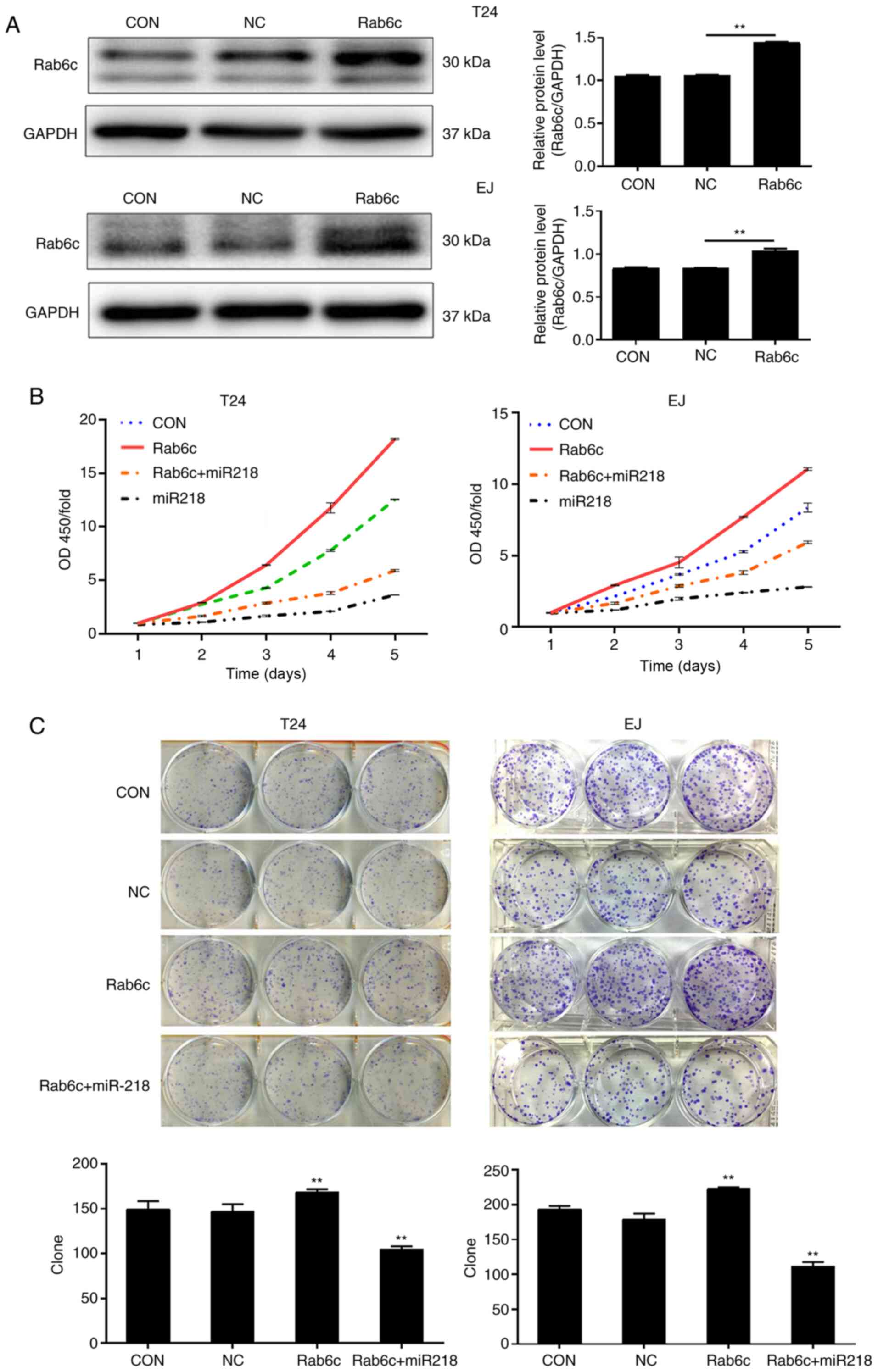
Overexpression of miR-218 reverses the
Rab6c-stimulated proliferation of bladder cancer cells
As the biological function of Rab6c in bladder
cancer cells has not been revealed, to the best of our knowledge,
proliferation was evaluated in T24 and EJ cells using CCK-8 and
colony formation assays. The result of western blotting indicated
that Rab6c expression was significantly upregulated following
transfection with Rab6c lentivirus particles (Fig. 4A). The CCK-8 assay demonstrated that
cell proliferation was notably promoted by Rab6c overexpression
(Fig. 4B). Consistent with the
results, Rab6c overexpression accelerated colony formation, as
indicated by an increased number of colonies (Fig. 4C). To elucidate whether the effects
of Rab6c overexpression were reversed by miR-218, restoration
experiments were performed. As shown in Fig. 4B and C, the cells overexpressing
Rab6c and miR-218 exhibited a lower proliferation rate and fewer
colonies compared with the cells overexpressing Rab6c alone.
Overexpression of miR-218 reduces
Rab6c-promoted invasion of bladder cancer cells
Bladder cancer cell invasion was examined using a
Transwell assays. Rab6c overexpression significantly promoted the
invasion of bladder cancer cells (Fig.
5). However, co-transfection with miR-218 overexpression
significantly reduced the invasion of T24 and EJ cells. Thus, it
was concluded that Rab6c may serve a stimulative role in the
proliferation and invasion of bladder cancer cells, which could be
reversed by miR-218.
Discussion
Bladder cancer affects ~430,000 individuals and
results in 165,000 deaths annually worldwide (32). Previous studies have reported
several genes relevant to the progression of bladder cancer
(33–35). For example, microarray data analysis
has shown that PCMT1 is more highly expressed in bladder cancer
than in normal urothelial tissue, and that it is positively
correlated with myometrial invasion, lymph node metastasis, distant
metastasis and clinical stage (33). Wnt7a activates canonical Wnt
signaling and promotes bladder cancer cell invasion, and Wnt7a is
associated with bladder cancer metastasis and predicts worse
clinical outcome (34). Forkhead
box M1 has been proposed to directly activate ATP binding cassette
subfamily G member 2 (Junior blood group) to increase the drug
efflux activation and drug resistance in bladder cancer cells
(35). These studies have expanded
the current knowledge on bladder cancer, providing a theoretical
foundation for a new treatment of bladder cancer.
Accumulating evidence has revealed that miR-218
affects the progression of various cancer types by interacting with
a variety of small molecules (36–40).
In acute promyelocytic leukemia, overexpression of miR-218
significantly inhibits cancer cell proliferation, arrests the cell
cycle in the G0/G1 phase and induces
apoptosis by targeting BMI1 (22).
High expression of RUNX family transcription factor 2 can restore
the inhibitory effects of miR-218 on malignant behavior of ovarian
cancer cells (17), while NEAT1
promotes cell invasion and proliferation by negatively regulating
miR-218 in breast cancer (41).
Moreover, in gastric cancer, miR-218 suppresses gastric cancer cell
cycle progression via the CDK6/Cyclin D1/E2F1 axis in a feedback
loop (42). Similarly, miR-218
functions as a tumor suppressor gene in cervical cancer (36). miR-218 suppresses the metastasis and
epithelial-mesenchymal transition of hepatocellular carcinoma cells
(43). Torres-Berrio et al
(44) demonstrated that miR-218 is
a molecular switch and potential biomarker of stress
susceptibility. However, the relationship between miR-218 and
bladder cancer remains unknown.
As an essential part of the vesicle transport
mechanism, specific Rab proteins coordinate with homologous
effectors to determine the destination of cargo proteins (45). Mutation of the Rab protein or
posttranslational modification leads to the destruction of the
regulatory network of vesicle transport, which impairs protein
secretion, endocytosis, recycling and degradation, and is
implicated in tumorigenesis (46,47).
Accordingly, the mechanism of vesicle transport serves an important
role in regulating the biological behavior of cancer cells. For
example, overexpression of Rab1a activates the mTOR complex 1
signaling pathway, which stimulates the progression and invasion of
colorectal cancer (48).
Additionally, Rab2a mediates the activation of Erk signaling to
drive the proliferation of breast cancer stem cells (12), and upregulation of Rab25 indicates a
poor prognosis in breast and ovarian cancer (49). Furthermore, phosphorylation of Rab
proteins is important for vesicle targeting and trafficking, and
phosphorylation of Rab5a by protein kinase C (PKC) facilitates
T-cell migration (50).
Mechanistically, Rab5a phosphorylation leads to the activation of
Rac1 to promote actin remodeling (51). Conventional PKC-mediated Rab11 and
Rab6 phosphorylation contributes to impaired endosomal recycling
and redistribution in the cytosolic fraction, respectively
(52,53). The present study reported a novel
role of Rab6c in bladder cancer progression and its regulation by
miR-218.
Rab6c, a newly identified member of the Rab family,
participates in the resistance of MCF7/AdrR cells (14). Moreover, Rab6c is a retrogene that
encodes a centrosome protein involved in cell cycle progression
(54). In addition, Rab6c is an
independent prognostic factor for estrogen receptor
positive/progesterone receptor negative breast cancer (55). The current study not only found that
Rab6c was upregulated in bladder cancer tissues, but also
identified that the upregulation of Rab6c enhanced the
proliferation and invasion of bladder cancer cells in vitro.
Thus, it was demonstrated that Rab6c exerted a tumor promoting role
in bladder cancer. Moreover, western blot analysis revealed
abnormally high expression of Rab6c protein in bladder cancer
cells. Overexpression of miR-218 in cultured bladder cancer cell
lines significantly inhibited Rab6c expression and reversed the
malignancy induced by Rab6c. This evidence suggested that Rab6c was
a target gene of miR-218 in bladder cancer. However, as the
collected bladder cancer tissue and cell types in the present study
were limited, additional detailed studies based on larger sample
sizes are required to further confirm the role of miR-218 and Rab6c
in the progression of bladder cancer.
In summary, the present study demonstrated that
Rab6c served a stimulative role in bladder cancer progression, and
that it was targeted and negatively regulated by miR-218.
Therefore, miR-218 may serve as a promising innovative therapeutic
target and Rab6c as a biomarker for bladder cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Military
Logistics Health Research Special General Project (grant no.
18BJZ17).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DH designed this project. LH, XP, YC, XW performed
the cell experiments. LH, PC and DH performed the rest of the
experiments. PC and CD conducted the data collection and analysis.
LH produced the manuscript, which was checked and revised by DH.
All authors read and approved the final manuscript. DH, LH and XP
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The collection and use of patient samples was
approved by the Ethics Committee of the General Hospital of
Shenyang Military (approval no. 201917), and written informed
consent was obtained from each patient prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoskin P and Dubash S: Bladder
conservation for muscle-invasive bladder cancer. Expert Rev
Anticancer Ther. 12:1015–1020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Hong YK, Zhuang DW, He XJ and Lin
ME: Bladder cancer survival nomogram: Development and validation of
a prediction tool, using the SEER and TCGA databases. Medicine
(Baltimore). 98:e177252019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu P, Zhang G, Zhao J, Chen J, Chen Y,
Huang W, Zhong J and Zeng J: Profiling the urinary microbiota in
male patients with bladder cancer in China. Front Cell Infect
Microbiol. 8:1672018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tzeng HT and Wang YC: Rab-mediated vesicle
trafficking in cancer. J Biomed Sci. 23:702016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Jia Q, Wang Y, Li F, Jia Z and Wan
Y: Rab40b upregulation correlates with the prognosis of gastric
cancer by promoting migration, invasion, and metastasis. Med Oncol.
32:1262015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas JD, Zhang YJ, Wei YH, Cho JH,
Morris LE, Wang HY and Zheng XF: Rab1A is an mTORC1 activator and a
colorectal oncogene. Cancer Cell. 26:754–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bin Z, Dedong H, Xiangjie F, Hongwei X and
Qinghui Y: The microRNA-367 inhibits the invasion and metastasis of
gastric cancer by directly repressing Rab23. Genet Test Mol
Biomarkers. 19:69–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang Y, Han Y, Sun C, Han C, Han N, Zhi W
and Qiao Q: Rab23 is overexpressed in human bladder cancer and
promotes cancer cell proliferation and invasion. Tumour Biol.
37:8131–8138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mitra S, Cheng KW and Mills GB: Rab25 in
cancer: A brief update. Biochem Soc Trans. 40:1404–1408. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu G, Niu M, Qin J, Wang Y and Tian J:
Inactivation of Rab27B-dependent signaling pathway by calycosin
inhibits migration and invasion of ER-negative breast cancer cells.
Gene. 709:48–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo ML, Gong C, Chen CH, Hu H, Huang P,
Zheng M, Yao Y, Wei S, Wulf G, Lieberman J, et al: The Rab2A GTPase
promotes breast cancer stem cells and tumorigenesis via Erk
signaling activation. Cell Rep. 11:111–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni
PH, Chen XH and Fan QS: Rab5a overexpression promoting ovarian
cancer cell proliferation may be associated with APPL1-related
epidermal growth factor signaling pathway. Cancer Sci.
101:1454–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian K, Jurukovski V, Yuan L, Shan J and
Xu H: WTH3, which encodes a small G protein, is differentially
regulated in multidrug-resistant and sensitive MCF7 cells. Cancer
Res. 65:7421–7428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuppusamy KT, Sperber H and Ruohola-Baker
H: MicroRNA regulation and role in stem cell maintenance, cardiac
differentiation and hypertrophy. Curr Mol Med. 13:757–764. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li N, Wang L, Tan G, Guo Z, Liu L, Yang M
and He J: MicroRNA-218 inhibits proliferation and invasion in
ovarian cancer by targeting Runx2. Oncotarget. 8:91530–91541. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng XJ, Wu YH, Luo M, Cong PG and Yu H:
Inhibition of pulmonary carcinoma proliferation or metastasis of
miR-218 via down-regulating CDCP1 expression. Eur Rev Med Pharmacol
Sci. 21:1502–1508. 2017.PubMed/NCBI
|
|
19
|
Wang P, Zhai G and Bai Y: Values of
miR-34a and miR-218 expression in the diagnosis of cervical cancer
and the prediction of prognosis. Oncol Lett. 15:3580–3585.
2018.PubMed/NCBI
|
|
20
|
Tatarano S, Chiyomaru T, Kawakami K,
Enokida H, Yoshino H, Hidaka H, Yamasaki T, Kawahara K, Nishiyama
K, Seki N and Nakagawa M: miR-218 on the genomic loss region of
chromosome 4p15.31 functions as a tumor suppressor in bladder
cancer. Int J Oncol. 39:13–21. 2011.PubMed/NCBI
|
|
21
|
Wang LL, Wang L, Wang XY, Shang D, Yin SJ,
Sun LL and Ji HB: MicroRNA-218 inhibits the proliferation,
migration, and invasion and promotes apoptosis of gastric cancer
cells by targeting LASP1. Tumour Biol. 37:15241–15252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Sun HH, Sui MH and Ma JJ: miR-218
inhibits acute promyelocytic leukemia cell growth by targeting
BMI-1. Oncol Lett. 14:8078–8083. 2017.PubMed/NCBI
|
|
23
|
Li P, Yang X, Cheng Y, Zhang X, Yang C,
Deng X, Li P, Tao J, Yang H, Wei J, et al: MicroRNA-218 increases
the sensitivity of bladder cancer to cisplatin by targeting Glut1.
Cell Physiol Biochem. 41:921–932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gulia C, Baldassarra S, Signore F, Rigon
G, Pizzuti V, Gaffi M, Briganti V, Porrello A and Piergentili R:
Role of non-coding RNAs in the etiology of bladder cancer. Genes
(Basel). 8:3392017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Chen L, Ju L, Qian K, Liu X, Wang
X and Xiao Y: Novel biomarkers associated with progression and
prognosis of bladder cancer identified by co-expression analysis.
Front Oncol. 9:10302019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choi W, Ochoa A, McConkey DJ, Aine M,
Hoglund M, Kim WY, Real FX, Kiltie AE, Milsom I, Dyrskjøt L and
Lerner SP: Genetic alterations in the molecular subtypes of bladder
cancer: Illustration in the cancer genome atlas dataset. Eur Urol.
72:354–365. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kumar P, Kanaujia SK, Singh A and Pradhan
A: In vivo detection of oral precancer using a fluorescence-based,
in-house-fabricated device: A Mahalanobis distance-based
classification. Lasers Med Sci. 34:1243–1251. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen X, Li J, Li CL and Lu X: Long
non-coding RNA ZFAS1 promotes nasopharyngeal carcinoma through
activation of Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci.
22:3423–3429. 2018.PubMed/NCBI
|
|
30
|
Li Y, Shi B, Dong F, Zhu X, Liu B and Liu
Y: LncRNA KCNQ1OT1 facilitates the progression of bladder cancer by
targeting miR-218-5p/HS3ST3B1. Cancer Gene Ther. 28:212–220. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shan J, Mason JM, Yuan L, Barcia M, Porti
D, Calabro A, Budman D, Vinciguerra V and Xu H: Rab6c, a new member
of the rab gene family, is involved in drug resistance in MCF7/AdrR
cells. Gene. 257:67–75. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bellmunt J, Powles T and Vogelzang NJ: A
review on the evolution of PD-1/PD-L1 immunotherapy for bladder
cancer: The future is now. Cancer Treat Rev. 54:58–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong L, Li Y, Xue D and Liu Y: PCMT1 is an
unfavorable predictor and functions as an oncogene in bladder
cancer. IUBMB Life. 70:291–299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang X, Zhu H, Gao Z, Li J, Zhuang J,
Dong Y, Shen B, Li M, Zhou H, Guo H, et al: Wnt7a activates
canonical Wnt signaling, promotes bladder cancer cell invasion, and
is suppressed by miR-370-3p. J Biol Chem. 293:6693–6706. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roh YG, Mun MH, Jeong MS, Kim WT, Lee SR,
Chung JW, Kim SI, Kim TN, Nam JK and Leem SH: Drug resistance of
bladder cancer cells through activation of ABCG2 by FOXM1. BMB Rep.
51:98–103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Z, Mao L, Wang L, Zhang H and Hu X:
miR218 functions as a tumor suppressor gene in cervical cancer. Mol
Med Rep. 21:209–219. 2020.PubMed/NCBI
|
|
37
|
Liu T, Zhang X, Du L, Wang Y, Liu X, Tian
H, Wang L, Li P, Zhao Y, Duan W, et al: Exosome-transmitted
miR-128-3p increase chemosensitivity of oxaliplatin-resistant
colorectal cancer. Mol Cancer. 18:432019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia C, Jiang H, Ye F and Zhuang Z: The
multifunction Of miR-218-5p-Cx43 axis in breast cancer. Onco
Targets Ther. 12:8319–8328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Setijono SR, Park M, Kim G, Kim Y, Cho KW
and Song SJ: miR-218 and miR-129 regulate breast cancer progression
by targeting Lamins. Biochem Biophys Res Commun. 496:826–833. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu L, Tu H, Liang Y and Tang D: miR-218
produces anti-tumor effects on cervical cancer cells in vitro.
World J Surg Oncol. 16:2042018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao D, Zhang Y, Wang N and Yu N: NEAT1
negatively regulates miR-218 expression and promotes breast cancer
progression. Cancer Biomark. 20:247–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Deng M, Zeng C, Lu X, He X, Zhang R, Qiu
Q, Zheng G, Jia X, Liu H and He Z: miR-218 suppresses gastric
cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis
in a feedback loop. Cancer Lett. 403:175–185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang T, Xu L, Jia R and Wei J: miR-218
suppresses the metastasis and EMT of HCC cells via targeting
SERBP1. Acta Biochim Biophys Sin (Shanghai). 49:383–391. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Torres-Berrio A, Nouel D, Cuesta S, Parise
EM, Restrepo-Lozano JM, Larochelle P, Nestler EJ and Flores C:
miR-218: A molecular switch and potential biomarker of
susceptibility to stress. Mol Psychiatry. 25:951–964. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Murray SS, Wong AW, Yang J, Li Y, Putz U,
Tan SS and Howitt J: Ubiquitin regulation of trk receptor
trafficking and degradation. Mol Neurobiol. 56:1628–1636. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie J, Yan Y, Liu F, Kang H, Xu F, Xiao W,
Wang H and Wang Y: Knockdown of Rab7a suppresses the proliferation,
migration, and xenograft tumor growth of breast cancer cells.
Biosci Rep. 39:BSR201804802019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Černochová R, Nekulová M and Holčaková J:
Rab proteins, intracellular transport and cancer. Klin Onkol. 29
(Suppl 4):S31–S39. 2016.(In Czech). View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan SJ, Snell C, Turley H, Li JL,
McCormick R, Perera SM, Heublein S, Kazi S, Azad A, Wilson C, et
al: PAT4 levels control amino-acid sensitivity of
rapamycin-resistant mTORC1 from the Golgi and affect clinical
outcome in colorectal cancer. Oncogene. 35:3004–3015. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yin C, Mou Q, Pan X, Zhang G, Li H and Sun
Y: miR-577 suppresses epithelial-mesenchymal transition and
metastasis of breast cancer by targeting Rab25. Thorac Cancer.
9:472–479. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ong ST, Freeley M, Skubis-Zegadlo J, Fazil
MH, Kelleher D, Fresser F, Baier G, Verma NK and Long A:
Phosphorylation of Rab5a protein by protein kinase C is crucial for
T-cell migration. J Biol Chem. 289:19420–19434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tan L, Zhang Y, Zhan Y, Yuan Y, Sun Y, Qiu
X, Meng C, Song C, Liao Y and Ding C: Newcastle disease virus
employs macropinocytosis and Rab5a-dependent intracellular
trafficking to infect DF-1 cells. Oncotarget. 7:86117–86133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pavarotti M, Capmany A, Vitale N, Colombo
MI and Damiani MT: Rab11 is phosphorylated by classical and novel
protein kinase C isoenzymes upon sustained phorbol ester
activation. Biol Cell. 104:102–115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fitzgerald ML and Reed GL: Rab6 is
phosphorylated in thrombin-activated platelets by a protein kinase
C-dependent mechanism: Effects on GTP/GDP binding and cellular
distribution. Biochem J. 342:353–360. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Young J, Menetrey J and Goud B: RAB6C is a
retrogene that encodes a centrosomal protein involved in cell cycle
progression. J Mol Biol. 397:69–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fohlin H, Bekkhus T, Sandström J,
Fornander T, Nordenskjöld B, Carstensen J and Stål O: RAB6C is an
independent prognostic factor of estrogen
receptor-positive/progesterone receptor-negative breast cancer.
Oncol Lett. 19:52–60. 2020.PubMed/NCBI
|