Introduction
Cervical cancer (CC) is the fourth most common type
of cancer among female patients worldwide (1) and is a major health problem,
particularly in developing countries (2). The introduction of a specific human
papillomavirus vaccine into clinical practice has led to a gradual
reduction in the incidence of CC (3). Over the past two decades, the
exploration of the molecular mechanisms underlying tumor
development has increased (4).
Although significant progress has been achieved, potential
prognostic and diagnostic biomarkers must be identified to
facilitate the management of patients with CC.
Cancer/testis antigens (CTAs) are a class of
tumor-related antigens with restrictive expression patterns that
are mostly located on the X chromosome or restricted to cancer and
reproductive tissues, and represent a set of extensively researched
targets of tumor immunotherapy (5).
Placenta-specific protein 1 (PLAC1) is a member of the CTAs; the
gene encoding PLAC1 is on the X chromosome (Xq26.3) near the
hypoxanthine-guanine phosphoribosyl transferase gene (6). PLAC1 is a small secreted cell-adhesion
protein, which has a predicted amino terminal transmembrane domain
and a divisible signal peptide of 23 amino acids (6). PLAC1 is normally expressed in
placental trophoblasts and the testis, but exhibits extremely low
expression in other normal tissues (7,8). In
recent decades, PLAC1 has been found to be localized on the surface
of tumor cells and be approachable to antibodies (9). High expression levels of PLAC1 have
been detected in a broad variety of solid tumors, including liver,
digestive tract, mammary, prostatic, ovarian and uterine tumors
(10–12). Devor et al (13) reported that PLAC1 was also expressed
in human papillomavirus-positive CC, including in four common
pathological types. Furthermore, Wang et al (8) revealed that PLAC1, which was elicited
by Epstein-Barr virus and expressed in human tumor cells, was an
important CTA. Moreover, PLAC1-knockout mice have been reported to
show considerable placental defects, with placental layers
revealing aberrant differentiation (14). Considering the inherent invasive
ability of the placental trophoblast, it has been hypothesized that
tumor cells may use the placental mechanisms to facilitate invasion
(15). Understanding this process
may lead to the development of methods for blocking PLAC1
expression in tumor cells. Currently, on account of its
immunogenicity and carcinogenicity, PLAC1 is on the National Cancer
Institute list of ‘cancer antigens’, which may act as a target for
developing vaccines to prevent breast carcinoma (16).
The present study investigated the role of PLAC1 in
the proliferation and invasion of CC cells, and proposed a
hypothesis that PLAC1 may be an important oncogenic and prognostic
factor in CC.
Materials and methods
Data preprocessing and survival
analysis
RNA-sequencing (RNA-seq) expression data and
clinicopathological information (including survival data) from
patients with CC were derived from The Cancer Genome Atlas (TCGA)
portal (https://portal.gdc.cancer.gov/). The methods used for
sample acquisition, RNA extraction and sequencing were previously
described by TCGA Research Network (17). Survival curves were estimated using
Kaplan-Meier methods and compared by the log-rank test.
Gene set enrichment analysis
(GSEA)
The aim of GSEA is to determine the different
expression levels of a predefined gene set in two different
phenotypes (18). In the present
study, GSEA was conducted to examine the significance of survival
differences between ‘high-PLAC1’ and ‘low-PLAC1’ expression groups
based on hallmark collections of gene sets in the Molecular
Signatures Database (19) using the
official Java GSEA tool (18)
(version: 4.1.0, number of permutations=1,000), the two groups were
divided by the median expression level. This method identifies
genes with large phenotype correlations showing the greatest
contribution of enrichment scores in multiple gene sets within a
molecular signatures database collection.
Immunohistochemistry (IHC)
analysis
The tissue microarray (HUteS154Su01) containing
patient clinical information was obtained from Shanghai Outdo
Biotech Company and was used to detect the expression levels of
PLAC1 in 101 cervical tumor tissues, 35 non-tumor tissues and 18
high-grade squamous intraepithelial lesions (HSILs). The tissue
microarray experiment was completed by Shanghai Outdo Biotech
Company. The slide was blocked in 10% goat serum (Sigma-Aldrich;
Merck KGaA) for 30 min at room temperature. Antibody against PLAC1
(1:100; cat. no. sc-365919, Santa Cruz Biotechnology Inc) was added
and incubated at 4°C overnight. The ready-to-use horseradish
peroxidase (HRP)-conjugated secondary antibody (cat. no. K5007;
Dako; Agilent Technologies, Inc.) was added and incubated for 30
min at room temperature. Slides were visualized under a light
microscope (Olympus Corporation; cat. no. CX31) at ×400
magnification. Semi-quantification of protein expression was
determined according to the percentage of positive cells (N) as
follows: 0, ≤5; 1, 5<N≤25; 2, 25<N≤50; 3, 50<N≤75 and 4,
N>75%. Staining intensity ranged from 0–3 (0 negative; 1, weak;
2, moderate; 3, strong)., The two scores were multiplied together
to calculate the total score, which ranged from 0–12. All IHC data
were interpreted by the same qualified pathologist for
consistency.
Cell culture
The human CC cell lines CaSki, MS751, C-33A and
HeLa, were obtained from the American Type Culture Collection. All
the cells were cultured in DMEM (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS (HyClone; Cytiva). All cells were
incubated in 5% CO2 at 37°C.
RNA silencing and cDNA
overexpression
Lentivirus harboring two short hairpin RNAs (shRNAs)
targeting PLAC1 gene (PLAC1 shRNA1 and PLAC1 shRNA2) or
PLAC1 cDNA were designed and produced by OriGene
Technologies, Inc. shRNA targeting firefly luciferase (shLuc) was
used as a negative control for RNA silencing, and empty vector was
used as a negative control for cDNA overexpression. HeLa cells at
70–80% confluence were exposed to the lentivirus with PLAC1
cDNA and empty vector (MOI=10), MS751 cells at 70–80% confluence
were exposed to the lentivirus harboring two shRNA (PLAC1 shRNA1
and PLAC1 shRNA2) and shLuc (MOI=10). Subsequently, 1 µg/ml
puromycin was used for selection for 10 days. The established cells
were then incubated at 37°C and 5% CO2.
Western blot analysis
The cells were harvested using an enzymatic
digestion method, then ice-cold lysis buffer (50 mM Tris, 150 mM
NaCl, 0.5% EDTA and 0.5% NP-40) was added and incubated for 20 min
at 4°C. Centrifugation was performed at 13,000 × g at 4°C for 15
min. Total protein concentration was determined using a BCA Protein
Quantification kit. Samples (30 µg total proteins) were separated
via SDS-PAGE on a 10% gel and subsequently transferred to a PVDF
membrane. Membranes were blocked with 5% non-fat dry milk for 1 h
at room temperature and incubated with the following primary
antibodies overnight at 4°C: PLAC1 (1:500; cat. no. sc-365919;
Santa Cruz Biotechnology, Inc.), Slug (1:1,000; cat. no. 9585; Cell
Signaling Technology), Snail (1:1,000; cat. no. 3879; Cell
Signaling Technology), E-cadherin (1:1,000; cat. no. 3195; Cell
Signaling Technology), Vimentin (1:1,000; cat. no. 5741; Cell
Signaling Technology) and MMP2 (1:1,000; cat. no. 4022; Cell
Signaling Technology) and β-actin (1:10,000; cat. no. MABT825;
Sigma-Aldrich; Merck KGaA). After secondary antibody incubation for
1 h at room temperature (both 1:3,000; HRP-conjugated anti-rabbit
and anti-mouse; cat. nos. 7074 and 7076, respectively; Cell
Signaling Technology, Inc.), protein expression was determined with
the Pierce ECL system (Thermo Fisher Scientific, Inc.). The protein
band density was determined using ImageJ (version: 1.53; National
Institutes of Health).
Wound healing assay
The established HeLa cells and MS751 cells were
seeded into 6-well plates at 80–90% confluence and grown to a
confluent monolayer overnight, then a scratch was made with a 200
µl pipette tip. DMEM without FBS was used for cell culture at 37°C.
The cell migratory ability was evaluated by recording the wound
area at 48 h and as percentage of initial wound area at 0 h. The
images were visualized under a light microscope. Experiments were
performed in triplicate.
Cell migration and invasion
assays
The in vitro cell migration and invasion
assays were conducted using cell culture chambers (24-well, pore
size, 8 µm; Corning, Inc.). For the invasion assay, the upper
chamber was pre-coated with 60 µl Matrigel (BD Biosciences) at 37°C
for 1 h. Cells were collected and resuspended in serum-free medium.
Subsequently, cells (2×104/well in migration assay or
5×104/well in invasion assay) were plated in the upper
chamber. DMEM containing 10% FBS was added to the lower chamber.
All samples were incubated for 24 or 36 h at 37°C for the cell
migration or cell invasion assay, respectively. In the lower
surface of the chamber membrane, migrating or invasive cells were
fixed with 4% formaldehyde for 15 min at room temperature and
stained with 0.1% crystal violet solution (Sigma-Aldrich; Merck
KGaA). All experiments were performed in triplicate, and numbers of
migrated or invaded cells in five random fields were visualized
under a light microscope.
Ethynyl-2′-deoxyuridine (EdU) cell
proliferation assay
Detection of EdU incorporation into the DNA was
performed with an EdU Apollo DNA in vitro kit (Guangzhou
RiboBio Co., Ltd.). According to the manufacturer's instructions,
50 µM EdU was used for incorporation. The established cells were
seeded in 96-well plates and incubated overnight at 37°C. The
supernatant was removed by aspiration, and the attached cells were
fixed with 100 µl fixing buffer (4% polyformaldehyde in PBS) for 30
min at room temperature. Following incubation with 2 mg/ml glycine
for 10 min at room temperature, the cells were washed with 1X PBS.
The cells were treated with 100 µl/well permeabilization buffer (1X
PBS containing 0.5% Triton X-100) for 10 min at room temperature
and incubated with 100 µl 1X Apollo solution for 30 min at room
temperature in the dark. Subsequently, cells were incubated with
100 µl 1X Hoechst 33342 solution for 30 min at room temperature in
the dark. Finally, the cells were observed under a fluorescence
microscope.
Colony formation assay
The established HeLa cells and MS751 cells were
plated in 6-well plates at the density of 1,000 cells per well and
cultured in DMEM supplemented with 10% FBS under the condition of
5% CO2 at 37°C for 10 days to form colonies consisting
of ≥50 cells. Subsequently, 100% methanol was used to fix cells for
30 min at room temperature and 0.1% crystal violet solution was
used to stain the cells for 20 min at room temperature. The number
of colonies was calculated using ImageJ (version no. 1.53)
(20). All experiments were
performed in triplicate, and representative results are shown.
Statistical analysis
Statistical analysis was performed using R software
(version: 4.0.2) (21). Univariate
Cox regression analysis was performed to evaluate survival
according to PLAC1 expression and clinicopathological parameters,
with median PLAC1 expression used as the boundary. Differences in
overall survival between the ‘high-PLAC1’ and ‘low-PLAC1’ groups
(based on median expression value of PLAC1) were analyzed via the
Kaplan-Meier method, with the P-value computed using the ‘survival’
package in R software (22).
Multivariate Cox analysis was performed to evaluate various
parameters associated with the hazard ratio, including age, BMI,
number of pregnancies, histology, clinical stage and PLAC1
expression using the ‘survival’ package in R software. In
multivariate Cox analysis, PLAC1 expression was converted into a
categorical variable based on the median value to interpret
relative risk of PLAC1 expression in CC. Unpaired Student's t,
χ2 or Fisher's exact test was performed to compare
variables between groups. For multiple comparisons, one-way ANOVA
and Tukey's test were used. The Shapiro-Wilk test was performed for
the assessment of data normality. Non-normally distributed data
were analyzed using Mann-Whitney U test. Normally distributed data
were presented as the mean ± SEM and non-normally distributed data
are presented as the median with quartile values (Q1, Q3).
P<0.05 was considered to indicate a statistically significant
difference.
Results
PLAC1 expression and its association
with clinicopathological variables
A total of 304 primary CC cases from TCGA database,
including both clinical information and RNA-seq data, were
analyzed. The median mRNA expression was 37.57 (range, 0–1059.43)
for PLAC1. The patient characteristics of TCGA cohort are
presented in Table I. The median
age of the patients with CC was 46 years (age range, 20–88 years).
In total, 28% of the patients had a history of smoking. Clinical
stages I, II, III and IV comprised 53, 23, 15 and 7% of the cases
(297/304), respectively. The pathological types included cervical
squamous cell carcinoma, cervical adenocarcinoma, cervical
endocervical mucinous adenocarcinoma and cervical adenosquamous
carcinoma, accounting for 83%, 10%, 5% and 2% of the cases,
respectively. The median follow-up time for surviving patients with
CC was 21.0 months (follow-up time range, 0–210.51 months).
 | Table I.Patient characteristics of The Cancer
Genome Atlas cohort. |
Table I.
Patient characteristics of The Cancer
Genome Atlas cohort.
|
|
| PLAC1 expression
level |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Total | Low | High | χ2 | P-value |
|---|
| Age at diagnosis,
years |
|
|
| 0.000 | >0.999 |
|
<46 | 145 | 72 | 73 |
|
|
|
≥46 | 159 | 80 | 79 |
|
|
| BMI |
|
|
| 0.096 | 0.757 |
|
<24 | 81 | 42 | 39 |
|
|
|
≥24 | 178 | 87 | 91 |
|
|
| Menopausal
status |
|
|
| 0.514 | 0.774 |
|
Premenopausal | 124 | 62 | 62 |
|
|
|
Perimenopausal | 28 | 13 | 15 |
|
|
|
Postmenopausal | 82 | 44 | 38 |
|
|
| Pregnancies count
total |
|
|
| 5.188 | 0.023 |
|
<5 | 191 | 105 | 86 |
|
|
| ≥5 | 73 | 28 | 45 |
|
|
| Pregnancies count
live birth |
|
|
| 4.760 | 0.029 |
|
<5 | 220 | 116 | 104 |
|
|
| ≥5 | 40 | 13 | 27 |
|
|
| Smoking status |
|
|
| 0.405 | 0.524 |
|
Yes | 86 | 46 | 40 |
|
|
| No | 218 | 106 | 112 |
|
|
| Smoking years |
|
|
| 0.000 | 1.000 |
|
<19 | 43 | 23 | 20 |
|
|
|
≥19 | 43 | 23 | 20 |
|
|
| Smoking pack
years |
|
|
| 2.616 | 0.106 |
|
<10 | 32 | 13 | 19 |
|
|
|
≥10 | 54 | 33 | 21 |
|
|
| ECOG score |
|
|
| – | 0.693a |
| 0 | 79 | 40 | 39 |
|
|
| 1 | 83 | 37 | 46 |
|
|
| 2 | 9 | 5 | 4 |
|
|
| 4 | 1 | – | 1 |
|
|
| Histology |
|
|
| – | 0.250a |
|
Cervical squamous cell
carcinoma | 252 | 121 | 131 |
|
|
|
Cervical adenocarcinoma | 30 | 20 | 10 |
|
|
|
Cervical endocervical mucinous
adenocarcinoma | 17 | 8 | 9 |
|
|
|
Cervical adenosquamous
carcinoma | 5 | 3 | 2 |
|
|
| HPV types
positive |
|
|
| – | 0.580a |
|
HPV16 | 12 | 5 | 7 |
|
|
|
HPV18 | 3 | 1 | 2 |
|
|
|
Others | 7 | 1 | 6 |
|
|
| Clinical stage |
|
|
| 1.709 | 0.635 |
| I | 161 | 85 | 76 |
|
|
| II | 69 | 32 | 37 |
|
|
|
III | 45 | 21 | 24 |
|
|
| IV | 22 | 13 | 9 |
|
|
| AJCC tumor
pathological score |
|
|
| – | 0.553a |
|
TX | 17 | 6 | 11 |
|
|
|
Tis | 1 | 0 | 1 |
|
|
|
T1 | 140 | 72 | 68 |
|
|
|
T2 | 71 | 40 | 31 |
|
|
|
T3 | 20 | 9 | 11 |
|
|
|
T4 | 10 | 6 | 4 |
|
|
| AJCC metastasis
pathological score |
|
|
| 7.167 | 0.028 |
|
MX | 128 | 56 | 72 |
|
|
|
M0 | 115 | 67 | 48 |
|
|
|
M1 | 11 | 8 | 3 |
|
|
| AJCC nodes
pathological score |
|
|
| 3.764 | 0.152 |
|
NX | 66 | 29 | 37 |
|
|
|
N0 | 133 | 76 | 57 |
|
|
|
N1 | 60 | 28 | 32 |
|
|
The expression levels of PLAC1 in CC tissues and
normal cervix tissues were subsequently compared, and the result
revealed that PLAC1 was highly expressed in CC tissues (Fig. 1A-E). The expression of PLAC1 was
significantly higher in all pathological types of cervical cancer
than that in normal cervical tissue. Consistent with this, higher
PLAC1 was also observed in cervical squamous cell carcinoma
samples. However, the increase of PLAC1 expression was not
statistically significance in other pathological types of cervical
cancer, including endocervical mucinous adenocarcinoma, cervical
adenosquamous carcinoma and cervical adenocarcinoma. According to
the median expression levels of PLAC1 (37.57), cases were divided
into ‘high-PLAC1’ and ‘low-PLAC1’ expression groups. χ2
or Fisher's exact test results showed that the two groups had
statistically significant differences in the following
characteristics: Number of pregnancies, number of live births and
American Joint Committee on Cancer metastasis pathological score
(Table I).
 | Figure 1.PLAC1 expression is significantly
upregulated in CC and high PLAC1 expression is associated with poor
survival. (A) PLAC1 expression was upregulated in CC tissues
compared with in normal cervical tissues, as revealed using
TCGA-CESC dataset. (B) PLAC1 expression was upregulated in cervical
squamous cell carcinoma tissues compared with that in normal
cervical tissues, as revealed using TCGA-CESC dataset. In other
pathological types of CC, (C) endocervical mucinous adenocarcinoma,
(D) adenosquamous carcinoma and (E) cervical adenocarcinoma, PLAC1
expression was upregulated, but did not reach a statistically
significant level. Data are presented as the mean ± SEM.
*P<0.05, **P<0.01. (F) Kaplan-Meier survival analysis showed
that CC cases with PLAC1-high expression had a worse prognosis than
those with PLAC1-low expression (P=0.0296). CC, cervical cancer;
TCGA-CESC, The Cancer Genome Atlas Cervical Squamous Cell Carcinoma
and Endocervical Adenocarcinoma; PLAC1, placenta-specific protein
1; ns, not significant; TPM, transcripts per million. |
GSEA based on PLAC1 expression
Based on the median expression value of PLAC1, all
subjects were split into two groups, ‘high-PLAC1’ and ‘low-PLAC1’
expression groups, and GSEA was performed. Enrichment results in
the ‘high-PLAC1’ phenotype group showed that 33/50 gene sets were
upregulated, 24 gene sets were significant at a false discovery
rate (FDR) <25% and 15 gene sets at FDR <5% (Table II), 14 gene sets were significantly
enriched at nominal P-value <1% and 16 gene sets were
significantly enriched at nominal P-value <5% (data not shown).
The normalized enrichment score of three gene sets was >2.0,
including ‘mTOR complex (mTORC)1 signaling’, ‘interferon α
response’ and ‘hypoxia’ (Fig.
2A-C). Enrichment results in the ‘low’ phenotype showed that
17/50 gene sets were upregulated and no gene sets were
significantly enriched at a FDR <25% (data not shown). The heat
map of the top 50 features for each phenotype is shown in Fig. 2D.
 | Table II.Gene sets enriched in ‘high-PLAC1’
phenotype (FDR<0.05). |
Table II.
Gene sets enriched in ‘high-PLAC1’
phenotype (FDR<0.05).
| Gene set | Size | NES | P-value | FDR | Rank at max |
|---|
| MTORC1
SIGNALING | 132 | 2.485 | <0.001 | <0.001 | 7,108 |
| INTERFERON ALPHA
RESPONSE | 48 | 2.381 | <0.001 | <0.001 | 7,782 |
| HYPOXIA | 127 | 2.130 | <0.001 | 0.001 | 4,222 |
| MYC TARGETS V1 | 135 | 1.993 | <0.001 | 0.002 | 7,970 |
| E2F TARGETS | 128 | 1.983 | <0.001 | 0.002 | 8,358 |
| GLYCOLYSIS | 130 | 1.968 | <0.001 | 0.002 | 5,077 |
| P53 PATHWAY | 126 | 1.949 | <0.001 | 0.002 | 5,817 |
| TNFA SIGNALING VIA
NFKB | 121 | 1.919 | <0.001 | 0.001 | 7,875 |
| PI3K AKT MTOR
SIGNALING | 66 | 1.899 | <0.001 | 0.001 | 7,224 |
| MYC TARGETS V2 | 32 | 1.832 | <0.001 | 0.004 | 8,037 |
| REACTIVE OXIGEN
SPECIES PATHWAY | 33 | 1.800 | 0.009 | 0.004 | 6,849 |
| INTERFERON GAMMA
RESPONSE | 124 | 1.783 | <0.001 | 0.005 | 7,379 |
| APOPTOSIS | 100 | 1.537 | <0.001 | 0.026 | 7,310 |
| ESTROGEN RESPONSE
LATE | 117 | 1.509 | <0.001 | 0.029 | 4,559 |
| CHOLESTEROL
HOMEOSTASIS | 49 | 1.474 | 0.021 | 0.038 | 6,553 |
PLAC1 expression in CC tissues and
cell lines
The expression levels of PLAC1 in CC tissues, HSILs
and normal exocervix were assessed via IHC analysis (Fig. 3A-F). A tissue microarray, comprising
101 CC tissues, 35 paired or unpaired non-cancer cervical tissues
and 18 HSILs, was analyzed. CC samples and HSILs exhibited
significantly higher expression levels of PLAC1 compared with those
in normal tissues (Fig. 3G and H).
Moreover, PLAC1 expression was detected in 100% (20/20) of CC
samples, compared with in 15% (3/20) of normal tissue (Data not
shown). The staining intensity of PLAC1 in CC tissues was more
intense than that in normal tissues. A similar trend was observed
in the HSILs (Data not shown). To the best of our knowledge, PLAC1
is expressed at low levels in normal cervical cells (23). Protein expression levels of PLAC1 in
CC lines, including CaSki, MS751, C-33A and HeLa, were notably
upregulated (Fig. 4A). Among these,
the expression of PLAC1 in MS751 cells was the highest and the
expression of PLAC1 in HeLa cells was relatively low. Therefore,
MS751 and HeLa cells were selected for further study.
 | Figure 4.PLAC1 promotes proliferation and
motility in CC. (A) Expression of PLAC1 in cervical cancer cell
lines CaSki, MS751, C-33A and HeLa. (B) PLAC1 was upregulated in
PLAC1-overexpressing HeLa cells and downregulated in PLAC1-silenced
MS751 cells. (C) Cell proliferation determined using colony
formation assay. (D and E) Cell proliferation measured using EdU
incorporation assay. Magnification, ×200. (F and G) Cell motility
examined using wound healing assay. Magnification, ×40. (H and I)
Cell motility detected using Transwell assay. Magnification, ×200.
Data are presented as the mean ± SEM. *P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. PLAC1, placenta-specific protein 1;
sh, short hairpin; EdU, ethynyl-2′-deoxyuridine. |
High PLAC1 expression is associated
with poor survival in CC
A total of 304 CC cases with PLAC1 expression data
and clinical information across all patient characteristics were
derived from TCGA. Three of these cases contained matched
paracancerous samples. A total of three normal cervical samples
were excluded from the survival analysis. It was demonstrated that
high expression of PLAC1 was a risk factor for the poor prognosis
of patients with CC (Fig. 1F).
Similarly, the clinical data contained in the tissue microarray
also suggested that patients with high expression of PLAC1 had a
poor survival (Fig. 3I).
Univariate Cox analysis suggested that four
characteristics were associated with prognosis, including BMI,
clinical stage, American Joint Committee on Cancer tumor
pathological score and PLAC1 expression (as a continuous
independent variable) (Table
III). Multivariate Cox analysis suggested age (hazard
ratio=1.02; P=0.047), clinical stage (hazard ratio=1.68;
P<0.001) and PLAC1 expression (hazard ratio=1.92; P=0.032) were
associated with the prognosis of patients with CC (Table III). Similarly, PLAC1 expression
as a binary variable (according to median expression value of
37.57) was associated with poor prognostic clinicopathological
characteristics (Table III). Age,
BMI, number of pregnancies, histology, clinical stage and PLAC1
expression were incorporated in the multivariate Cox model. All six
variables were consistent with the proportional hazard assumption
(P-value of each component was <0.05, and Global Schoenfeld test
P>0.05).
 | Table III.Overall survival and associations
with clinicopathological characteristics determined using Cox
regression analysis. |
Table III.
Overall survival and associations
with clinicopathological characteristics determined using Cox
regression analysis.
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age at diagnosis,
years | 1.02 | 1.00–1.03 | 0.078 | 1.02 | 1.00–1.05 | 0.047 |
| BMI | 0.95 | 0.91–1.00 | 0.033 | 0.99 | 0.94–1.03 | 0.533 |
| Pregnancies count
total | 1.03 | 0.94–1.12 | 0.594 | 0.98 | 0.88–1.09 | 0.692 |
| Pregnancies count
live birth | 1.05 | 0.94–1.17 | 0.407 | – | – | – |
| Menopausal
status | 1.08 | 0.81–1.43 | 0.612 | – | – | – |
| Smoking years | 0.98 | 0.92–1.05 | 0.635 | – | – | – |
| Smoking pack
years | 1.01 | 0.98–1.04 | 0.565 | – | – | – |
| ECOG score | 1.32 | 0.76–2.30 | 0.329 | – | – | – |
| Histology | 1.11 | 0.75–1.64 | 0.603 | 1.25 | 0.81–1.93 | 0.308 |
| HPV types
positive | 1.37 | 0.60–3.12 | 0.455 | – | – | – |
| Clinical stage | 1.47 | 1.18–1.83 | <0.001 | 1.68 | 1.27–2.23 | <0.001 |
| AJCC tumor
pathological score | 1.34 | 1.00–1.78 | 0.047 | – | – | – |
| AJCC metastasis
pathological score | 0.77 | 0.47–1.25 | 0.295 | – | – | – |
| AJCC nodes
pathological score | 0.78 | 0.52–1.17 | 0.233 | – | – | – |
| PLAC1
expression | 1.70 | 1.05–2.74 | 0.031a | 1.92 | 1.06–3.51 | 0.032b |
PLAC1 promotes cell proliferation,
migration and invasion in CC
Since the expression of PLAC1 is associated with
metastasis and cell proliferation in CC, the impact of
overexpression or knockdown of PLAC1 on proliferation and
invasion in CC cell lines was examined. PLAC1-overexpressing HeLa
cells and PLAC1-silenced MS751 cells with shRNA2 (Fig. 4B) were employed for functional
studies. Colony formation and EdU incorporation assays were used to
investigate the effect of PLAC1 on cell proliferation. The results
demonstrated that overexpression of PLAC1 significantly promoted
cell proliferation, whereas PLAC1 silencing inhibited proliferation
(Fig. 4C-E). Furthermore, as
confirmed by wound healing assay (Fig.
4F and G), the motility of cells was significantly increased in
PLAC1-overexpressing group and decreased in PLAC1-silenced group
compared with that in control group. Transwell migration and
invasion assays were performed to estimate the effect of PLAC1 on
cell migration and invasion. The results revealed that
overexpression of PLAC1 facilitated cell migration and invasion
(Fig. 4H), whereas PLAC1 silencing
in MS751 cells suppressed migration and invasion (Fig. 4I). These results suggested that
PLAC1 may exert a critical role in regulating cell proliferation
and invasion in CC.
PLAC1 enhances invasion via the
epithelial-mesenchymal transition (EMT) signaling pathway
Since EMT is an essential process in the
pathogenesis of metastasis in tumor cells (24), studies were subsequently conducted
to investigate whether EMT was a causative factor for PLAC1-induced
changes in cell motility. Compared with control cells, the
expression of E-cadherin, one of most common epithelial markers,
was decreased in the PLAC1-overexpressing HeLa cells, accompanied
by an increase in the expression of the mesenchymal markers, Slug,
MMP2 and Vimentin (Fig. 5A and
B).
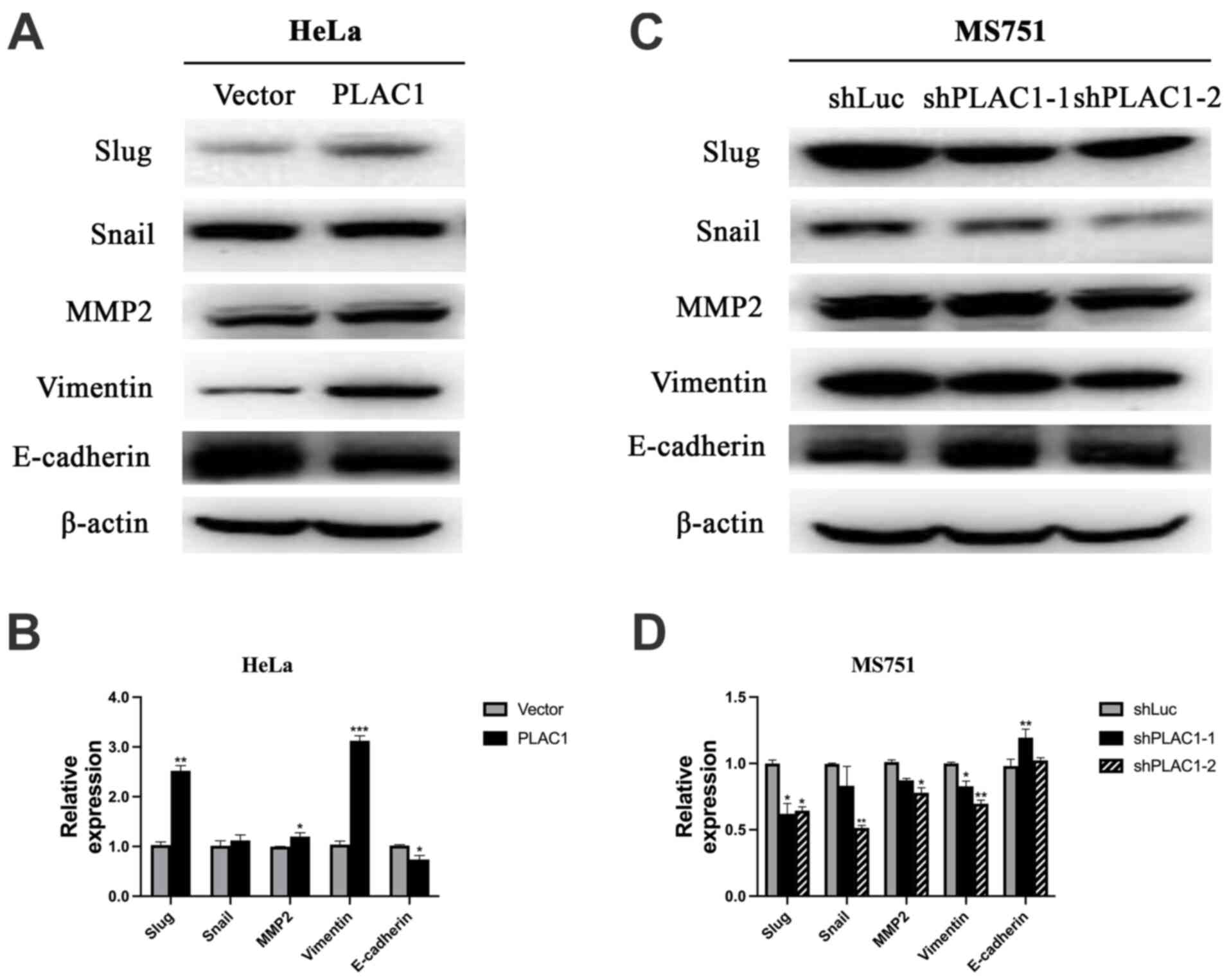 | Figure 5.Overexpression of PLAC1 significantly
increases the expression of EMT-associated protein and knockdown of
PLAC1 significantly decreases the expression of EMT-associated
protein. (A and B) Overexpression of PLAC1 in HeLa cells
significantly increased the expression of Slug, MMP2 and Vimentin,
but decreased the expression of E-cadherin, as detected via western
blot analysis. (C and D) Knockdown of PLAC1 in MS751 cells
significantly decreased the expression of Slug, Snail, MMP2, and
Vimentin but increased the expression of E-cadherin, as detected
via western blot analysis. The results were normalized to the
expression of β-actin. *P<0.05, **P<0.01, ***P<0.001.
PLAC1, placenta-specific protein 1; sh, short hairpin; EMT,
epithelial-mesenchymal transition. |
As Slug and Snail are transcriptional regulators
that have been substantiated as crucial EMT regulators through
inhibiting the epithelial phenotype-related genes and activating
mesenchymal markers (25), we then
evaluated whether E-cadherin, Snail and Slug were affected in the
MS751 cells infected with PLAC1-shRNA. The results showed that
silencing of PLAC1 inhibited the expression of Slug and Snail, but
upregulated E-cadherin (Fig. 5C and
D), suggesting that decreased expression of PLAC1 may increase
the expression of E-cadherin by suppressing Slug and Snail.
Mesenchymal marker Vimentin was downregulated in PLAC1-silenced
cells. Taken together, these findings indicated that PLAC1 may
serve a crucial role in carcinogenesis by activating the
EMT-related signaling pathways.
Discussion
The present study analyzed the clinical information
and RNA-seq data from 304 patients with CC in TCGA database.
Verification studies using IHC confirmed that the expression levels
of PLAC1 were significantly increased in CC tissues and HSILs. It
was identified that high PLAC1 expression indicated a poor
prognosis. Furthermore, multivariate survival analysis revealed
that high PLAC1 expression was a poor prognostic parameter for
overall survival. Functional studies demonstrated that
overexpression of PLAC1 promoted cell proliferation and motility.
Collectively, these data indicated that PLAC1 may be a prognostic
marker and therapeutic target in CC.
Previous studies examining PLAC1 in other tumor
types suggested that higher relative expression of PLAC1 had an
association with poor survival (26,27).
Inhibition of PLAC1 in breast cancer cells markedly affected
motility, migration and invasion, and induced a G1-S
cell cycle block that almost completely suppressed proliferation
(10). Additionally, patients with
gastric adenocarcinoma exhibiting relatively low PLAC1 expression
had a longer survival time compared with those exhibiting higher
PLAC1 expression (26).
Furthermore, numerous studies related to hepatocellular carcinoma
(11), colorectal cancer (28), ovarian cancer (29), uterine cancer (30) and prostate cancer (31) have reported that PLAC1 expression
was positively correlated with clinical parameters, including
clinical stage, grade and survival outcome. All these previous
reports showed similar results as those obtained in the present
study, as high PLAC1 expression levels indicated poor prognosis in
CC.
Based on the GSEA, 33 gene sets exhibited
significant differences in the ‘high-PLAC1’ phenotype group;
however, no gene set enrichment exhibited significant differences
in the ‘low-PLAC1’ phenotype group. ‘mTORC1 signaling’, ‘interferon
α response’ and ‘hypoxia’ were differentially enriched in the
‘high-PLAC1’ expression phenotype. mTORC1 is sensitive to
rapamycin, and is typically activated by numerous stimuli and
essential signaling pathways, such as PI3K, MAPK and AMP-activated
protein kinase, to control cell proliferation and survival
(32). mTORC1 pathways have been
reported be involved in various types of cancer (33). Kremer et al (34) revealed that activation of mTOR
signaling led to the survival of CC cells. Moreover, microRNA-99
has been reported to inhibit CC cell proliferation and invasion by
targeting the mTOR signaling pathway (35), whereas AKT inhibitors may promote
cell death in CC by disrupting mTOR signaling and glucose uptake
(36). Improving the anti-tumoral
interferon α response can be used to treat hematological
malignancies and solid tumors in cancer immunotherapy (37,38).
Hypoxia, an important regulator in tumor growth (39), has been shown to be associated with
poor survival and radiotherapy resistance, and numerous molecules
in the hypoxia-response pathway are candidates as therapeutic
targets (40,41). Furthermore, Pilch et al
(42) reported that hypoxia
stimulated the expression of tumor angiogenesis factors in CC cells
and derived fibroblasts. Therefore, most gene sets enriched in the
high expression group have previously been associated with tumor
pathogenesis.
To investigate the carcinogenic characteristics of
PLAC1 in the tumor process, we overexpressed PLAC1 in HeLa cells
and used PLAC1-specific shRNA to knockdown PLAC1 in MS751 cells.
Subsequently, the effects of PLAC1 on cell proliferation, migration
and invasion were assessed. The present results suggested that cell
proliferation was inhibited by PLAC1 silencing in MS751 cells.
Furthermore, overexpression of PLAC1 in HeLa cells increased cell
proliferation, which indicated a role for PLAC1 in enhancing tumor
cell proliferation. Furthermore, the current results indicated that
PLAC1 facilitated migration and invasion in tumor cells, as shown
by the fact that PLAC1-overexpressing HeLa cells had a stronger
cell motility, and PLAC1 silencing inhibited migration and invasion
in MS751 cells, as determined using in vitro wound healing
and Transwell assays.
The present study also investigated whether the
changes in tumor cell migration and invasion were due to EMT. EMT
is a crucial physiological process affecting the invasion and
migration of cancer cells (43).
Epithelial cells gain mesenchymal characteristics via
dedifferentiation processes that result in the loss of epithelial
characteristics. In this process, downregulation of E-cadherin, an
epithelial cell adhesion marker, is considered to be an essential
step (44). Vimentin, a mesenchymal
marker, and the most well-studied inhibitors of E-cadherin, such as
Snail, Slug, Twist, ZEB-1 and ZEB-2, which act by binding to
E-boxes of E-cadherin promoter and inhibiting its transcription,
actively participate in the EMT process (45). In the present study, PLAC1 silencing
enhanced E-cadherin expression, along with a noticeable reduction
in vimentin and MMP2 expression. Moreover, the expression levels of
Slug and Snail were downregulated, which supported the
aforementioned findings. Taken together, these results suggested
that PLAC1 served an oncogenic role via EMT, thus promoting
metastasis in CC. Moreover, it was confirmed that PLAC1 may be a
candidate oncogene for CC. Future studies will be conducted to
investigate the underlying molecular mechanisms by which PLAC1
regulates EMT in CC.
In conclusion, the present study demonstrated that
PLAC1 was a key regulator for CC progression by promoting cell
proliferation, migration and invasion via activation of the EMT
process, and high expression of PLAC1 was revealed to be a marker
of poor prognosis in patients with CC. RNA-seq data from CC
samples, as reported by TCGA, were analyzed; however, matched
protein data were not available to support the elevation of PLAC1
transcriptome expression. Therefore, a tissue microarray was used
to validate the higher expression levels of PLAC1 in CC tissues
compared with in non-tumor tissues. However, the expression levels
of PLAC1 were not verified in the serum of patients with CC. Thus,
further studies are required to evaluate the prognostic value and
feasibility of serum PLAC1 as a predictive marker in a larger,
prospective cohort of patients with CC, and to elucidate the
specific molecular mechanisms of PLAC1 in CC.
Acknowledgements
The tissue microarray experiment was completed by
Shanghai Outdo Biotech Company.
Funding
The project was supported by Health Profession
Clinical Research Funds of Shanghai Municipal Health Commission
(grant no. 20194Y0259), Natural Science Research Funds of Minhang
District, Shanghai (grant no. 2019MHZ038) and Key Medical Specialty
funded by Shanghai Fifth People's Hospital, Fudan University (grant
no. 2020WYZDZK13)
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The results shown here are in part based upon data
generated by TCGA Research Network: https://www.cancer.gov/tcga
Authors' contributions
LY and YC conceived and designed the project. RM,
LZ, CS and RC completed the research work. CS and RC collected
data, performed statistical analyses and drafted the manuscript.
All authors read and approved the final manuscript. RM and RC
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Use of the tissue microarray (HUteS154Su01) in this
study has been approved by the Ethics Committee of the Shanghai
Outdo Biotech Company (approval no. YB M-05-01). The tissue
microarray experiment was completed by Shanghai Outdo Biotech
Company.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay JEM, Lam F, Colombet M, Mery L,
Piñeros M, Znaora, Soerjomataram I and Bray F: Global Cancer
Observatory: Cancer Today. International Agency for Research on
Cancer; Lyon, France: 2020
|
|
2
|
Arbyn M, Weiderpass E, Bruni L, de Sanjosé
S, Saraiya M, Ferlay J and Bray F: Estimates of incidence and
mortality of cervical cancer in 2018: A worldwide analysis. Lancet
Glob Health. 8:e191–e203. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang R, Pan W, Jin L, Huang W, Li Y, Wu D,
Gao C, Ma D and Liao S: Human papillomavirus vaccine against
cervical cancer: Opportunity and challenge. Cancer Lett.
471:88–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hermyt E, Zmarzły N, Grabarek B,
Kruszniewska-Rajs C, Gola J, Jęda-Golonka A, Szczepanek K, Mazurek
U and Witek A: Interplay between miRNAs and Genes Associated with
Cell Proliferation in Endometrial Cancer. Int J Mol Sci.
20:E60112019. View Article : Google Scholar
|
|
5
|
Scanlan MJ, Gure AO, Jungbluth AA, Old LJ
and Chen YT: Cancer/testis antigens: An expanding family of targets
for cancer immunotherapy. Immunol Rev. 188:22–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cocchia M, Huber R, Pantano S, Chen EY, Ma
P, Forabosco A, Ko MS and Schlessinger D: PLAC1, an Xq26 gene with
placenta-specific expression. Genomics. 68:305–312. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fant ME, Fuentes J, Kong X and Jackman S:
The nexus of prematurity, birth defects, and intrauterine growth
restriction: A role for plac1-regulated pathways. Front Pediatr.
2:82014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang X, Baddoo MC and Yin Q: The placental
specific gene, PLAC1, is induced by the Epstein-Barr virus and is
expressed in human tumor cells. Virol J. 11:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koslowski M, Sahin U, Mitnacht-Kraus R,
Seitz G, Huber C and Türeci O: A placenta-specific gene ectopically
activated in many human cancers is essentially involved in
malignant cell processes. Cancer Res. 67:9528–9534. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Devor EJ: Placenta-specific protein 1
(PLAC1) is a unique onco-fetal-placental protein and an
underappreciated therapeutic target in cancer(J). Integr Cancer Sci
Ther. 3:479–483. 2016. View Article : Google Scholar
|
|
11
|
Dong XY, Peng JR, Ye YJ, Chen HS, Zhang
LJ, Pang XW, Li Y, Zhang Y, Wang S, Fant ME, et al: Plac1 is a
tumor-specific antigen capable of eliciting spontaneous antibody
responses in human cancer patients. Int J Cancer. 122:2038–2043.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Pang XW, Liu FF, Dong XY, Wang HC,
Wang S, Zhang Y and Chen WF: PLAC1/CP1 gene expression and
autologous humoral immunity in gastric cancer patients. Beijing Da
Xue Xue Bao. 38:124–127. 2006.PubMed/NCBI
|
|
13
|
Devor EJ, Reyes HD, Gonzalez-Bosquet J,
Warrier A, Kenzie SA, Ibik NV, Miller MD, Schickling BM, Goodheart
MJ, Thiel KW, et al: Placenta-specific protein 1 expression in
human papillomavirus 16/18-positive cervical cancers is associated
with tumor histology. Int J Gynecol Cancer. 27:784–790. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jackman SM, Kong X and Fant ME: Plac1
(placenta-specific 1) is essential for normal placental and
embryonic development. Mol Reprod Dev. 79:564–572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Old LJ: Cancer is a somatic cell
pregnancy. Cancer Immun. 7:192007.PubMed/NCBI
|
|
16
|
Cheever MA, Allison JP, Ferris AS, Finn
OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL,
Weiner LM, et al: The prioritization of cancer antigens: A national
cancer institute pilot project for the acceleration of
translational research. Clin Cancer Res. 15:5323–5337. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
GDC Data User's Guide(EB/OL):
(2021.08.19), . https://docs.gdc.cancer.gov/Data/PDF/Data_UG.pdf
|
|
18
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The Molecular Signatures
Database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
R Core Team R, . A Language And
Environment For Statistical Computing. R Foundation For Statistical
Computing; Vienna: 2018
|
|
22
|
Goel MK, Khanna P and Kishore J:
Understanding survival analysis: Kaplan-Meier estimate. Int J
Ayurveda Res. 1:274–278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bradley SD, Talukder AH, Lai I, Davis R,
Alvarez H, Tiriac H, Zhang M, Chiu Y, Melendez B, Jackson KR, et
al: Vestigial-like 1 is a shared targetable cancer-placenta antigen
expressed by pancreatic and basal-like breast cancers. Nat Commun.
11:53322020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goossens S, Vandamme N, Van Vlierberghe P
and Berx G: EMT transcription factors in cancer development
re-evaluated: Beyond EMT and MET. Biochim Biophys Acta Rev Cancer.
1868:584–591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu F, Shen D, Kang X, Zhang C and Song Q:
New tumour antigen PLAC1/CP1, a potentially useful prognostic
marker and immunotherapy target for gastric adenocarcinoma. J Clin
Pathol. 68:913–916. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, Lin X, Di X, Chen Y, Zhao H and Wang
X: Oncogenic function of Plac1 on the proliferation and metastasis
in hepatocellular carcinoma cells. Oncol Rep. 37:465–473. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu FF, Dong XY, Pang XW, Xing Q, Wang HC,
Zhang HG, Li Y, Yin YH, Fant M, Ye YJ, et al: The specific immune
response to tumor antigen CP1 and its correlation with improved
survival in colon cancer patients. Gastroenterology. 134:998–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tchabo NE, Mhawech-Fauceglia P, Caballero
OL, Villella J, Beck AF, Miliotto AJ, Liao J, Andrews C, Lele S,
Old LJ, et al: Expression and serum immunoreactivity of
developmentally restricted differentiation antigens in epithelial
ovarian cancer. Cancer Immun. 9:62009.PubMed/NCBI
|
|
30
|
Devor EJ and Leslie KK: The oncoplacental
gene placenta-specific protein 1 is highly expressed in endometrial
tumors and cell lines. Obstet Gynecol Int. 2013:8078492013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ghods R, Ghahremani MH, Madjd Z, Asgari M,
Abolhasani M, Tavasoli S, Mahmoudi AR, Darzi M, Pasalar P,
Jeddi-Tehrani M, et al: High placenta-specific 1/low
prostate-specific antigen expression pattern in high-grade prostate
adenocarcinoma. Cancer Immunol Immunother. 63:1319–1327. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pópulo H, Lopes JM and Soares P: The mTOR
signalling pathway in human cancer. Int J Mol Sci. 13:1886–1918.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Laplante M and Sabatini DM: mTOR signaling
in growth control and disease. Cell. 149:274–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kremer CL, Klein RR, Mendelson J, Browne
W, Samadzedeh LK, Vanpatten K, Highstrom L, Pestano GA and Nagle
RB: Expression of mTOR signaling pathway markers in prostate cancer
progression. Prostate. 66:1203–1212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Chang L, Li Z, Gao Q, Cai D, Tian
Y, Zeng L and Li M: miR-99a and −99b inhibit cervical cancer cell
proliferation and invasion by targeting mTOR signaling pathway. Med
Oncol. 31:9342014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rashmi R, DeSelm C, Helms C, Bowcock A,
Rogers BE, Rader JL, Grigsby PW and Schwarz JK: AKT inhibitors
promote cell death in cervical cancer through disruption of mTOR
signaling and glucose uptake. PLoS One. 9:e929482014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kirkwood J: Cancer immunotherapy: The
interferon-alpha experience. Semin Oncol. 29:18–26. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kretzmann NA, Chiela E, Matte U, Marroni N
and Marroni CA: N-acetylcysteine improves antitumoural response of
Interferon alpha by NF-κB downregulation in liver cancer cells.
Comp Hepatol. 11:42012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nat Rev Cancer. 2:38–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Thomas GV, Tran C, Mellinghoff IK, Welsbie
DS, Chan E, Fueger B, Czernin J and Sawyers CL: Hypoxia-inducible
factor determines sensitivity to inhibitors of mTOR in kidney
cancer. Nat Med. 12:122–127. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wilson WR and Hay MP: Targeting hypoxia in
cancer therapy. Nat Rev Cancer. 11:393–410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pilch H, Schlenger K, Steiner E,
Brockerhoff P, Knapstein P and Vaupel P: Hypoxia-stimulated
expression of angiogenic growth factors in cervical cancer cells
and cervical cancer-derived fibroblasts. Int J Gynecol Cancer.
11:137–142. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aiello NM, Maddipati R, Norgard RJ, Balli
D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, et al:
EMT subtype influences epithelial plasticity and mode of cell
migration. Dev Cell. 45:681–695.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wong SHM, Fang CM, Chuah LH, Leong CO and
Ngai SC: E-cadherin: Its dysregulation in carcinogenesis and
clinical implications. Crit Rev Oncol Hematol. 121:11–22. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee JY and Kong G: Roles and epigenetic
regulation of epithelial-mesenchymal transition and its
transcription factors in cancer initiation and progression. Cell
Mol Life Sci. 73:4643–4660. 2016. View Article : Google Scholar : PubMed/NCBI
|



















