Introduction
Bladder cancer (BC) is one of the most common
malignancies in the genitourinary tract and has high morbidity and
mortality rates (1). According to
the statistics, >400,000 new cases of BC are diagnosed annually
and >2 million patients currently suffer from this disease
(2,3). Despite therapeutic interventions, such
as surgery, chemotherapy and radiotherapy, the 5-year survival rate
of BC remains low, which is a significant economic burden (4–6). Thus,
it is of great importance to identify novel biomarkers of BC and
investigate the genetic regulatory networks involved in BC
progression to improve patient prognosis and clinical outcomes.
Circular RNA (circRNA), a newly identified member of
the non-coding RNA family, is characterized by a covalently closed
loop without a 5′cap and 3′polyadenylated tail (7). Accumulating evidence has revealed that
circRNAs exert key effects on multiple biological and pathological
processes, such as proliferation, apoptosis, migration and
metastasis, indicating that they may be involved in the occurrence
and development of numerous types of disease, including cancer
(8–10). circRNAs serve as competitive
endogenous RNAs to modulate the activity of microRNAs (miRNAs/miRs)
and restore miRNA-mediated suppression of target genes (11,12).
circRNA-baculoviral IAP repeat-containing 6 (circ-BIRC6) is
generated by the back-splicing of the BIRC6 transcript (NM_016252)
and has been reported to be associated with oncogenesis (13,14).
Previous studies have reported that BIRC6 expression was
significantly upregulated in several human cancer types, such as
hepatocellular carcinoma, prostate cancer and lung cancer (15–17).
However, to the best of our knowledge, the role of circ-BIRC6 in BC
remains to be elucidated and is therefore worthy of further
investigations.
The present study analyzed the expression levels of
circ-BIRC6 in BC cell lines. Then, the effects of circ-BIRC6 on the
biological functions of BC cells and on tumor growth were explored
in vitro and in vivo. The potential underlying
mechanisms of the effects of circ-BIRC6 in BC were also
investigated. These results may offer novel insight into potential
biomarkers for diagnosing and predicting the prognosis of patients
with BC.
Materials and methods
Cell lines and culture
Several BC cell lines (SW780, T24, J82 and 5637) and
a human immortalized uroepithelium cell line (SV-HUC-1) were
purchased from the American Type Culture Collection. SV-HUC-1,
SW780 and T24 cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.), while J82 and 5637 cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). All media
were supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.). Cells were maintained in an incubator with 5% CO2
at 37°C.
Cell transfection
The pLVX lentiviral short hairpin RNA (shRNA/sh)
circ-BIRC6 (sh-circ-BIRC6#1 and sh-circ-BIRC6#2) and sh-negative
control (NC) vectors were synthesized by Shanghai GeneChem Co.,
Ltd. The miR-495-3p mimic (5′-AAACAAACATGGTGCACTTCTT-3′; 50 nM),
miR-495-3p inhibitor (5′-GCTTTATATGTGACGAAACAA-3′; 50 nM) and their
controls [mimic NC (5′-ATCGTGCTAGTCGATGCTAGCT-3′; 50 nM) and NC
inhibitor (5′-CGATCGCAGCGGTGCAGTGCG-3′; 50 nM)] were obtained from
Shanghai GenePharma Co., Ltd. T24 cells (2×105 cells per
well) were plated and incubated in 6-well plates for 24 h. The
transfection procedure was conducted at 37°C using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
transfection efficiency was evaluated using reverse
transcription-quantitative PCR (RT-qPCR) following 48 h of
transfection. The transfected cells were used for subsequent
experiments at 48 h after transfection.
Cell proliferation assay
Briefly, 5×103 T24 cells/well were plated
into 96-well culture plates and cultured at 37°C. Following 24, 48
or 72 h of incubation, 10 µl Cell Counting Kit-8 (CCK-8) reagent
(Shanghai Yeasen Biotechnology Co. Ltd.) was added into each well
and the plates were cultured for a further 4 h at 37°C. At 0 h, T24
cells would have just been seeded into a 96-well plate and would
not reflect the activity of adherent cells. Therefore, the 0 h time
point was omitted from the measurements. The optical density of
each sample was determined at a wavelength of 450 nm using a
microplate reader.
Colony formation assay
Following transfection, T24 cells were plated into
6-well plates (500 cells/well). After incubation at 37°C for 7–10
days, the cell colonies were washed with PBS, fixed with 4%
paraformaldehyde for 10 min at room temperature and stained with
0.1% crystal violet for 15 min at room temperature. Images of the
colonies were captured using a light microscope (Olympus
Corporation; magnification, ×10).
Transwell assay
The invasive ability of T24 cells was determined
using Transwell plates with 8-µm pore inserts; the membranes were
precoated with Matrigel (BD Biosciences) at 37°C for 6 h. A total
of 2×105 cells/well were resuspended in 200 µl
serum-free DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) and
plated into the upper chambers of the Transwell plates. The lower
chambers were filled with 600 µl DMEM supplemented with 10% FBS.
After incubation for 24 h at 37°C, the invasive cells were fixed
with 4% paraformaldehyde 20 min at 37°C and stained with 0.1%
crystal violet 10 min at 37°C for subsequent imaging and counting.
Stained cells were visualized using an inverted light microscope
(Olympus Corporation; magnification, ×100).
Wound healing assay
For the wound healing assay, T24 cells were plated
into 6-well plates (5×105 cells/well) and cultured until
they reached 90% confluence. Then, cells were incubated overnight
in serum-free DMEM prior to initiating the experiment. The cell
monolayer was subsequently scratched with a sterilized 100-µl
pipette tip. Following 24 h of incubation at 37°C, the migration of
cells was visualized using an inverted light microscope (Olympus
Corporation; magnification, ×100). Semi-quantitative analysis of
the wound healing area was performed using ImageJ software (version
1.52r; National Institutes of Health).
Dual luciferase reporter assay
To study the mechanism via which circ-BIRC6 promotes
BC progression, the potential miRNAs binding to circ-BIRC6 were
predicted using the StarBase database (starbase.sysu.edu.cn). The wild-type (WT) and mutant
(MUT) binding sites of miR-495-3p in the circ-BIRC6 or X-box
binding protein 1 (XBP1) 3′-untranslated region (UTR) were
sub-cloned into a pmirGLO dual luciferase reporter vector (Promega
Corporation) to construct circ-BIRC6 WT/MUT and XBP1-WT/MUT
vectors. Briefly, 5×103 T24 cells were seeded into
24-well plates and cultured for 24 h at 37°C. The plasmids were
then co-transfected with miR-495-3p mimic or mimic NC into T24
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Following 48 h of transfection, the
relative luciferase activities were analyzed using a Dual
Luciferase Reporter assay kit (Promega Corporation). Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Immunofluorescence assay
Transfected cells were fixed with 4%
paraformaldehyde at room temperature for 20 min, followed by the
addition of 0.05% Triton X-100 solution at room temperature for 10
min. After blocking with 3% BSA (Sigma-Aldrich; Merck KGaA) at 37°C
for 90 min, cells were incubated with a primary antibody against
Ki-67 (cat. no. 11882S; 1:1,000; Cell Signaling Technology, Inc.)
at 4°C overnight, followed by probing with the DyLight™
488-conjugated secondary antibody (cat. no. ab96899; 1:250; Abcam)
at 37°C for 1.5 h in the dark. The nuclei were stained with DAPI
(Roche Diagnostics) in the dark for 5 min at room temperature.
Images were captured under a fluorescence microscope (Olympus
Corporation; magnification, ×200) and the relative fluorescence
intensity was used to quantify the Ki-67-postive cells in three
randomly selected fields of view. The relative fluorescence
intensity was normalized to the average optical density of the
control group.
In vivo xenograft experiments
A total of 12 BALB/c nude mice (age, 5–6 weeks;
weight, ~18–22 g) were provided by the Shanghai Slac Animal
Laboratory, and the animal experiments were approved by the Animal
Care and Use Committee of The Third Xiangya Hospital, Central South
University (Changsha, China). Mice were housed under pathogen-free
conditions with a 12-h light/dark cycle, constant temperature of
25–27°C and constant humidity of 45–50%. Mice had free access to
food and water. Animals were randomly allocated into four groups (3
mice/group): sh-NC, sh-circ-BIRC6#1, sh-circ-BIRC6#1+NC inhibitor
and sh-circ-BIRC6#1+miR-495-3p inhibitor groups. T24 cells
transfected with lentiviral sh-circ-BIRC6#1, miR-495-3p or both
were subcutaneously injected into the flank of nude mice
(5×106 cells/mouse). Each mouse (7 weeks) was injected
with 30 µl cell suspension. Subsequently, the mice were maintained
for 3 weeks. Tumor volume was recorded every 3 days. Tumor volume
was calculated using the formula: Length × width2/2. At
the end of the experiments, all animals were sacrificed with an
intraperitoneal injection of 100 mg/kg sodium pentobarbital (body
weight). The tumor tissues were obtained for further investigation.
The maximum tumor diameter and volume observed in the study were 19
mm and 1,519 mm3, respectively.
Immunohistochemistry analysis
Tumor tissues were fixed in 10% buffered formalin 24
h at room temperature and then embedded in paraffin.
Paraffin-embedded tissue sections (4 µm thick) were deparaffinized
with xylene and then rehydrated with a graded descending series of
ethanol (100, 95 and 80%). Following antigen retrieval in boiling
water with a 10 mM citrate buffer, endogenous hydrogen peroxidase
activity was blocked by incubation with 10% hydrogen peroxide for
30 min at room temperature. The tissues samples were incubated with
a primary rabbit anti-Ki-67 antibody (cat. no. 9027T; 1:500; Cell
Signaling Technology, Inc.) at 4°C overnight, then with an
HRP-conjugated anti-rabbit IgG secondary antibody (cat. no.
ab181658; 1:1,000; Abcam) for 2 h at room temperature.
Subsequently, the tissue sections were treated with
3,3′-diaminobenzidine solution at room temperature for 3–5 min and
counterstained with hematoxylin at room temperature for 5 min.
Sections were viewed under an inverted light microscope (Olympus
Corporation; magnification, ×100).
RT-qPCR
Total RNA was extracted from cells or tumor tissues
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA using HiScript II (Vazyme
Biotech Co., Ltd.) according to the manufacturer's protocol. qPCR
to determine circ-BIRC6, miR-495-3p and XBP1 expression levels was
subsequently performed using SYBR Select Master mix (Tiangen
Biotech Co., Ltd.) on an ABI 7300 Real-Time PCR detection system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Initial denaturation at 95°C
for 10 min; followed by 40 cycles of denaturation at 95°C for 15
sec and annealing at 60°C for 1 min; and a final extension of 10
min at 72°C. The primers used were: circ-BIRC6 forward,
5′-TGAAAGGTTCTTGCACGCAT-3′ and reverse, 5′-GCTGGGGTTCGTTCACAATC-3′;
miR-495-3p forward, 5′-AAACAAACAUGGUGCACUUCUU-3′ and reverse,
5′-GAAGUGCACCAUGUUUGUUUUU-3′; XBP1s forward,
5′-ATGGATGCCCTGGTTGCTGAAGA-3′ and reverse,
5′-TGCACCTGCTGCGGACTCA-3′; XBP1u forward,
5′-AGCACTCAGACTACGTGCACCTCT-3′ and reverse,
5′-CCAGAATGCCCAACAGGATATCAG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′. U6 levels were used to normalize
miR-495-3p expression. GAPDH the was endogenous control for
circ-BIRC6. The relative expression levels were quantitatively
analyzed using the 2−ΔΔCq method (18).
Western blotting
Total protein was extracted from cells or tumor
tissues using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Protein concentration was quantified using a BCA
assay kit (Beyotime Institute of Biotechnology) and equal amounts
of protein (40 µg protein/lane) were separated via 10% SDS-PAGE.
The proteins were subsequently transferred onto PVDF membranes
(Thermo Fisher Scientific, Inc.) and blocked with 5% skimmed milk
at room temperature for 2 h. The membranes were then incubated with
specific primary antibodies at 4°C overnight. The following primary
antibodies were used: anti-E-cadherin (cat. no. 3195T; 1:1,000;
Cell Signaling Technology, Inc.), anti-N-cadherin (cat. no. 13116T;
1:1,000; Cell Signaling Technology, Inc.), anti-Vimentin (cat. no.
5741T; 1:1,000; Cell Signaling Technology, Inc.), anti-Snail (cat.
no. 3879T; 1:1,000; Cell Signaling Technology, Inc.), anti-XBP1s/u
(cat. no. bs-1668R; 1:1,000; Bioss) and anti-GAPDH (cat. no. 5174T;
1:1,000; Cell Signaling Technology, Inc.). Following the primary
antibody incubation, the membranes were incubated with an
HRP-conjugated secondary antibody for 1.5 h at room temperature.
Protein bands were visualized using an ECL reagent (MilliporeSigma)
and the bands were semi-quantified using ImageJ software (version
1.52r; National Institutes of Health). The gray value of the target
protein was normalized to that of GAPDH.
Statistical analysis
Data are presented as the mean ± SD and statistical
analysis was performed using GraphPad Prism 8.0 software (GraphPad
Software, Inc.). All experiments were repeated independently in
triplicate. One way ANOVA followed by a Tukey's post hoc test was
used to analyze the significant differences among multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
circ-BIRC6 expression levels are
upregulated in BC cell lines and circ-BIRC6 knockdown inhibits the
proliferation of BC cells
Firstly, the expression levels of circ-BIRC6 in
several BC cell lines (SW780, T24, J82 and 5637) and a human
immortalized uroepithelium cell line (SV-HUC-1) were detected using
RT-qPCR. As presented in Fig. 1A,
circ-BIRC6 expression was found to be upregulated in BC cell lines
compared with the SV-HUC-1 cells, especially in the T24 cells,
which were used for the subsequent experiments. Next, circ-BIRC6
was silenced in T24 cells via transfection with sh-circ-BIRC6#1 or
sh-circ-BIRC6#2. The results revealed that BIRC6 expression was
significantly downregulated in the sh-circ-BIRC6 groups compared
with the sh-NC group, and T24 cells transfected with
sh-circ-BIRC6#1 decreased the expression of circ-BIRC6 to a greater
extent than sh-circ-BIRC6#2 (Fig.
1B). Therefore, sh-circ-BIRC6#1 was selected for use in further
experiments. The results demonstrated that cell proliferation was
markedly reduced following circ-BIRC6 knockdown compared with the
sh-NC group (Fig. 1C). Moreover,
the colony formation assay results revealed that the proliferative
ability of T24 cells was markedly suppressed in the sh-circ-BIRC6#1
group compared with the sh-NC group (Fig. 1D). Simultaneously, the expression
levels of the proliferation-related protein, Ki-67, were
significantly decreased following circ-BIRC6 knockdown compared
with the sh-NC group (Fig. 1E).
These findings suggested that circ-BIRC6 may be upregulated in BC
cells and that circ-BIRC6 knockdown may decrease the proliferation
of BC cells.
 | Figure 1.circ-BIRC6 is highly expressed in BC
cell lines and circ-BIRC6 knockdown inhibits the proliferation of
BC cells. (A) BIRC6 expression was detected using RT-qPCR in
several BC cell lines (SW780, T24, J82 and 5637) and a human
immortalized uroepithelium cell line (SV-HUC-1). ***P<0.001 vs.
SV-HUC-1. (B) BIRC6 expression in T24 cells was measured using
RT-qPCR after transfection. (C) Cell proliferation was determined
using a Cell Counting Kit-8 kit. (D) Colony formation assay of T24
cells. (E) Ki-67 expression was assessed using immunofluorescence.
(F) Fluorescence intensity from part (E). Magnification, ×200.
*P<0.05, **P<0.01, ***P<0.001 vs. sh-NC. NC, negative
control; sh, short hairpin RNA; RT-qPCR, reverse
transcription-quantitative PCR; circ-BIRC6, circular RNA
baculoviral IAP repeat-containing 6; BC, bladder cancer. |
circ-BIRC6 knockdown prevents the
invasion, migration and epithelial-mesenchymal transition (EMT) of
BC cells
Subsequently, the effects of BIRC6 knockdown on cell
invasion were measured using a Transwell assay. As shown in
Fig. 2A and B, the invasive ability
of T24 cells was significantly inhibited in the sh-circ-BIRC6#1
group compared with the sh-NC group. Consistently, the results from
the wound healing assay showed the same trend with cell migration
(Fig. 2C and D). In addition, the
expression levels of EMT-related proteins were analyzed using
western blotting. The expression levels of E-cadherin were
significantly upregulated in the sh-circ-BIRC6#1 group compared
with the sh-NC group, while the expression levels of N-cadherin,
vimentin and snail family transcriptional repressor 1 (snail) were
downregulated in T24 cells following circ-BIRC6 knockdown (Fig. 2E). These results indicated that
circ-BIRC6 knockdown may block the invasion, migration and EMT of
BC cells.
 | Figure 2.circ-BIRC6 silencing suppresses the
invasion, migration and EMT of bladder cancer cells. (A and B)
Transwell assay was used to evaluate the effects of BIRC6 silencing
on the invasion of T24 cells. Magnification, ×100. (C and D) Cell
migration was evaluated using a wound healing assay. (E) Expression
levels of EMT-associated proteins, including E-cadherin,
N-cadherin, vimentin and snail, were determined using western
blotting. **P<0.01, ***P<0.001 vs. sh-NC. NC, negative
control; sh, short hairpin RNA; circ-BIRC6, circular RNA
baculoviral IAP repeat-containing 6; EMT, epithelial-mesenchymal
transition; snail, snail family transcriptional repressor 1. |
circ-BIRC6 acts as a sponge for
miR-495-3p
To study the mechanism via which circ-BIRC6 promotes
BC progression, the potential miRNAs binding to circ-BIRC6 were
predicted using the StarBase database (http://starbase.sysu.edu.cn), which identified
miR-495-3p as a candidate molecule. A putative binding site between
circ-BIRC6 and miR-495-3p was identified and is presented in
Fig. 3A. The expression levels of
miR-495-3p were significantly downregulated in BC cells compared
with SV-HUC-1 cells (Fig. 3B).
After successful transfection of T24 cells with the miR-495-3p
mimic (Fig. 3C), the relative
luciferase activity of the circ-BIRC6 WT vector was reduced
compared with the mimic NC group (Fig.
3C and D). Moreover, circ-BIRC6 knockdown significantly
upregulated the expression levels of miR-495-3p in T24 cells
compared with the sh-NC group (Fig.
3E), further suggesting that circ-BIRC6 may act as a sponge for
miR-495-3p.
 | Figure 3.circ-BIRC6 acts as a sponge for
miR-495-3p. (A) Binding region between circ-BIRC6 and miR-495-3p is
shown. (B) Expression levels of miR-495-3p in several BC cell lines
were determined using RT-qPCR. *P<0.05, ***P<0.001 vs.
SV-HUC-1. (C) RT-qPCR was employed to detect the expression levels
of miR-495-3p in T24 cells. ***P<0.001 vs. mimic NC. (D)
Relative luciferase activities were detected in T24 cells.
***P<0.001 vs. circ-BIRC6 WT + mimic NC. (E) miR-495-3p
expression was assessed using RT-qPCR in T24 cells following
circ-BIRC6 silencing. ***P<0.001 vs. sh-NC. NC, negative
control; sh, short hairpin RNA; circ-BIRC6, circular RNA
baculoviral IAP repeat-containing 6; miR, microRNA; RT-qPCR,
reverse transcription-quantitative PCR; Luc, luciferase; R,
Renilla; WT, wild-type; MUT, mutant. |
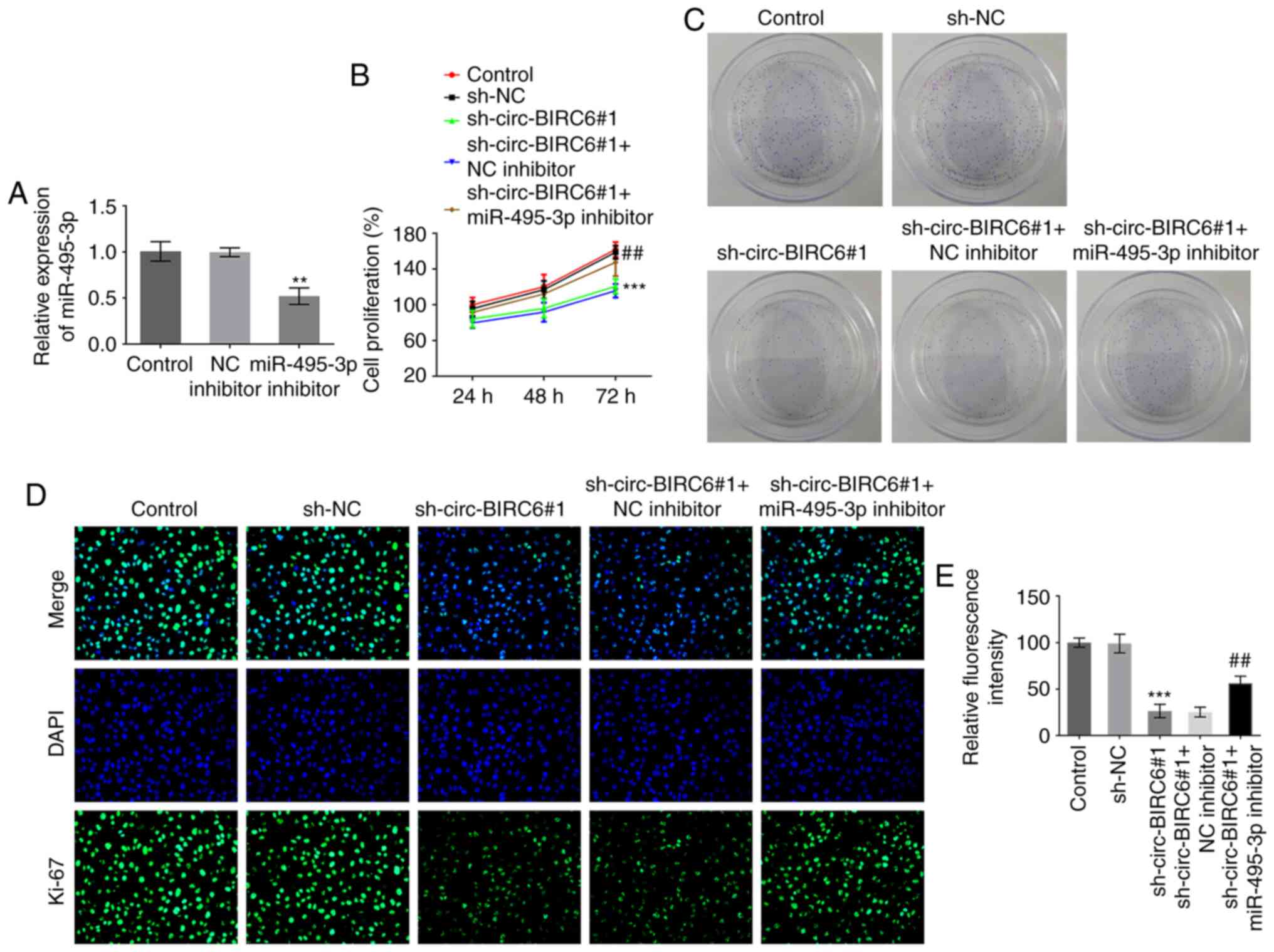
miR-495-3p inhibitor rescues the
effects induced by circ-BIRC6 knockdown in BC cells
To further elucidate the regulatory association
between circ-BIRC6 and miR-495-3p in BC, the mRNA expression levels
of miR-495-3p were successfully silenced via transfection with a
miR-495-3p inhibitor (Fig. 4A).
Co-transfection with miR-495-3p inhibitor and sh-circ-BIRC6#1
enhanced the proliferative ability of T24 cells compared with those
in cells transfected with sh-circ-BIRC6#1 alone, which was
accompanied by an increase in Ki-67 expression (Fig. 4B-E). Additionally, the invasive and
migratory abilities of cells were significantly elevated following
miR-495-3p silencing in T24 cells with sh-circ-BIRC6#1 transfection
compared with the sh-circ-BIRC6#1+NC inhibitor group (Fig. 5A-D). Moreover, co-transfection with
the miR-495-3p inhibitor and sh-circ-BIRC6#1 markedly downregulated
the expression levels of E-cadherin and upregulated those of
N-cadherin, vimentin and snail in T24 cells compared with cells
transfected with sh-circ-BIRC6#1 alone (Fig. 5E). These findings provide evidence
to suggest that circ-BIRC6 may regulate the progression of BC cells
by targeting miR-495-3p.
 | Figure 5.miR-495-3p inhibitor attenuates the
effects of circ-BIRC6 knockdown on the invasion, migration and EMT
of bladder cancer cells. T24 cell (A and B) invasion and (C and D)
migration were detected using Transwell and wound healing assays,
respectively. Magnification, ×100. (E) Western blotting was used to
determine the expression levels of EMT-related proteins, including
E-cadherin, N-cadherin, vimentin and snail. ***P<0.001 vs.
sh-NC; #P<0.05, ##P<0.01,
###P<0.001 vs. sh-circ-BIRC6#1 + NC inhibitor. NC,
negative control; sh, short hairpin RNA; circ-BIRC6, circular RNA
baculoviral IAP repeat-containing 6; miR, microRNA; EMT,
epithelial-mesenchymal transition. snail, snail family
transcriptional repressor 1. |
circ-BIRC6 regulates the expression
levels of XBP1 via modulating miR-495-3p
The bioinformatics results identified that XBP1 was
a potential target gene of miR-495-3p, which was confirmed using a
dual luciferase reporter assay (Fig. 6A
and B). Furthermore, compared with SV-HUC-1 cells, the mRNA and
protein expression levels of XBP1s/u were upregulated in BC cell
lines when compared with SV-HUC-1 cells (Fig. 6C and D). Notably, T24 cells
transfected with sh-circ-BIRC6#1 exhibited a decreased expression
level of XBP1s/u compared with the sh-NC group, which was blocked
by the miR-495-3p inhibitor (Fig. 6E
and F). Taken together, these data provide evidence to indicate
that circ-BIRC6 may regulate the expression level of XBP1 via
modulating miR-495-3p.
 | Figure 6.circ-BIRC6 regulates the expression
of XBP1 via miR-495-3p. (A) Binding region between XBP1 and
miR-495-3p is shown. (B) Relative luciferase activities were
detected in T24 cells. ***P<0.001 vs. XBP1 WT+ mimic NC. Protein
and mRNA expression levels of XBP1 were examined using (C) western
blotting and (D) RT-qPCR, respectively. **P<0.01, ***P<0.001
SV-HUC-1. (E) Western blotting and (F) RT-qPCR were employed to
measure the protein and mRNA expression levels, respectively, of
XBP1 after co-transfection with sh-circ-BIRC6#1 and miR-495-3p
inhibitor. ***P<0.001 vs. sh-NC; ##P<0.01 vs.
sh-circ-BIRC6#1 + NC inhibitor. NC, negative control; sh, short
hairpin RNA; circ-BIRC6, circular RNA baculoviral IAP
repeat-containing 6; miR, microRNA; XBP1, X-box binding protein 1;
WT, wild-type; MUT, mutant; Luc, luciferase; R, Renilla;
RT-qPCR, reverse transcription-quantitative PCR. |
circ-BIRC6 silencing suppresses the
growth of BC xenografts by regulating the miR-495-3p/XBP1 signaling
axis
Next, to analyze the role of circ-BIRC6 in BC, a
BALB/c nude mice model was established. The xenograft results
demonstrated that circ-BIRC6 knockdown notably inhibited tumor
weight and volume compared with the sh-NC group (Fig. 7A-D). By contrast, the miR-495-3p
inhibitor partially counteracted the inhibitory effects of
circ-BIRC6 knockdown on the growth of tumors. Additionally, notably
downregulated XBP1s/u expression was observed in tumor tissues of
xenograft mice in the sh-circ-BIRC6#1 group compared with the sh-NC
group, while miR-495-3p knockdown markedly reversed their
expression levels (Fig. 7E and F).
Moreover, immunohistochemical detection of Ki-67 staining
demonstrated that silencing of circ-BIRC6 inhibited Ki-67
expression in tumor tissues compared with the sh-NC group (Fig. 7G). It was also found that the tumor
tissues that were transplanted with T24 cells (silenced for
circ-BIRC6 and miR-495-3p expression) exhibited an elevated
expression of Ki-67 when compared with sh-circ-BIRC6#1+NC inhibitor
group. Moreover, it was identified that circ-BIRC6 knockdown
notably upregulated the expression levels of E-cadherin, but
downregulated those of N-cadherin, vimentin and snail compared with
the sh-NC group (Fig. 7H). The
results also indicated that the miR-495-3p inhibitor partially
abrogated the effect of circ-BIRC6 silencing on the expression
levels of the aforementioned proteins. These data suggested that
circ-BIRC6 knockdown may suppress the growth of BC cancer
xenografts by regulating the miR-495-3p/XBP1 signaling axis.
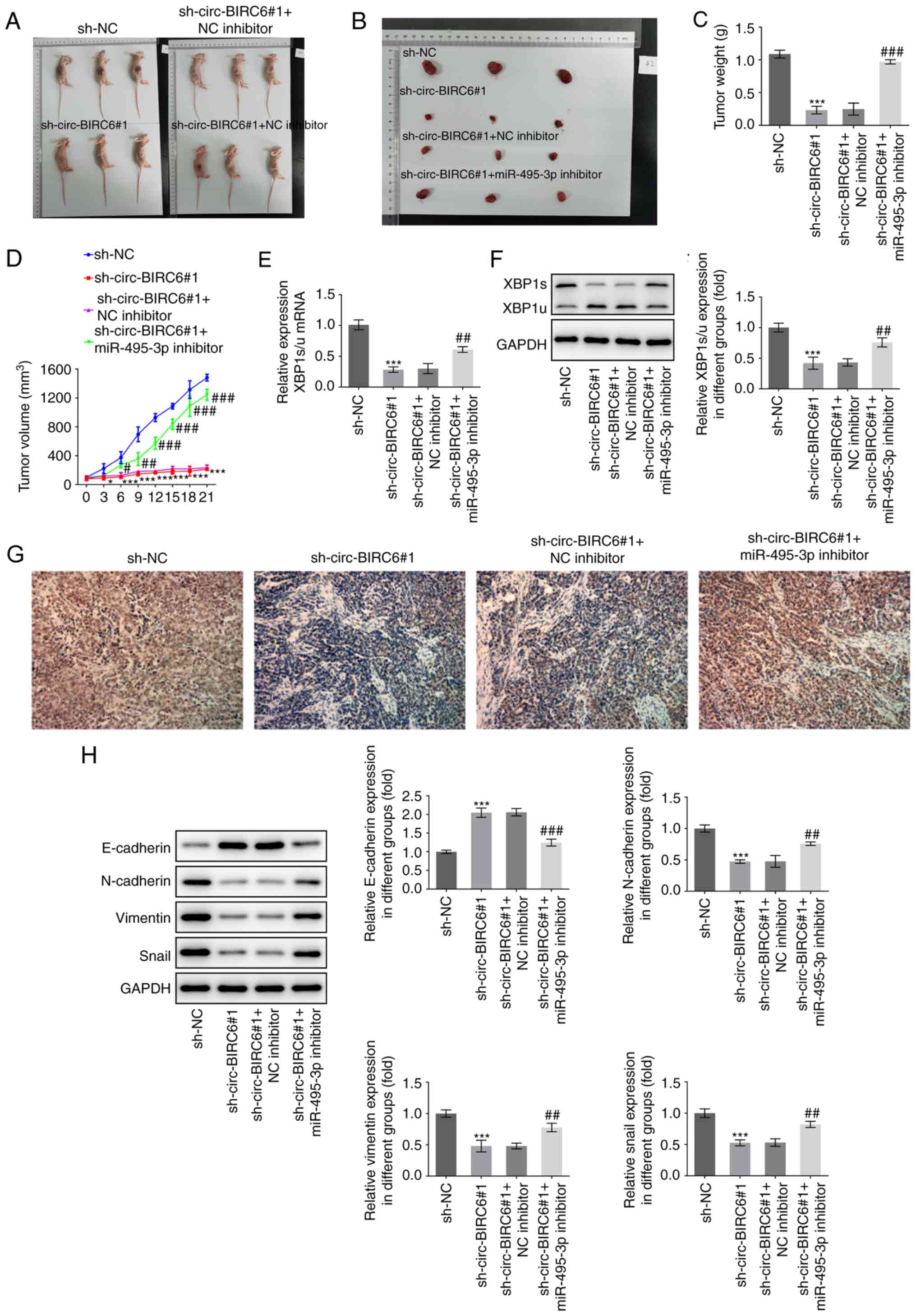 | Figure 7.circ-BIRC6 silencing suppresses the
growth of bladder cancer xenografts via regulating the
miR-495-3p/XBP1 signaling axis. (A and B) Photographs of tumors.
(C) Tumor weights. (D) Growth curves of T24 tumor volume. mRNA and
protein expression of XBP1 in tumor tissues of nude mice were
examined using (E) reverse transcription-quantitative PCR and (F)
western blotting, respectively. (G) Ki-67 expression was evaluated
using immunohistochemical staining. Magnification, ×200. (H)
Expression levels of epithelial-mesenchymal transition-related
proteins, including E-cadherin, N-cadherin, vimentin and snail,
were examined using western blotting. ***P<0.001 vs. sh-NC;
##P<0.01, ###P<0.001 vs.
sh-circ-BIRC6#1 + NC inhibitor. NC, negative control; sh, short
hairpin RNA; circ-BIRC6, circular RNA baculoviral IAP
repeat-containing 6; miR, microRNA; XBP1, X-box binding protein 1;
snail, snail family transcriptional repressor 1. |
Discussion
Previous studies have reported that circRNAs
participate in the occurrence and progression of numerous cancer
types, such as glioma, gastric and bladder cancer, by acting as
sponges of miRNAs and keeping target genes away from miRNAs
(19–21). There is an urgent requirement to
increase the current understanding of the molecular mechanisms
underlying BC development. In the present study, it was first
demonstrated that circ-BIRC6 expression was upregulated in BC cell
lines. Functional experiments identified that circ-BIRC6 silencing
suppressed the progression of BC by mediating the behaviors of
cancer cells (proliferation, invasion, migration and EMT). In
addition, the results of the mechanistic studies revealed that
circ-BIRC6 acted as a competing endogenous RNA (ceRNA) for
miR-495-3p to promote XBP1 expression. The present findings
highlighted the vital roles of the circ-BIRC6/miR-495-3p/XBP1
signaling axis in the progression of BC.
Accumulating evidence has shown that abnormal
proliferation is recognized as a feature of BC cells (22–24).
Moreover, invasion and metastasis are the most challenging
obstacles to successful tumor treatment and are the leading cause
for the resultant mortality of patients with BC (25,26).
EMT, a crucial driver of tumor progression, is characterized by
loss of cell-cell adhesion and cell polarity that increases cell
invasion and migration (27). The
EMT process is accompanied by the downregulation of epithelial
markers (E-cadherin) and upregulation of mesenchymal markers
(N-cadherin, vimentin and snail) (28,29).
In this regard, interruption of the aforementioned processes is an
effective method to inhibit cancer development. In the present
study, circ-BIRC6 knockdown upregulated E-cadherin expression and
downregulated N-cadherin, Vimentin and Snail expression.
circRNAs have been found to participate in
tumorigenesis by regulating multiple biological processes, such as
growth, migration and invasion (30–32).
circ-BIRC6 was reported to accelerate the progression of non-small
cell lung cancer by potently promoting the proliferation, invasion
and migration of cancer cells (33). In addition, it was revealed that
circ-BIRC6 knockdown suppressed hepatocellular carcinoma cell
proliferation, invasion and migration (13). However, to date, there is no
previous literature regarding the expression and/or role of
circ-BIRC6 in BC tissues and/or animal and cell models. The results
of the present study identified that circ-BIRC6 was upregulated in
BC cell lines, and silencing of circ-BIRC6 notably suppressed the
proliferation, invasion, migration and EMT process of BC cells,
suggesting the inhibitory effects of circ-BIRC6 on the progression
of BC.
circRNAs can function as ceRNAs to regulate
biological activity by sponging miRNAs to completely or partially
relieve their suppression of target mRNAs (34). With regards to their underlying
mechanism, the current study bioinformatics analysis predicted that
miR-495-3p was a potential target miRNA of circ-BIRC6, which was
verified using a dual luciferase reporter assay. According to
previous studies, miR-495-3p exerted a tumor suppressive effect in
several cancer types. For instance, miR-495-3p expression was found
to be downregulated in colorectal cancer (35). Moreover, miR-495-3p restrained the
proliferation, invasion and migration of osteosarcoma cells by
directly targeting complement C1q/TNF-related protein 3 (36). It is also worth noting that
miR-495-3p expression was reported to be downregulated in glioma
tissues and cells, and that miR-495-3p overexpression markedly
inhibited the proliferation, invasion and EMT of glioma cells
(37). In the present study,
miR-495-3p expression was markedly downregulated in BC cell lines.
Notably, transfection with the miR-495-3p inhibitor partially
restored the inhibitory effects on BC progression caused by
circ-BIRC6 knockdown in BC cells.
Subsequently, the present study identified that XBP1
was a direct target of miR-495-3p. XBP1 is a key factor involved in
endoplasmic reticulum stress. In general, XBP1 encodes two
subtypes: XBP1s contains nuclear localization signals and
transcriptional activation domains, while XBP1u contains nuclear
rejection signals (38). XBP1s/u is
an indicator of XBP1s activity, which, as an independent prognostic
indicator, can be used to predict the overall survival of patients
with myeloma (39). The loss of
XBP1 was discovered to prevent glioma growth and promote cellular
apoptosis (40). Furthermore, XBP1
was found to accelerate proliferation and invasion in human
esophageal squamous cell carcinoma by upregulating the expression
levels of MMP9 (41). Additionally,
it was reported that XBP1 induced snail expression to promote the
EMT and invasion of breast cancer cells (42). Emerging evidence supports the notion
that elevated XBP1s/u is associated with a poor prognosis in
patients with BC (43). In the
current study, it was demonstrated that XBP1s/u expression was
markedly upregulated in BC cell lines, and that XBP1 expression was
regulated by the circ-BIRC6/miR-495-3p signaling axis. The
xenograft results further verified the inhibitory effects of
circ-BIRC6 knockdown on the growth and EMT of BC via interacting
with miR-495-3p/XBP1.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that circ-BIRC6 was
highly expressed in BC cells. Functionally, circ-BIRC6 knockdown
inhibited the proliferation, invasion, migration and EMT of BC
cells. Mechanistically, circ-BIRC6 knockdown suppressed BC
progression in vitro and in vivo via a novel
circ-BIRC6/miR-495-3p/XBP1 signaling regulatory network. These
findings highlight a novel regulatory mechanism
(circ-BIRC6/miR-495-3p/XBP1 signaling axis) in BC for researchers
in this field to explore further, thereby identifying a new
theoretical basis for targeted therapy. However, the lack of
studies verifying the clinical value of circ-BIRC6 in clinical BC
tissue samples and experiments showing any dose effect of
circ-BIRC6 silencing (such as initial cell cycle arrest that would
lead to inhibition of cell proliferation) as well as the usage of
only one BC cell line to clarify the effects of the
circ-BIRC6/miR-495-3p/XBP1 signaling axis in BC are potential
limitations of the present research. Additionally, whether the
overexpression of circ-BIRC6 would enhance the tumorigenesis of
normal cells (SV-HUC-1) should also be further investigated.
Therefore, comprehensive and in-depth analyses are required in the
future to validate the findings of the current study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, BW and YZ searched the literature and designed
and performed the experiments. LZ, KY and BL analyzed and
interpreted the data and wrote the manuscript. BW revised the
manuscript. LZ and BL confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of The Third Xiangya Hospital, Central South University
(Changsha, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ploeg M, Aben KK and Kiemeney LA: The
present and future burden of urinary bladder cancer in the world.
World J Urol. 27:289–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roupret M, Babjuk M, Comperat E, Zigeuner
R, Sylvester RJ, Burger M, Cowan NC, Gontero P, Van Rhijn BWG,
Mostafid AH, et al: European association of urology guidelines on
upper urinary tract urothelial carcinoma: 2017 Update. Eur Urol.
73:111–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burger M, Grossman HB, Droller M,
Schmidbauer J, Hermann G, Drăgoescu O, Ray E, Fradet Y, Karl A,
Burgués JP, et al: Photodynamic diagnosis of non-muscle-invasive
bladder cancer with hexaminolevulinate cystoscopy: A meta-analysis
of detection and recurrence based on raw data. Eur Urol.
64:846–854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia
M, Zhan Y, Lin J, Chen Z, He A, et al: Tetracycline-inducible shRNA
targeting antisense long non-coding RNA HIF1A-AS2 represses the
malignant phenotypes of bladder cancer. Cancer Lett. 376:155–164.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang W, Liu T, Li T and Zhao X:
Hsa_circRNA_102002 facilitates metastasis of papillary thyroid
cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther.
28:229–293. 2021. View Article : Google Scholar
|
|
9
|
Jia YJ, Liu M and Wang S: CircRNA
hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis
in endometrial cancer via downregulating LRIG2 by sponging miR-182.
Cancer Cell Int. 20:4122020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun Z, Niu S, Xu F, Zhao W, Ma R and Chen
M: CircAMOTL1 promotes tumorigenesis through miR-526b/SIK2 axis in
cervical cancer. Front Cell Dev Biol. 8:5681902020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan D, Dong W, He Q, Yang M, Huang L, Kong
J, Qin H, Lin T and Huang J: Circular RNA circPICALM sponges
miR-1265 to inhibit bladder cancer metastasis and influence FAK
phosphorylation. EBioMedicine. 48:316–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen B, Wei W, Huang X and Xie X, Kong Y,
Dai D, Yang L, Wang J, Tang H and Xie X: circEPSTI1 as a prognostic
marker and mediator of triple-negative breast cancer progression.
Theranostics. 8:4003–4015. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P,
Liu Z and Li J: circ-BIRC6, a circular RNA, promotes hepatocellular
carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell
Cycle. 18:976–989. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luk IS, Shrestha R, Xue H, Wang Y, Zhang
F, Lin D, Haegert A, Wu R, Dong X, Collins CC, et al: BIRC6
targeting as potential therapy for advanced, enzalutamide-resistant
prostate cancer. Clin Cancer Res. 23:1542–1551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang W, Xue R, Weng S, Wu J, Fang Y, Wang
Y, Ji L, Hu T, Liu T, Huang X, et al: BIRC6 promotes hepatocellular
carcinogenesis: Interaction of BIRC6 with p53 facilitating p53
degradation. Int J Cancer. 136:E475–E487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luk SU, Xue H, Cheng H, Lin D, Gout PW,
Fazli L, Collins CC, Gleave ME and Wang Y: The BIRC6 gene as a
novel target for therapy of prostate cancer: Dual targeting of
inhibitors of apoptosis. Oncotarget. 5:6896–6908. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong X, Lin D, Low C, Vucic EA, English
JC, Yee J, Murray N, Lam WL, Ling V, Lam S, et al: Elevated
expression of BIRC6 protein in non-small-cell lung cancers is
associated with cancer recurrence and chemoresistance. J Thorac
Oncol. 8:161–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H,
Cai H, Ma J, Liu L, Wang P, et al: circ-SHKBP1 regulates the
angiogenesis of U87 glioma-exposed endothelial cells through
miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids.
10:331–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bi J, Liu H, Dong W, Xie W, He Q, Cai Z,
Huang J and Lin T: Circular RNA circ-ZKSCAN1 inhibits bladder
cancer progression through miR-1178-3p/p21 axis and acts as a
prognostic factor of recurrence. Mol Cancer. 18:1332019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li M, Liu Y, Liu J, Li W, Li N, Xue D,
Zhang X and Wang P: Circ_0006332 promotes growth and progression of
bladder cancer by modulating MYBL2 expression via miR-143. Aging
(Albany NY). 11:10626–10643. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dai R, Jiang Q, Zhou Y, Lin R, Lin H,
Zhang Y, Zhang J and Gao X: Lnc-STYK1-2 regulates bladder cancer
cell proliferation, migration, and invasion by targeting
miR-146b-5p expression and AKT/STAT3/NF-kB signaling. Cancer Cell
Int. 21:4082021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He H, Yi L, Zhang B, Yan B, Xiao M, Ren J,
Zi D, Zhu L, Zhong Z, Zhao X, et al: USP24-GSDMB complex promotes
bladder cancer proliferation via activation of the STAT3 pathway.
Int J Biol Sci. 17:2417–2429. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abdollah F, Gandaglia G, Thuret R,
Schmitges J, Tian Z, Jeldres C, Passoni NM, Briganti A, Shariat SF,
Perrotte P, et al: Incidence, survival and mortality rates of
stage-specific bladder cancer in United States: A trend analysis.
Cancer Epidemiol. 37:219–225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen M, Li J, Zhuang C and Cai Z:
Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of
bladder cancer. Oncotarget. 8:28176–28186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Wan B, Liu L, Zhou L and Zeng Q:
Circular RNA circMTO1 suppresses bladder cancer metastasis by
sponging miR-221 and inhibiting epithelial-to-mesenchymal
transition. Biochem Biophys Res Commun. 508:991–996. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Georgakopoulos-Soares I, Chartoumpekis DV,
Kyriazopoulou V and Zaravinos A: EMT factors and metabolic pathways
in cancer. Front Oncol. 10:4992020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ko JH, Yang MH, Baek SH, Nam D, Jung SH
and Ahn KS: Theacrine attenuates epithelial mesenchymal transition
in human breast cancer MDA-MB-231 cells. Phytother Res.
33:1934–1942. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang W, Lu Y, Wang F, Huang X and Yu Z:
Circular RNA circRNA_103809 accelerates bladder cancer progression
and enhances chemo-resistance by activation of miR-516a-5p/FBXL18
Axis. Cancer Manag Res. 12:7561–7568. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan C, Qu H, Xiong F, Tang Y, Tang T,
Zhang L, Mo Y, Li X, Guo C, Zhang S, et al: CircARHGAP12 promotes
nasopharyngeal carcinoma migration and invasion via ezrin-mediated
cytoskeletal remodeling. Cancer Lett. 496:41–56. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin X, Huang C, Chen Z, Wang H and Zeng Y:
CircRNA_100876 is upregulated in gastric cancer (GC) and promotes
the GC cells' growth, migration and invasion via miR-665/YAP1
signaling. Front Genet. 11:5462752020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang H, Zhao M, Zhao L, Li P, Duan Y and
Li G: CircRNA BIRC6 promotes non-small cell lung cancer cell
progression by sponging microRNA-145. Cell Oncol. 43:477–488. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L and Liu J: Research progress of
competing endogenous RNA. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi.
34:967–971. 2017.(In Chinese). PubMed/NCBI
|
|
35
|
Qian J, Garg A, Li F, Shen Q and Xiao K:
lncRNA LUNAR1 accelerates colorectal cancer progression by
targeting the miR-495-3p/MYCBP axis. Int J Oncol. 57:1157–1168.
2020.PubMed/NCBI
|
|
36
|
Zhao G, Zhang L, Qian D, Sun Y and Liu W:
miR-495-3p inhibits the cell proliferation, invasion and migration
of osteosarcoma by targeting C1q/TNF-related protein 3. OncoTargets
Ther. 12:6133–6143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mutalifu N, Du P, Zhang J, Akbar H, Yan B,
Alimu S, Tong L and Luan X: Circ_0000215 Increases the Expression
of CXCR2 and promoted the progression of glioma cells by sponging
miR-495-3p. Technol Cancer Res Treat. 19:1533033820957026.
2020.doi:10.1177/1533033820957026. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yoshida H, Oku M, Suzuki M and Mori K:
pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded
protein response activator pXBP1(S) in mammalian ER stress
response. J Cell Biol. 172:565–575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bagratuni T, Wu P, Gonzalez de Castro D,
Davenport EL, Dickens NJ, Walker BA, Boyd K, Johnson DC, Gregory W,
Morgan GJ and Davies FE: XBP1s levels are implicated in the biology
and outcome of myeloma mediating different clinical outcomes to
thalidomide-based treatments. Blood. 116:250–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Romero-Ramirez L, Cao H, Nelson D, Hammond
E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, et
al: XBP1 is essential for survival under hypoxic conditions and is
required for tumor growth. Cancer Res. 64:5943–5947. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xia T, Tong S, Fan K, Zhai W, Fang B, Wang
SH and Wang JJ: XBP1 induces MMP-9 expression to promote
proliferation and invasion in human esophageal squamous cell
carcinoma. Am J Cancer Res. 6:2031–2040. 2016.PubMed/NCBI
|
|
42
|
Li H, Chen X, Gao Y, Wu J, Zeng F and Song
F: XBP1 induces snail expression to promote epithelial-
to-mesenchymal transition and invasion of breast cancer cells. Cell
Signal. 27:82–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen W, Zhou J, Wu K, Huang J, Ding Y, Yun
EJ, Wang B, Ding C, Hernandez E, Santoyo J, et al: Targeting
XBP1-mediated β-catenin expression associated with bladder cancer
with newly synthetic oridonin analogues. Oncotarget. 7:56842–56854.
2016. View Article : Google Scholar : PubMed/NCBI
|