Introduction
Colon cancer is a common, highly malignant type of
cancer characterized by a high morbidity rate and a poor prognosis
(1). According to statistics
obtained from the American Cancer Society, human colon cancer has
an incidence rate of 10.2% and a mortality rate of 9.2% worldwide,
ranking it as the fourth most common cancer (2,3).
Although advances have been made in the current treatment options
available for colon cancer, the associated mortality rates continue
to increase and the current 5-year survival rate remains low
(4,5). Risk factors associated with colon
cancer include age and diet; however, the overexpression of
oncogenes and the inactivation of tumor suppressor genes remain the
most important contributors (6).
At present, conventional therapies, such as surgery, chemotherapy
and antibody therapies have been adopted to protect against colon
cancer (7). However, these
methods exhibit low levels of effectiveness in clinical practice
(8). Thus, the present study
aimed to investigate the mechanisms underlying the development and
progression of colon cancer, which may help to determine effective
therapeutic options.
GINS complex subunit 2 (GINS2), also known as PSF2,
belongs to the GINS family and acts as a vital participant in both
DNA duplication and cell cycle progression (9,10).
In addition, GINS2 has been found to serve a key role in the
tumorigenesis of various cancers (11). For example, GINS2 is overexpressed
in breast cancer cell lines and GINS2 knockdown inhibited breast
cancer growth and metastasis (12). In addition, Yan et al
(13) demonstrate that
GINS2-regulated cell proliferation and apoptosis in human
epithelial ovarian cancer. GINS2 is highly expressed in colon
cancer (14,15), but the specific role is yet to be
fully elucidated.
The results of previous studies demonstrated that
phosphatase of regenerating liver 1 (PTP4A1) is upregulated in
numerous tumor cells and is involved in promoting both cell
migration and invasion (16–18). The analysis of BioGrid (https://thebiogrid.org/), a protein-protein
interaction database, suggested that PTP4A1 could interact with
GINS2. Results from a previous study demonstrate that PTP4A1 is
highly expressed in colon cancer, but not in normal colon tissues
or colonic adenomas (19). In
addition, GINS2 inhibition regulates the proliferation and
apoptosis of non-small cell lung cancer cells through p53 (20). PTP4A1 is considered as a novel p53
target and PTP4A1 inhibition increases p53 expression level
(21).
Therefore, the present study aimed to explore the
role of GINS2 in the proliferation and apoptosis of colon cancer
and its potential regulatory mechanism on the PTP4A1/p53
pathway.
Materials and methods
Cell culture, treatment and
transfection
Normal human intestinal epithelial cell line
(HIEC-6) and colon cancer cell lines (HCT116, LS174T, HCT8 and
SW620) were purchased from the American Type Culture Collection.
Cells were incubated in DMEM supplemented with 10% FBS (both from
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere with 5% CO2.
To knockdown GINS2, short hairpin RNA
(shRNA)-GINS2-1/2 and its negative control (shRNA-NC) were designed
and synthesized by Shanghai GenePharma Co., Ltd. and shRNA
fragments were cloned into a lentiviral GV493 vector (Shanghai
GeneChem Co., Ltd.). The target sequences were as follows:
shRNA-GINS2-1, 5′-GATTAACCTGAAACAAAGA-3′; shRNA-GINS2-2,
5′-ATCAACACCAGCGGGACTTTC-3′; shRNA-NC, 5′-TTTCTCCGAACGTGTCACGT-3′.
To overexpression PTP4A1, the PTP4A1 coding sequence was
synthesized and cloned into a lentiviral GV492 vector. The empty
GV492 vector was considered as a negative control (Ov-NC). When
293T cells were cultured to 60–70% confluency, according to the
manufacturer's protocol, a total of 1 µg lentiviral plasmid, 1 µg
3rd generation viral packaging vectors and 5 µl
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) in serum-free DMEM was incubated
for 15 min at room temperature and then added into 293T cells to
amplify at 37°C for 6 h. After transfection for 48 h, the
supernatant containing virus was collected by centrifugation at
10,000 × g for 4 h at 4°C. HCT116 cells at the 3rd passage were
infected with lentiviral vectors at an MOI of 50 in the presence of
5 µg/ml polybrene (MilliporeSigma) for 24 h at 37°C. Subsequently,
the medium was replaced with fresh medium and the stable cells were
selected with 2 µg/ml puromycin for 3 days. The transfection
efficiency was evaluated by reverse transcription-quantitative
(RT-q) PCR and western blotting.
RT-qPCR
Total RNA was extracted from 1×106 cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and subsequently reverse transcribed into cDNA
using the PrimeScript RT Reagent kit (Takara Biotechnology Co.,
Ltd.) according to the manufacturer's protocols under the following
conditions: 37°C for 15 min and at 85°C for 5 sec. The synthesized
cDNA was used as a template for PCR, which was performed using an
ABI 7000 quantitative PCR instrument (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using the SYBR Green PCR kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 6 min; followed by 40 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec and
extension at 72°C for 45 sec and a final extension at 72°C for 5
min. The sequences of the gene primers were as follows: GINS2
forward, 5′-CAGAAATGTCGCCTGCTCC-3′ and reverse,
5′-GGATTTCGTCTGCCTTCG-3′; PTP4A1 forward,
5′-ATTGAAGGTGGAATGAAATACGAAG-3′ and reverse,
5′-TACTTCTCCAAATACAGAAGTTGCT-3′; and GAPDH forward,
5′-ACCTGACCTGCCGTCTAGAAAA-3′ and reverse,
5′-TTGAAGTCAGAGGAGACCACCTG-3′. Relative mRNA expression levels were
quantified using the 2−ΔΔCq method (22) and normalized to the internal
reference gene GAPDH.
Western blotting
Total protein was extracted from the cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
subsequently quantified using BCA kits (Thermo Fisher Scientific,
Inc.). Proteins (30 µg/lane) were separated on a 10% gel by
SDS-PAGE and subsequently transferred onto PVDF membranes. The
membranes were blocked with 5% non-fat milk for 1 h at room
temperature and the proteins were incubated with primary antibodies
against GINS2 (1:500; cat. no. ab197123; Abcam), PTP4A1 (1:400;
cat. no. ab121185; Abcam), p21 (1:1,200; cat. no. ab109520; Abcam),
cyclin D1 (1:200; cat. no. ab16663; Abcam), Bcl2 (1:1,500; cat. no.
ab182858; Abcam), Bax (1:1,500; cat. no. ab182733; Abcam), poly
(ADP-ribose) polymerase (PARP) (1:1,000; cat. no. ab191217; Abcam),
p53 (1:5,000; cat. no. ab32389; Abcam) and GAPDH (1:2,500; cat. no.
ab9485; Abcam) at 4°C overnight. Following primary incubation, the
membranes were incubated with an HRP-conjugated goat anti-rabbit
secondary antibody (1:20,000; cat. no. ab205718; Abcam) for 2 h at
room temperature. Protein bands were visualized using enhanced
chemiluminescence (MilliporeSigma) and imaging system (Tanon-5200;
Tanon Science and Technology Co., Ltd.). The gray values of the
bands were semi-quantified using ImageJ software (version 1.0;
National Institutes of Health) and the protein expression was
normalized against GAPDH.
Cell Counting Kit-8 (CCK-8)
Transfected cells (5×103 cells/well) were
inoculated into 96-well plates. CCK-8 reagent (10 µl/well; Beyotime
Institute of Biotechnology) was added and the cells were incubated
at 37°C for 2 h. Subsequently, cell viability at 24, 48 or 72 h was
evaluated using a microplate reader at 450 nm.
5-ethynyl-2′-deoxyuridine (EdU)
staining
Transfected cells (5×104 cells/well) were
inoculated into 96-well plates and incubated with 20 µM EdU (Thermo
Fisher Scientific, Inc.) at 37°C for 2 h. Subsequently, cells were
fixed and permeated using 4% paraformaldehyde for 30 min and 0.5%
Triton X-100 for 10 min at room temperature, respectively. The
cells were stained with Cell-Light™ EdU Apollo® 488
In Vitro Imaging kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Cells were observed using
a fluorescence microscope (magnification, ×200; Nikon
Corporation).
Flow cytometry
The harvested cells were fixed overnight with 75%
ethanol at 4°C and subsequently cultured with propidium iodide
(PI)/RNase staining buffer at 37°C for 30 min in the dark. The cell
cycle analysis was performed using a BD FACSCalibur flow cytometer
(BD Biosciences). The distribution of cell cycle, that is, the cell
percentages at the G0/G1, S and
G2/M phases, were quantified using FlowJo software
(version 7.0; FlowJo LLC).
TUNEL staining
Cell apoptosis was determined using the TUNEL
Apoptosis Assay kit (Beyotime Institute of Biotechnology). Briefly,
fixation and permeabilization of HCT116 cells were performed using
4% paraformaldehyde for 20 min and 0.1% Triton X-100 for 10 min at
room temperature, respectively. Subsequently, cells were incubated
with the TUNEL reaction solution at 37°C for 1 h in the dark and
the nuclei were stained with DAPI (5 µg/ml) for 5 min at room
temperature. A total of five visual fields were randomly selected
and apoptotic cells were observed on glass coverslips under a
fluorescence microscope (magnification, ×200; Nikon
Corporation).
Co-immunoprecipitation (Co-IP)
assay
The interaction between GINS2 and PTP4A1 was
determined using a Co-IP assay. Briefly, the harvested cells were
fully lysed using 1 ml Co-RIPA buffer (Applygen Technologies, Inc.)
and then lysates were pre-cleared with 30 µl Protein A/G
PLUS-Agarose (Santa Cruz Biotechnology, Inc.) at 4°C for 1 h. After
centrifuging the lysate, taking the supernatant and removing the
beads in the supernatant, immunoprecipitation with antibodies
against GINS2 (1:50; cat. no. sc-376595; Santa Cruz Biotechnology,
Inc.), PTP4A1 (1:30; cat. no. sc-365659; Santa Cruz Biotechnology,
Inc.) and IgG (1:50; cat. no. sc-69786; Santa Cruz Biotechnology,
Inc.) was performed with shaking at 4°C overnight. The protein
A/G-Sepharose beads were added to the samples and shaken at 4°C for
4 h. Following centrifugation at 800 × g for 5 min at 4°C, the
supernatant was removed and the beads were washed three times (800
µl/time) with pre-cooled Co-RIPA buffer to obtain protein samples
for western blotting analysis.
Statistical analysis
All experiments were performed in triplicate. All
data are presented as the mean ± standard deviation and statistical
analysis was performed using SPSS 20.0 (IBM Corp.). Unpaired
Student's t-tests were performed to compare the differences between
two groups and one-way ANOVA followed by Tukey's post hoc test was
used for multiple comparisons. P<0.05 was considered to indicate
a statistically significant difference.
Results
GINS2 is overexpressed in colon cancer
cell lines
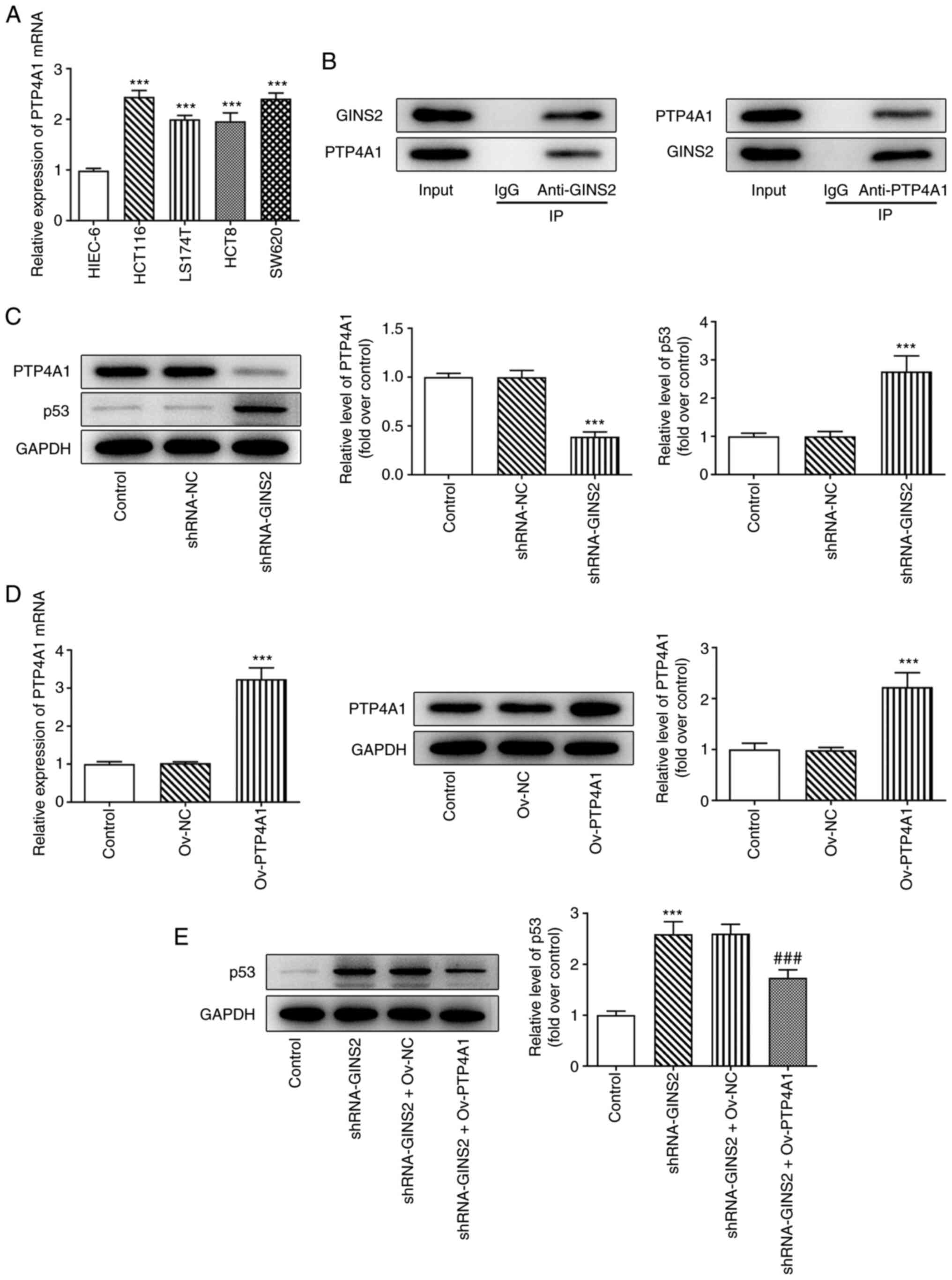
The expression levels of GINS2 were measured in all
colon cancer cells. As depicted in Fig. 1A and B, compared with HIEC-6, the
GINS2 mRNA and protein expression levels were increased in colon
cancer cell lines and the levels were highest in HCT116 cells.
Subsequently, the HCT116 cell line was selected for the following
experiments.
GINS2 knockdown inhibits the
proliferation of HCT116 cells
HCT116 cells were transfected with shRNA-GINS2
plasmids for GINS2 knockdown and this was subsequently detected
using RT-qPCR and western blotting. The results displayed in
Fig. 2A and B demonstrated that
the mRNA and protein expression levels of GINS2 were significantly
reduced in shRNA-GINS2-1 and shRNA-GINS2-2 cells compared with
shRNA-NC. GINS2 expression levels were lowest in HCT116 cells
transfected with the shRNA-GINS2-1 plasmid. Thus, the shRNA-GINS2-1
plasmid was used for subsequent experiments. The results of the
CCK-8 assay demonstrated that, compared with cells transfected with
shRNA-NC, the viability of HCT116 cells was markedly reduced
following GINS2 knockdown (Fig.
2C). Similarly, the results of the EdU staining assay
demonstrated that transfection with the shRNA-GINS2 plasmid notably
inhibited the levels of cell proliferation, compared with HCT116
cells transfected with shRNA-NC (Fig.
2D). In summary, GINS2 promoted colon cancer cell
proliferation.
GINS2 knockdown promotes HCT116 cell
cycle arrest
The cell cycle was evaluated using flow cytometry.
As demonstrated in Fig. 3A, the
number of cells in the G0/G1 phase was
increased, while those in S phase and G2 phase were
decreased following GINS2 knockdown, compared with cells
transfected with shRNA-NC. In addition, western blotting was
performed to determine the expression levels of cell
cycle-associated proteins and the results demonstrated that
shRNA-GINS2 reduced the levels of cyclin D1 expression, while p21
expression was increased, compared with cells transfected with
shRNA-NC (Fig. 3B). Collectively,
these results demonstrated that GINS2 promoted colon cancer cell
cycle progression.
GINS2 knockdown promotes colon cancer
cell apoptosis
TUNEL staining was performed to determine the
effects of GINS2 on HCT116 cell apoptosis. As demonstrated in
Fig. 4A, cell apoptosis was
markedly increased following transfection with the shRNA-GINS2
plasmids. In addition, the levels of apoptosis-associated proteins,
such as Bcl2, Bax and cleaved PARP, were measured using western
blotting. The results of the present study demonstrated that the
expression levels of Bcl2 were notably reduced following GINS2
knockdown, but the expression levels of Bax and cleaved PARP/PARP
were increased, compared with cells transfected with shRNA-NC
(Fig. 4B).
GINS2 knockdown activates the p53
pathway through PTP4A1
To determine the expression levels of PTP4A1 in
colon cancer cells, RT-qPCR was performed. The results demonstrated
that the mRNA expression levels of PTP4A1 were increased in HCT116,
LS174T, HCT8 and SW620 cells, compared with the with HIEC-6 cells
(Fig. 5A). In addition, a Co-IP
assay was performed to verify the targeted binding of GINS2 and
PTP4A1. The results of Co-IP demonstrated that GINS2 could interact
with PTP4A1 (Fig. 5B). In
addition, compared with shRNA-NC, shRNA-GINS2 reduced PTP4A1
expression level and increased p53 expression level (Fig. 5C).
Following transfection with Ov-PTP4A1 plasmid, the
mRNA and protein levels of PTP4A1 were detected using RT-qPCR and
western blotting, respectively. Compared with cells transfected
with Ov-NC, PTP4A1 expression levels were increased in Ov-PTP4A1
(Fig. 5D). In addition, the
results displayed in Fig. 5E
demonstrated that shRNA-GINS2 upregulated the expression levels of
p53, which were subsequently reversed following transfection with
the Ov-PTP4A1 plasmid.
GINS2 knockdown regulates cell
proliferation, cycle arrest and apoptosis in colon cancer cells
through PTP4A1/p53 pathway
As displayed in Fig.
6A and B, the shRNA-GINS2-mediated decrease in cell
proliferation was increased following PTP4A1 overexpression. In
addition, shRNA-GINS2 markedly decreased the number of cells in the
G0/G1 phase and decreased the number of cells
in the S phase and G2 phase. Notably, this effect was
inhibited following PTP4A1 overexpression (Fig. 6C), which highlighted that the cell
cycle arrest induced by shRNA-GINS2 was subsequently restored
following PTP4A1 overexpression. The results of the TUNEL assay
revealed that apoptosis was increased in shRNA-GINS2; however, this
effect was inhibited following transfection with Ov-PTP4A1
(Fig. 6D). Furthermore, the
western blotting analysis results demonstrated that the
shRNA-GINS2-mediated reduction in the protein expression levels of
cyclin D1 and Bcl2 and the increased protein expression levels of
p21, Bax, cleaved PARP/PARP, were rescued following PTP4A1
overexpression (Fig. 6E and F).
These results indicated that GINS2 modulated colon cancer cell
proliferation, cycle arrest and apoptosis through the PTP4A1/p53
pathway (Fig. 6G).
 | Figure 6.GINS2 knockdown regulates the
proliferation, cycle arrest and apoptosis through the PTP4A1/p53
pathway. (A) Cell viability was detected using a Cell Counting
Kit-8 assay. (B) Cell proliferation was detected using EdU
staining. (C) Flow cytometry was performed to examine cell cycle
distribution. (D) Cell apoptosis was detected using a TUNEL assay.
(E) Western blotting analyses were performed and (F) quantified to
detect the protein expression levels of cyclin D1, p21, Bcl2, Bax,
cleaved PARP and PARP. (G) Schematic depiction of regulatory
mechanism underlying colon cancer proliferation, cycle arrest and
apoptosis via GINS2/PTP4A1/p53 pathway. ***P<0.001 vs. Control;
##P<0.01, ###P<0.001 vs. shRNA-GINS2 +
Ov-NC. GINS2, GINS complex subunit 2; PTP4A1, protein tyrosine
phosphatase 4A1; shRNA, short hairpin RNA; NC, negative control;
Ov, overexpression; PARP, poly (ADP-ribose) polymerase. |
Discussion
Novel indicators for determining the prognosis of
patients with colon cancer are required for the development of
effective treatment options (1).
GINS2 has been identified as a crucial regulator in cell cycle
progression (23). It has also
been reported that GINS2 promotes cell proliferation and apoptosis
desensitization (24). The
present results demonstrated that GINS2 knockdown inhibited the
proliferation and induced cell cycle arrest and apoptosis in colon
cancer. GINS2 may serve a key role in colon cancer.
The cell cycle is a complex process that provides
the tumor cell with the opportunity to repair its damaged DNA
(25). This process is regulated
by numerous protein families, including cyclin-dependent kinases
(CDKs), which drive cell cycle progression (26,27). Cyclin D1, in association with
CDK4/6, functions as an important regulator of the cell cycle
(28). As a CDK inhibitor, p21
mainly serves a suppressive role in cell cycle progression
(29). Apoptosis, also known as
programmed cell death, is a physiological process that occurs in
multicellular organisms (30).
Chen et al (31)
demonstrate that apoptosis acts as a critical participant in
numerous biological processes, including tissue development and
organ formation. Downregulation of GINS2 induces cell cycle arrest
and apoptosis in lung cancer A549 cells (32). Silencing of GINS2, which is
overexpressed in melanoma, inhibits cell proliferation and
increases apoptosis in A375 cells (33). GINS2 interference induces cell
cycle arrest and apoptosis of pancreatic cancer via the MAPK/ERK
pathway (34). The results of the
present study demonstrated that GINS2 knockdown induced cell cycle
arrest and promoted apoptosis in colon cancer. In addition, the
results of the present study also demonstrated that GINS2 knockdown
reduced the expression levels of the anti-apoptotic protein Bcl2,
but increased the expression levels of the pro-apoptotic proteins
Bax and cleaved PARP.
PTP4A1 expression is increased in numerous cancers,
including intrahepatic cholangiocarcinoma (18), non-small cell lung cancer
(35) and cervical cancer
(36). According to the Biogrid
database, GINS2 has the ability to interact with PTP4A1, which was
subsequently confirmed using a Co-IP assay. In addition, PTP4A1
expression levels were increased in colon cancer cells and the
increased levels of PTP4A1 expression were decreased following
transfection with the shRNA-GINS2 plasmids.
p53, a tumor suppressor, is described as a
co-immunoprecipitating protein by Kress et al (37). Through the transcription
regulation of downstream target genes involved in cell cycle
arrest, apoptosis, DNA repair and metabolism, p53 exerts multiple
biological functions in human diseases (38,39). In addition, p53 influences the
sensitivity of colon cancer cells to bleomycin (40). Mutant p53 is shown to promote
angiogenesis in colon cancer, leading to a poor prognosis (41). As a regulatory target gene of p53,
PTP4A1 activates p53 expression (21). The results of the present study
demonstrated that GINS2 knockdown led to the activation of the p53
pathway through PTP4A1. Additionally, rescue experiments confirmed
that the GINS2 knockdown-mediated suppression in cell proliferation
and the increased levels of cell cycle arrest and apoptosis in
colon cancer cells, were restored following PTP4A1 overexpression.
These results highlighted that GINS2 knockdown exerted its
regulatory effects on the proliferation, cycle arrest and apoptosis
of colon cancer cells via regulation of the PTP4A1/p53 pathway.
However, a key limitation of the present study was the lack of
detection of GINS2 and PTP4A1 expression in clinical samples.
Therefore, future studies should validate the results of the
present study by performing some in-vivo experiments.
In conclusion, it was demonstrated that GINS2
regulated the proliferation, cell cycle progression and apoptosis
of colon cancer cells through PTP4A1, indicating that GINS2 may
serve as a biomarker for the development of novel therapies for
colon cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study and supervised the project. HH
and LY performed the experiments and analyzed data. HH drafted the
manuscript. All authors participated in the revision of the
manuscript. ZL and HH confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen H, Luo J and Guo J: Development and
validation of a five-immune gene prognostic risk model in colon
cancer. BMC Cancer. 20:3952020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edwards BK, Ward E, Kohler BA, Eheman C,
Zauber AG, Anderson RN, Jemal A, Schymura MJ, Lansdorp-Vogelaar I,
Seeff LC, et al: Annual report to the nation on the status of
cancer, 1975–2006, featuring colorectal cancer trends and impact of
interventions (risk factors, screening, and treatment) to reduce
future rates. Cancer. 116:544–573. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rupnarain C, Dlamini Z, Naicker S and
Bhoola K: Colon cancer: Genomics and apoptotic events. Biol Chem.
385:449–464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao C, Yan TD, Black D and Morris DL: A
systematic review and meta-analysis of cytoreductive surgery with
perioperative intraperitoneal chemotherapy for peritoneal
carcinomatosis of colorectal origin. Ann Surg Oncol. 16:2152–2165.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yaghoubi A, Khazaei M, Avan A, Hasanian SM
and Soleimanpour S: The bacterial instrument as a promising therapy
for colon cancer. Int J Colorectal Dis. 35:595–606. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kubota Y, Takase Y, Komori Y, Hashimoto Y,
Arata T, Kamimura Y, Araki H and Takisawa H: A novel ring-like
complex of Xenopus proteins essential for the initiation of DNA
replication. Genes Dev. 17:1141–1152. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
MacNeill SA: Structure and function of the
GINS complex, a key component of the eukaryotic replisome. Biochem
J. 425:489–500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stamova BS, Apperson M, Walker WL, Tian Y,
Xu H, Adamczy P, Zhan X, Liu DZ, Ander BP, Liao IH, et al:
Identification and validation of suitable endogenous reference
genes for gene expression studies in human peripheral blood. BMC
Med Genomics. 2:492009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng L, Song Z, Chen D, Linghu R, Wang Y,
Zhang X, Kou X, Yang J and Jiao S: GINS2 regulates matrix
metallopeptidase 9 expression and cancer stem cell property in
human triple negative Breast cancer. Biomed Pharmacother.
84:1568–1574. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan T, Liang W, Jiang E, Ye A, Wu Q and Xi
M: GINS2 regulates cell proliferation and apoptosis in human
epithelial ovarian cancer. Oncol Lett. 16:2591–2598.
2018.PubMed/NCBI
|
|
14
|
Wei HB, Wen JZ, Wei B, Han XY and Zhang S:
Expression and clinical significance of GINS complex in colorectal
cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 14:443–447. 2011.(In
Chinese). PubMed/NCBI
|
|
15
|
Kabir MF, Mohd Ali J and Haji Hashim O:
Microarray gene expression profiling in colorectal (HCT116) and
hepatocellular (HepG2) carcinoma cell lines treated with Melicope
ptelefolia leaf extract reveals transcriptome profiles exhibiting
anticancer activity. PeerJ. 6:e52032018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Achiwa H and Lazo JS: PRL-1 tyrosine
phosphatase regulates c-Src levels, adherence, and invasion in
human lung cancer cells. Cancer Res. 67:643–650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Kirby CE and Herbst R: The
tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum
and the mitotic spindle and is required for normal mitosis. J Biol
Chem. 277:46659–46668. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu LZ, He YZ, Dong PP, Ma LJ, Wang ZC,
Liu XY, Duan M, Yang LX, Shi JY, Zhou J, et al: Protein tyrosine
phosphatase PTP4A1 promotes proliferation and
epithelial-mesenchymal transition in intrahepatic
cholangiocarcinoma via the PI3K/AKT pathway. Oncotarget.
7:75210–75220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Li ZF, He J and Li YL, Zhu GB,
Zhang LH and Li YL: Expression of the human phosphatases of
regenerating liver (PRLs) in colonic adenocarcinoma and its
correlation with lymph node metastasis. Int J Colorectal Dis.
22:1179–1184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chi F, Wang Z, Li Y and Chang N: Knockdown
of GINS2 inhibits proliferation and promotes apoptosis through the
p53/GADD45A pathway in non-small-cell lung cancer. Biosci Rep.
40:BSR201939492020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Min SH, Kim DM, Heo YS, Kim YI, Kim HM,
Kim J, Han YM, Kim IC and Yoo OJ: New p53 target, phosphatase of
regenerating liver 1 (PRL-1) downregulates p53. Oncogene.
28:545–554. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang YH, Galal WC, Farina A, Tappin I and
Hurwitz J: Properties of the human Cdc45/Mcm2-7/GINS helicase
complex and its action with DNA polymerase epsilon in rolling
circle DNA synthesis. Proc Natl Acad Sci USA. 109:6042–6047. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Zhong L, Liu BZ, Gao YJ, Gao YM
and Hu XX: Effect of GINS2 on proliferation and apoptosis in
leukemic cell line. Int J Med Sci. 10:1795–1804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwartz GK and Shah MA: Targeting the
cell cycle: A new approach to cancer therapy. J Clin Oncol.
23:9408–9421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Swanton C: Cell-cycle targeted therapies.
Lancet Oncol. 5:27–36. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hydbring P, Malumbres M and Sicinski P:
Non-canonical functions of cell cycle cyclins and cyclin-dependent
kinases. Nat Rev Mol Cell Biol. 17:280–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tchakarska G and Sola B: The double
dealing of cyclin D1. Cell Cycle. 19:163–178. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karimian A, Ahmadi Y and Yousefi B:
Multiple functions of p21 in cell cycle, apoptosis and
transcriptional regulation after DNA damage. DNA Repair (Amst).
42:63–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kerr JF: History of the events leading to
the formulation of the apoptosis concept. Toxicology.
181-182:471–474. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen M, Wu W, Liu D, Lv Y, Deng H, Gao S,
Gu Y, Huang M, Guo X, Liu B, et al: Evolution and Structure of API5
and Its Roles in Anti-Apoptosis. Protein Pept Lett. 28:612–622.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun D, Zong Y, Cheng J, Li Z, Xing L and
Yu J: GINS2 attenuates the development of lung cancer by inhibiting
the STAT signaling pathway. J Cancer. 12:99–110. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hao YQ, Liu KW, Zhang X, Kang SX, Zhang K,
Han W, Li L and Li ZH: GINS2 was regulated by lncRNA
XIST/miR-23a-3p to mediate proliferation and apoptosis in A375
cells. Mol Cell Biochem. 476:1455–1465. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang M, He S, Ma X, Ye Y, Wang G, Zhuang
J, Song Y and Xia W: GINS2 affects cell viability, cell apoptosis,
and cell cycle progression of pancreatic cancer cells via MAPK/ERK
pathway. J Cancer. 11:4662–4670. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang T, Shi X, Wang Z, Liu X, Zhang G, Zhu
Q, Mi L and Wang R: Overexpression of PTP4A1 is associated with
poor overall survival in non-small cell lung cancer. Int J Clin Exp
Pathol. 11:3583–3590. 2018.PubMed/NCBI
|
|
36
|
Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang
X, Li X, Wu D, Wang D, Yang Z and Zhang S: Circular RNA circNRIP1
promotes migration and invasion in cervical cancer by sponging
miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death
Dis. 11:3992020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kress M, May E, Cassingena R and May P:
Simian virus 40-transformed cells express new species of proteins
precipitable by anti-simian virus 40 tumor serum. J Virol.
31:472–483. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lane D and Levine A: p53 Research: The
past thirty years and the next thirty years. Cold Spring Harb
Perspect Biol. 2:a0008932010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Levav-Cohen Y, Goldberg Z, Tan KH,
Alsheich-Bartok O, Zuckerman V, Haupt S and Haupt Y: The p53-Mdm2
loop: A critical juncture of stress response. Subcell Biochem.
85:161–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee YS, Yoon S, Park MS, Kim JH, Lee JH
and Song CW: Influence of p53 expression on sensitivity of cancer
cells to bleomycin. J Biochem Mol Toxicol. 24:260–269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Takahashi Y, Bucana CD, Cleary KR and
Ellis LM: p53, vessel count, and vascular endothelial growth factor
expression in human colon cancer. Int J Cancer. 79:34–38. 1998.
View Article : Google Scholar : PubMed/NCBI
|