Introduction
Colon cancer is a highly invasive and metastatic
cancer. Colon cancer cells can spread to several organs in the body
via the blood, where they colonize the new site and form a
metastatic lesion. Metastasis underlies the death of 90% of
patients with colon cancer (1).
Epithelial-mesenchymal transition (EMT) is a key factor that endows
a potent invasive ability on colon cancer cells (2). In this process, colon cancer cells
gradually acquire the characteristics of mesenchymal cells and the
increased expression of EMT-related proteins, such as vimentin and
N-cadherin, promote the development of EMT in colon cancer cells
(3). The occurrence of EMT is
indicative of a poor prognosis in patients with colon cancer
(4). Therefore, there is an
urgent need to develop novel therapeutic options for the inhibition
of the metastasis of colon cancer in the body and therefore
alleviate the symptoms of colon cancer.
Chaperonin containing TCP1 subunit 6A (CCT6A)
belongs to type II chaperone, which is a heteromorphic oligomeric
protein widely existing in the cytoplasm and serves an important
role in the assembly and folding of actin and tubulin (5). CCT6A is a protein that is associated
with the development of several types of cancer (6). The expression of CCT6A is
upregulated in the tissues of patients with Ewing sarcoma compared
with that of the pericarcinomatous tissues (7). Overexpression of CCT6A can promote
the transition from the G1 to S phase and therefore
enhance the proliferation of hepatocellular carcinoma cells
(8). Moreover, the expression of
CCT6A has been revealed to be associated with a poor prognosis in
patients with an adenocarcinoma of the colon (9). In addition, CCT6A has also been
revealed to be associated with the depth of colorectal cancer
invasion, tumor size and the occurrence of tumors (10). However, the effect of CCT6A on the
development of colon cancer and the specific mechanism has not been
studied thus far.
The homeobox proteins encoded by HOX gene are
basically some transcriptional regulatory factors, most of which
play an important role in the physiological and pathological
changes such as embryo development, cell differentiation and cell
carcinogenesis (11). HOXB2 is a
member of the HOX family, which is one of the critical genes
involved in regulation of cell differentiation (12). However, the expression of HOXB2 is
also related to the development of lung (13) and pancreatic cancer (14), and the upregulated expression of
HOXB2 can induce the development of colorectal cancer (15). However, to the best of our
knowledge, whether HOXB2 affects the proliferation and invasion of
colon cancer cells by regulating the expression of CCT6A has not
been determined.
Therefore, the relationship between HOXB2 and CCT6A
was detected. Additionally, the effect of CCT6A on the development
of colon cancer was assessed.
Materials and methods
Cell culture and treatment
Normal intestinal epithelial cells (HIEC cells) and
colon cancer cell lines (Caco-2, LoVo, HCT116 and SW480 cells) were
obtained from the American Type Culture Collection. All cell lines
were cultured in RPMI-1640 medium (HyClone; Cytiva) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Cells were
maintained in a humidified incubator at 37°C with 5%
CO2. Cells were transfected with 20 nM CCT6A knockdown
[short hairpin (sh)RNA-CCT6A#1 and shRNA-CCT6A#2] and interference
control (shRNA-NC), HOXB2 overexpression (Ov-HOXB2) and
overexpressed control group (Ov-NC), which were purchased from
Shanghai GeneChem, Co., Ltd. Polybrene (Shanghai GeneChem, Co.,
Ltd.) was used as a transfection reagent. Transfections were
performed using Lipofectamine® 2000 according to the
manufacturer's protocol. After transfection for 48 h at 37°C with
5% CO2, transfection efficiencies were assessed via
reverse transcription-quantitative PCR (RT-qPCR) and western
blotting after 48 h.
Databases
The GEPIA (http://gepia.cancer-pku.cn/) database predicted the
relationship between HOXB2 and CCT6A. The CCLE database
(portals.broadinstitute.org/ccle) detected the expression of CCT6A
in colon cancer cells. JASPAR (jaspar.genereg.net/) was used to
predict the relationship between HOXB2 and CCT6A.
Cell Counting Kit-8 (CCK-8)
assays
Cell suspensions were plated into 96 well plates
(8×103 cells/well). After cells had adhered, 10 µl CCK-8
solution (Dojindo Molecular Technologies, Inc.) was diluted using
culture medium and added to the 96-well plates and cells were
incubated for 1 h at 37°C. Finally, the absorbance at a wavelength
of 450 nm was detected using a spectrophotometer (Thermo Fisher
Scientific, Inc.).
Colony formation assays
Cell suspensions were plated in 60-mm culture dishes
(300 cells per dish) and cultured for 2 weeks at 37°C.
Subsequently, cells were fixed using 70% ethanol solution at room
temperature for 15 min, after which, cells were stained using
0.005% crystal violet at room temperature for 30 min (Thermo Fisher
Scientific, Inc.). The number of colonies with >50 cells was
counted by an inverted phase contrast light microscope
(magnification, ×200; Olympus Corporation).
Wound healing assays
Cells were plated (1×106 cells/well) in
6-well plates and cultured with serum-free medium for 12 h. Once
cells reached 80% confluence,, a tweezer was used to create a
scratch in the monolayer of cells and the images of the scratch
were captured using a light microscope (magnification, ×200;
Olympus Corporation). After a 24 h incubation in serum-free medium
at 37°C, images of the scratch were captured and the width was
measured using ImageJ software (version 146; National Institutes of
Health).
Transwell assays
Cells were cultured in serum-free medium for 12 h,
then suspended in culture medium. Matrigel (BD Biosciences) was
diluted with serum-free RPMI-1640 medium at 37°C for 30 min and
added to the upper chamber of a Boyden insert (8-µm pores; Corning,
Inc.). Subsequently, 600 µl RPMI-1640 medium supplemented with 10%
FBS was added to the lower chamber. The suspended cells
(5×105 cells/well) were added to the upper chamber and
incubated for 24 h at 37°C. The cells which had invaded through the
Matrigel and migrated across the membrane were fixed with 4%
paraformaldehyde for 15 min at room temperature and stained using
0.1% crystal violet at room temperature for 45 min and observed
using a light microscope (magnification, ×200; Olympus
Corporation).
Chromatin immunoprecipitation
(ChIP)
Total RNA was extracted using TRIzol®
buffer (Thermo Fisher Scientific, Inc.). RNA was reverse
transcribed into cDNA using the High Capacity cDNA Reverse
Transcription kit (cat. no. R022B; Takara Bio, Inc.), according to
the manufacturer's protocol (cat. no. R022B; Takara Bio, Inc.). DNA
(8–40 µg) was diluted with DNase-free water (cat. no. R0021;
Beyotime Institute of Biotechnology) and incubated with the primary
antibody (1–5 µg) at 4°C overnight. The primary antibody used was
an anti-HOXB2 antibody (1:200, cat. no. ab220390; Abcam). Next, the
DNA that had bound to HOXB2 was collected using DNA extraction
buffer in DNase-free water and amplified using qPCR to detect
CCT6A.
Luciferase reporter assay
Cell were plated in 6-well plates and after the
cells had adhered, 0.5 µg vectors containing the 3′-untranslated
region (UTR) of wild-type (WT) CCT6A or mutant (MUT) 3′-UTR CCT6A,
with control vector or HOXB2 overexpression vector and
pMIR-Renilla vector (Shanghai GeneChem Co., Ltd.) were
co-transfected with the aforementioned kit (Polybrene Shanghai
GeneChem, Co., Ltd.) into the cells (1×106 cells/well)
and cells were incubated for 48 h at 37°C. Finally, the luciferase
activity was detected using a Renilla-Glo®
Luciferase Assay System (cat. no. E2710; Promega Corporation) at
room temperature and a spectrophotometer at 490 nm (Thermo Fisher
Scientific, Inc.). Renilla luciferase activity was used to
normalize the firefly luciferase activity.
Immunocytochemistry
Cells (1×106 cells/well) were plated in
6-well plates and after they had adhered, 4% paraformaldehyde was
used to fix the cells at 4°C for 24 h. Subsequently, 0.5% Triton
X-100 (Beyotime Institute of Biotechnology) was used to
permeabilize the cells and 5% BSA (Beyotime Institute of
Biotechnology) was used to block non-specific binding at room
temperature for 1 h. A primary antibody was diluted with 5% BSA and
incubated with the cells at 4°C overnight. The primary antibody
used was an anti-Ki-67 (1:200; product code ab16667; Abcam). Next,
the cells were incubated at 37°C for 1.5 h with the goat polyclonal
secondary antibody to Rabbit IgG (heavy chain and light chain)
Alexa Fluor® 488 (1:3,000; cat. no. ab150077; Abcam) in
a dark room for 1.5 h. Finally, the nucleus of these cells was
stained with 1 mg DAPI for 10 min at room temperature (Invitrogen;
Thermo Fisher Scientific, Inc.). The fluorescence was observed
using a laser scanning confocal microscope (magnification, ×200;
Olympus Corporation).
RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). Subsequently, RNA was
reverse transcribed into cDNA using a commercial kit, according to
the manufacturer's protocol (cat. no. R022B; Takara Bio, Inc.).
cDNA was amplified using an ABI 7500 system (Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for qPCR: The thermocycling conditions were as follows: 95°C for 10
min, 40 cycles of 95°C for 10 sec, 55°C for 10 sec, and 72°C for 30
sec.qPCR was performed using a SYBR-Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). And the results were
analyzed with the 2−∆∆Cq method (16). The sequences of the primers used
were: CCT6A forward, 5′-TGACGACCTAAGTCCTGACTG-3′ and reverse,
5′-ACAGAACGAGGGTTGTTACATTT-3′; HOXB2 forward,
5′-CGAGTGACAAGGTGTAGCC-3′ and reverse, 5′-GTTGACCTTCTCTGGTAGG-3′;
GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Western blotting
Protein samples were collected using RIPA lysis
buffer (Beyotime Institute of Biotechnology). Next, the
concentration of these samples was determined using the BCA
(Beyotime Institute of Biotechnology) method. Proteins (30 µg/lane)
were resolved using 10% SDS-PAGE (Beyotime Institute of
Biotechnology) and subsequently transferred to a PVDF membrane
(MilliporeSigma). Next, these membranes were blocked with 5% BSA at
37°C for 1.5 h and incubated with primary antibodies at 4°C
overnight. The primary antibodies (1:1,000) used in the present
study were CCT6A (cat. no. ab110905), HOXB2, E-cadherin (cat. no.
ab40772), N-cadherin (cat. no. ab18203), vimentin (cat. no. ab8978)
and GAPDH (cat. no. ab8245) (all purchased from Abcam). The
following day, the membranes were incubated at 37°C for 1.5 h with
the secondary antibody HRP-conjugated anti-mouse antibody (goat
IgG; 1:5000; cat. no. ab150113; Abcam). Finally, the bands were
developed using ECL substrate (MilliporeSigma). Protein expression
levels were semi-quantified using ImageJ software (version 1.46;
National Institutes of Health).
Statistical analysis
Analysis was performed using GraphPad Prism version
7.0 (GraphPad Software, Inc.). Data are presented as the mean ±
standard deviation of three repeats. Differences between groups
were compared using an unpaired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
CCT6A expression is upregulated in
colon cancer tissues
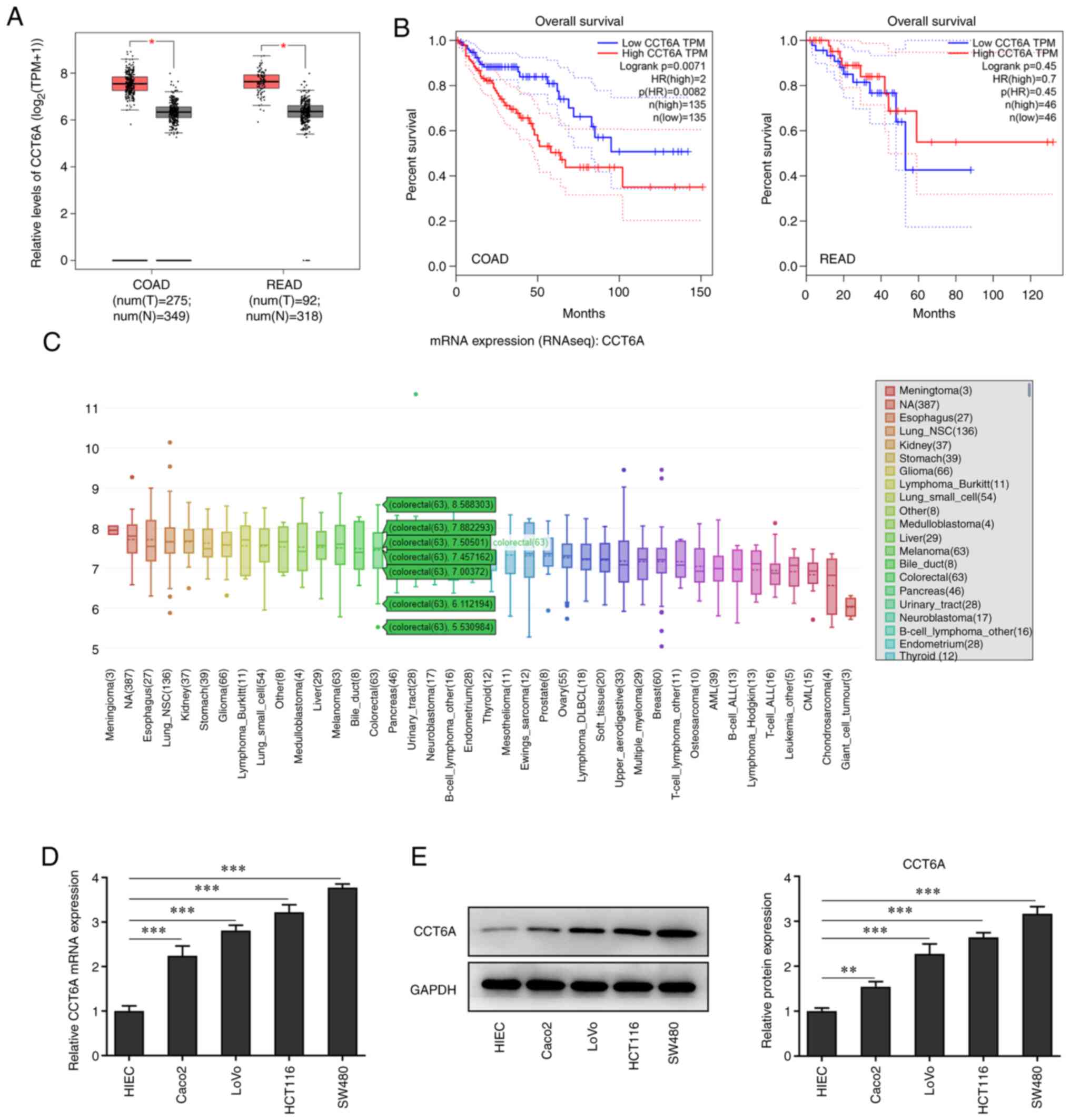
The expression of CCT6A in colon cancer tissues was
detected. Data from GEPIA revealed that the expression of CCT6A in
colorectal cancer tissues was higher than that in the
para-carcinoma tissues (Fig. 1A).
Furthermore, the data from GEPIA also revealed that higher levels
of CCT6A were associated with lower survival rates in these
patients (Fig. 1B). Data from the
CCLE database (portals.broadinstitute.org/ccle) showed that the
expression of CCT6A was also higher in colorectal cancer cells
compared with normal cells (Fig.
1C). Next, the expression of CCT6A in colon cancer cells was
detected using RT-qPCR (Fig. 1D)
and western blotting (Fig. 1E).
The results revealed that the expression level of CCT6A in colon
cancer cell lines (Caco2, LoVo, HCT116 and SW480 cells) was higher
compared with the HIEC cells.
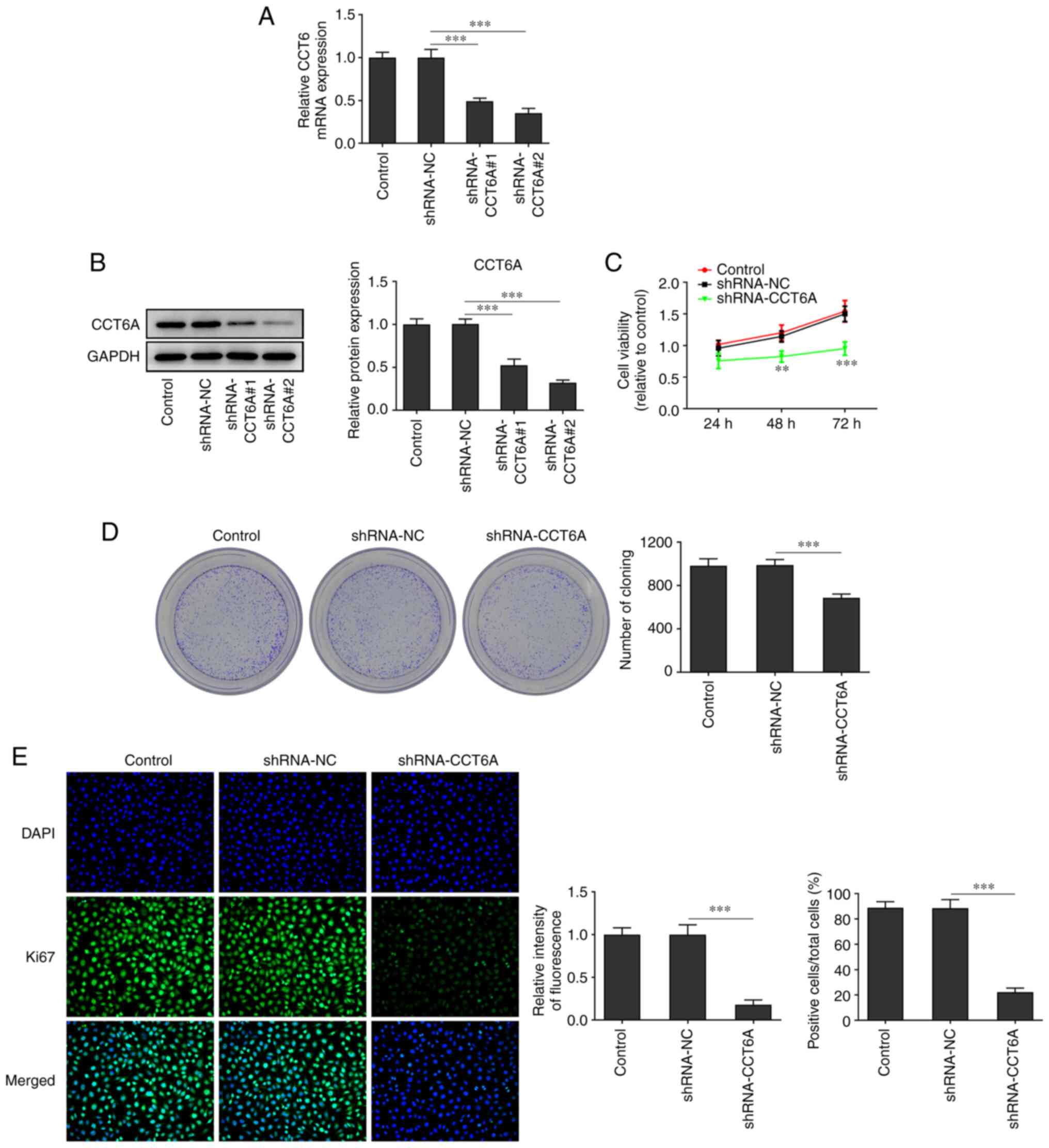
Knockdown of CCT6A suppresses the
proliferation of colon cancer cells
The expression of CCT6A in SW480 cells was higher
than that in the other colon cancer cells. Therefore, SW480 cells
were selected for subsequent experiments. CCT6A-knockdown SW480
cells were established. As revealed in Fig. 2A and B, the expression of CCT6A in
SW480 cells in the knockdown group was lower compared with the
control cells. The inhibitory effect of shRNA-CCT6A#2 was greater
than that of shRNA-CCT6A#1. Therefore, shRNA-CCT6A#2 was selected
for subsequent experiments. Next, the viability and proliferation
of these cells was detected using CCK-8 and colony formation
assays, respectively. The results revealed that knockdown of CCT6A
induced a decrease in viability and restricted the formation of
colonies of SW480 cells compared with the control cells (Fig. 2C and D). Next, the expression of
Ki-67 in these cells was also determined using immunofluorescence
analysis. The results revealed that knockdown of CCT6A suppressed
the expression of Ki-67 in SW480 cells compared with the control
cells (Fig. 2E).
Knockdown of CCT6A inhibits the
migration and invasion of colon cancer cells
Changes in the migratory and invasive abilities of
SW480 cells after the knockdown of CCT6A were next determined. The
wound healing and Transwell invasion assays revealed that the
migration and invasion of SW480 cells were reduced following
knockdown of CCT6A compared with the control cells (Fig. 3A-C). Furthermore, the expression
of N-cadherin and vimentin was also suppressed, whereas that of
E-cadherin was increased in the CCT6A-knockdown SW480 cells
compared with the control cells (Fig.
3D).
HOXB2 activates the expression of
CCT6A in colon cancer cells
Using JASPAR (jaspar.genereg.net/), it was predicted
that HOXB2 could bind to the promoter of CCT6A and affect the
expression of CCT6A (Fig. 4A and
B). According to the results of RT-qPCR and western blotting,
the expression of HOXB2 was higher in colon cancer cells (SW480
cells) compared with the HIEC cells (Fig. 4C and D). Next, HOXB2 was
overexpressed in SW480 cells, and the results revealed that the
expression of HOXB2 in SW480 cells in the overexpression group was
higher than that in cells of the negative control group (Fig. 4E and F). Moreover, luciferase
reporter assays revealed that the luciferase activity was highest
in the CCT6A WT and HOXB2 overexpression system (Fig. 4G). ChIP analysis demonstrated that
HOXB2 bound to the promoter region of CCT6A (Fig. 4H), and western blotting and
RT-qPCR analysis also revealed that overexpression of HOXB2 rescued
the expression of CCT6A in the CCT6A-knockdown SW480 cells compared
with the shRNA-CCT6A + OV-NC group (Fig. 4I and J).
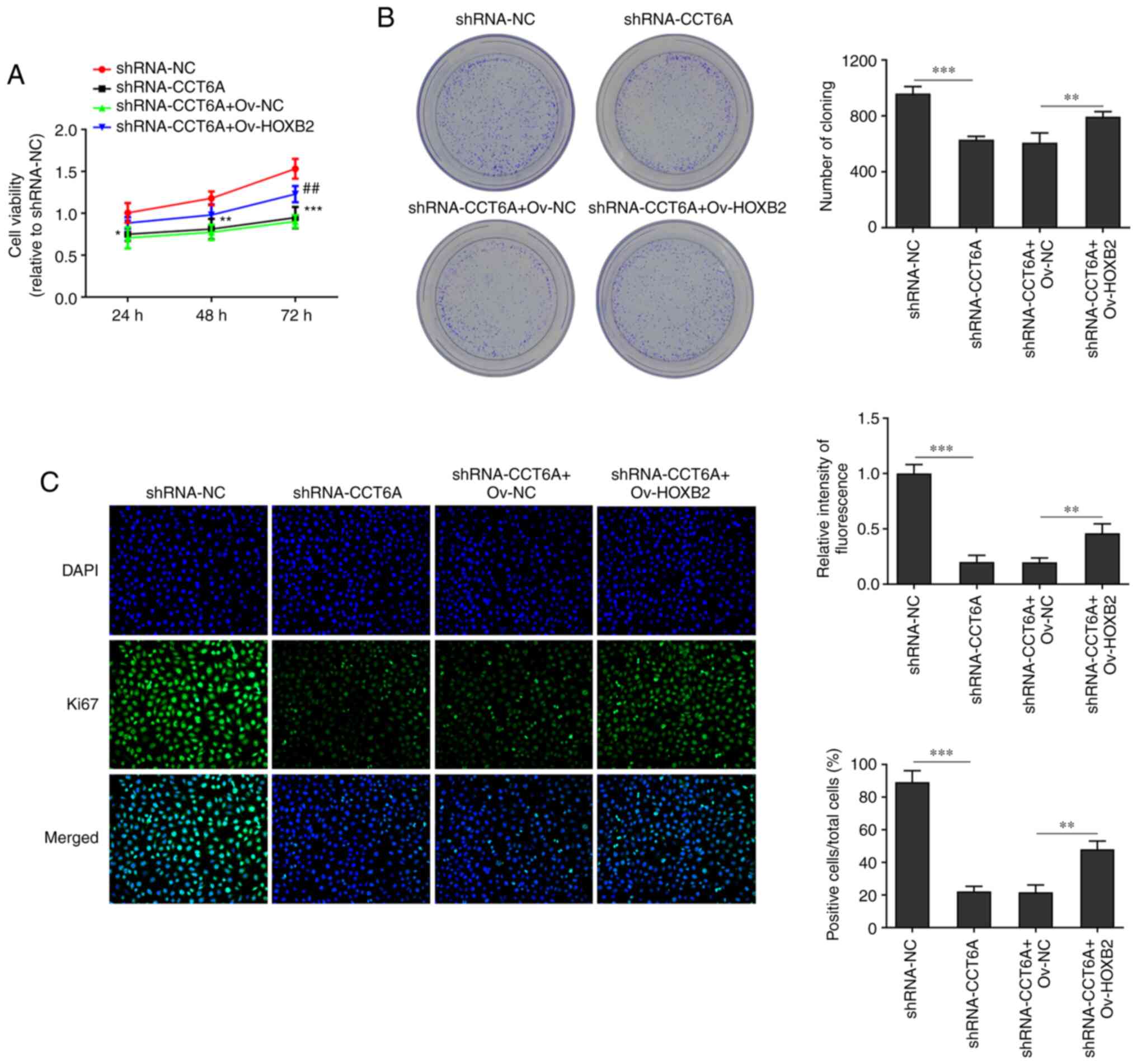
Overexpression of HOXB2 abolishes the
effects of CCT6A knockdown on the proliferation, migration and
invasion of colon cancer cells
The effect of HOXB2 on the proliferation, migration
and invasion of SW480 cells was next determined. The viability of
CCT6A-knockdown SW480 cells and number of colonies formed were
rescued following overexpression of HOXB2 compared with the
shRNA-CCT6A + OV-NC group (Fig. 5A
and B). Moreover, the expression of Ki-67 in CTT6A-knockdown
SW480 cells was rescued following overexpression of HOXB2 compared
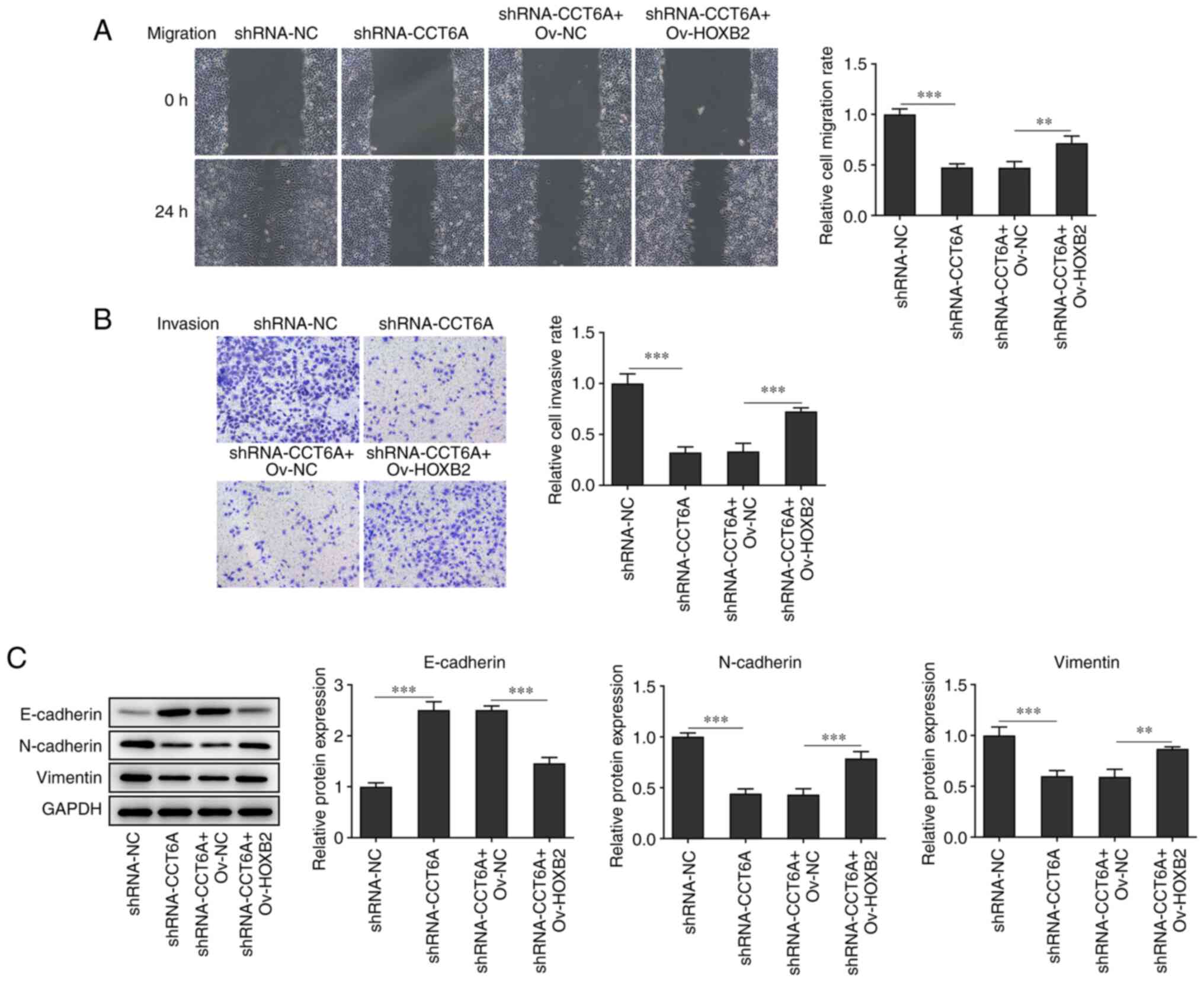
with the shRNA-CCT6A + OV-NC group (Fig. 5C). Furthermore, the wound healing
and Transwell assays revealed that overexpression of HOXB2
abolished the inhibitory effect of CCT6A knockdown on the migration
and invasion of SW480 cells compared with the shRNA-CCT6A + OV-NC
group (Fig. 6A and B). The
expression levels of N-cadherin and vimentin were rescued, whereas
the levels of E-cadherin in SW480 cells were decreased following
overexpression of HOXB2 compared with the shRNA-CCT6A + OV-NC group
(Fig. 6C).
Discussion
Colon cancer is a common malignant tumor of the
digestive tract and the incidence and mortality rates of colon
cancer have increased in recent years (17). With changes in lifestyle and
eating habits, the incidence of colon cancer in China is also
increasing and there is a trend of a decreasing age of onset
(18). Thus far, the molecular
mechanisms of colon cancer have not been fully elucidated. It has
been suggested that the onset of colon cancer is the result of a
combination of genetic and environmental factors (19). Moreover, environmental factors,
intestinal homeostasis, dietary choices, tobacco, alcohol and
physical exercise are crucial factors influencing the onset of
colon cancer (20). At present,
the clinical treatment of colon cancer is still based on surgery,
while chemotherapy and radiotherapy are used as adjuvant treatments
(21). However, the effect of
this treatment strategy is extremely limited for patients with
advanced colon cancer. The postoperative metastasis of colon cancer
cells is the critical cause of poor prognosis (22). Blood, peritoneal and distant lymph
node metastases are the primary means of postoperative colon cancer
metastasis (23). Through
invasion of cancer cells, new tumors may be formed and this will
lead to the deterioration of the condition. Therefore, there is an
urgent need to identify novel targets to suppress the metastasis of
colon cancer cells.
CCT6A is a protein that enhances the development of
multiple types of cancer (24).
Higher levels of CCT6A can promote the proliferation of
hepatocellular carcinoma cells by inducing the transition from the
G1 phase to the S phase (8). In addition, the expression of CCT6A
has been revealed to be upregulated in breast cancer tissues and is
associated with a poor prognosis of patients with breast cancer
(24). Additionally, the
expression of CCT6A has also been revealed to increase drug
resistance of melanoma cells and promote the proliferation of these
cells (25). Furthermore, CCT6A
was revealed to be an inhibitor of SMAD2; and it promoted the
proliferation and invasion of non-small cell lung cancer cells by
inhibiting the expression of SMAD2 (26). In the present study, it was
demonstrated that knockdown of CCT6A expression reduced the
proliferation, migration and invasion of colon cancer cells by
promoting the expression of EMT-related proteins (N-cadherin and
vimentin). These results also indicated that CCT6A is a promoting
factor of colon cancer.
HOXB2 is a transcription factor that has been
revealed to enhance the occurrence and development of ovarian
cancer (27). Aberrant expression
of HOXB2 has been observed in multiple types of cancer, such as
leukemia, breast and liver cancer and gastric carcinoma (28). Additionally, another study
indicated that upregulated expression of HOXB2 can induce the
occurrence and development of bladder cancer (11). At present, there has been no
literature report on the specific relationship between HOXB2 and
CCT6A, which is also the innovation of our study. The transcription
factor HOXB2 binding to the CCT6A promoter was revealed in the
JASPAR database. By querying UniProt and using a dual luciferase
assay, it was revealed that HOXB2 binds to the promoter region of
CCT6A and upregulates the expression of CCT6A in colon cancer
cells. Furthermore, overexpression of HOXB2 abolished the
inhibitory effect of CCT6A knockdown on the proliferation,
migration and invasion of colon cancer cells. These results
indicated that HOXB2 could promote the proliferation and
invasiveness of colon cancer cells by promoting the expression of
CCT6A.
The present study also has certain limitations. In
our experiments, SW480 cells were only selected and selecting
another cell line for verification will render the results more
rigorous. Another cell line will be selected for verification in a
future study. In addition, the results should be further verified
in animal experiments in the future. Finally, the downstream
regulation mechanism of HOXB2 and CCT6A should be further
explored.
In conclusion, the effect of CCT6A on the
proliferation, migration and invasion of colon cancer cells was
assessed. The results revealed that HOXB2 promoted the
proliferation and invasion of colon cancer cells by directly
targeting and activating the expression of CCT6A in colon cancer
cells. These results highlighted CC6TA and HOXB2 as novel
potentially druggable targets.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed generated during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
XY and LC wrote the manuscript and analyzed the
data. YT and WY carried out the experiments, supervised the present
study, searched the literature and revised the manuscript. All
authors read and approved the final manuscript. XY and LC confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang
J, Xia L, Yin Q, Zou B, Zheng J, et al: A novel protein encoded by
circFNDC3B inhibits tumor progression and EMT through regulating
Snail in colon cancer. Mol Cancer. 19:712020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Spaderna S, Schmalhofer O, Hlubek F, Berx
G, Eger A, Merkel S, Jung A, Kirchner T and Brabletz T: A
transient, EMT-linked loss of basement membranes indicates
metastasis and poor survival in colorectal cancer.
Gastroenterology. 131:830–840. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi M, Liu Y, Feng L, Cui Y, Chen Y, Wang
P, Wu W, Chen C, Liu X and Yang W: Protective effects of
scutellarin on human cardiac microvascular endothelial cells
against hypoxia-reoxygenation injury and its possible
target-related proteins. Evid Based Complement Alternat Med.
2015:2780142015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hallal S, Russell BP, Wei H, Lee MY, Toon
CW, Sy J, Shivalingam B, Buckland ME and Kaufman KL: Extracellular
vesicles from neurosurgical aspirates identifies chaperonin
containing TCP1 subunit 6A as a potential glioblastoma biomarker
with prognostic significance. Proteomics. 19:e18001572019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang J, Liu C, Xu G, Liang T, Yu C, Liao
S, Zhang Z, Lu Z, Wang Z, Chen J, et al: CCT6A, a novel prognostic
biomarker for Ewing sarcoma. Medicine (Baltimore). 100:e244842021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeng G, Wang J and Huang Y, Lian Y, Chen
D, Wei H, Lin C and Huang Y: Overexpressing CCT6A contributes to
cancer cell growth by affecting The G1-To-S phase transition and
predicts a negative prognosis in hepatocellular carcinoma. Onco
Targets Ther. 12:10427–10439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu M, Fu X, Si Z, Li C, Sun J, Du X and
Zhang H: Identification of differently expressed genes associated
with prognosis and growth in colon adenocarcinoma based on
integrated bioinformatics analysis. Front Genet. 10:12452019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Myung JK, Afjehi-Sadat L,
Felizardo-Cabatic M, Slavc I and Lubec G: Expressional patterns of
chaperones in ten human tumor cell lines. Proteome Sci. 2:82004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J, Li S, Cheng X, Du P, Yang Y and
Jiang WG: HOXB2 is a Putative tumour promotor in human bladder
cancer. Anticancer Res. 39:6915–6921. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inamura K, Togashi Y, Ninomiya H, Shimoji
T, Noda T and Ishikawa Y: HOXB2, an adverse prognostic indicator
for stage I lung adenocarcinomas, promotes invasion by
transcriptional regulation of metastasis-related genes in HOP-62
non-small cell lung cancer cells. Anticancer Res. 28:2121–2127.
2008.PubMed/NCBI
|
|
13
|
Clemenceau A, Boucherat O, Landry-Truchon
K, Lamontagne M, Biardel S, Joubert P, Gobeil S, Secco B, Laplante
M, Morissette M, et al: Lung cancer susceptibility genetic variants
modulate HOXB2 expression in the lung. Int J Dev Biol. 62:857–864.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Segara D, Biankin AV, Kench JG, Langusch
CC, Dawson AC, Skalicky DA, Gotley DC, Coleman MJ, Sutherland RL
and Henshall SM: Expression of HOXB2, a retinoic acid signaling
target in pancreatic cancer and pancreatic intraepithelial
neoplasia. Clin Cancer Res. 11:3587–3596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Zhu G, Xing Y, Zhu Y and Piao D:
MiR-4324 functions as a tumor suppressor in colorectal cancer by
targeting HOXB2. J Int Med Res. 48:3000605198837312020.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Walter V, Boakye D, Weberpals J, Jansen L,
Haefeli WE, Martens UM, Knebel P, Chang-Claude J, Hoffmeister M and
Brenner H: Decreasing use of chemotherapy in older patients with
stage iii colon cancer irrespective of comorbidities. J Natl Compr
Canc Netw. 17:1089–1099. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Shi J, Huang H, Ren J, Li N and
Dai M: Burden of colorectal cancer in China. Zhonghua Liu Xing Bing
Xue Za Zhi. 36:709–714. 2015.(In Chinese). PubMed/NCBI
|
|
19
|
Cheng B, Rong A, Zhou Q and Li W: LncRNA
LINC00662 promotes colon cancer tumor growth and metastasis by
competitively binding with miR-340-5p to regulate CLDN8/IL22
co-expression and activating ERK signaling pathway. J Exp Clin
Cancer Res. 39:52020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
AlMutairi M, Parine NR, Shaik JP, Aldhaian
S, Azzam NA, Aljebreen AM, Alharbi O, Almadi MA, Al-Balbeesi AO and
Alanazi M: Association between polymorphisms in PRNCR1 and risk of
colorectal cancer in the Saudi population. PLoS One.
14:e02209312019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhai X, Xue Q, Liu Q, Guo Y and Chen Z:
Colon cancer recurrence-associated genes revealed by WGCNA
co-expression network analysis. Mol Med Rep. 16:6499–6505. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Q, Meng WY, Jie Y and Zhao H: LncRNA
MALAT1 induces colon cancer development by regulating
miR-129-5p/HMGB1 axis. J Cell Physiol. 233:6750–6757. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang K, Zeng Y, Xie Y, Huang L and Wu Y:
Bioinformatics analysis of the prognostic value of CCT6A and
associated signalling pathways in breast cancer. Mol Med Rep.
19:4344–4352. 2019.PubMed/NCBI
|
|
25
|
Tanic N, Brkic G, Dimitrijevic B,
Dedovic-Tanic N, Gefen N, Benharroch D and Gopas J: Identification
of differentially expressed mRNA transcripts in drug-resistant
versus parental human melanoma cell lines. Anticancer Res.
26:2137–2142. 2006.PubMed/NCBI
|
|
26
|
Ying Z, Tian H, Li Y, Lian R, Li W, Wu S,
Zhang HZ, Wu J, Liu L, Song J, et al: CCT6A suppresses SMAD2 and
promotes prometastatic TGF-β signaling. J Clin Invest.
127:1725–1740. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu HY and Pan SS: MiR-202-5p suppressed
cell proliferation, migration and invasion in ovarian cancer via
regulating HOXB2. Eur Rev Med Pharmacol Sci. 24:2256–2263.
2020.PubMed/NCBI
|
|
28
|
Lee JY, Kim JM, Jeong DS and Kim MH:
Transcriptional activation of EGFR by HOXB5 and its role in breast
cancer cell invasion. Biochem Biophys Res Commun. 503:2924–2930.
2018. View Article : Google Scholar : PubMed/NCBI
|