Introduction
Breast cancer is the most common cancer among women
and is the second most frequently occurring newly diagnosed cancer
worldwide (1). The cause of
mortality in patients with breast cancer is often not the primary
tumor, but metastasis to distant organs (2). Since 1989, female breast cancer has
increased by 41%; the incidence of breast cancer is still
increasing at an annual rate of ~0.5% (3). Studies have shown that
chemotherapeutic drugs may promote tumor metastasis (4,5).
Therefore, there is a need for novel markers to screen patients at
high risk of breast cancer metastasis and novel targets for the
treatment of breast cancer metastasis should be explored.
REC8 is a member of the structural maintenance of
chromosomes protein complex adhesion (6). Adhesin serves an important role in
regulating chromosome separation and homologous recombination in
mitosis and meiosis, as well as in proliferation of eukaryotic
cells (7). It was previously
demonstrated that abnormal chromosome segregation is associated
with a variety of diseases; in particular, it serves a key role in
the occurrence and development of tumors (8). For example, in thyroid cancer, REC8
inhibits tumor cell proliferation and migration by targeting the
PI3K pathway (9). REC8
overexpression was shown to significantly suppress metastasis of
gastric cancer cells by downregulating early growth response (EGR)1
(10). Although REC8 serves as a
tumor suppressor in most types of tumor, to the best of our
knowledge, its role in breast cancer has not yet been studied.
Similar to REC8, cell division cycle (CDC)20 serves
an important role in chromosome separation and mitosis exit.
Activation of CDC20 promotes activation of anaphase-promoting
complex/cyclosome, which is a key regulator of mitotic duration
(11,12). Previous studies have shown that
CDC20 is highly expressed in a variety of malignant tumors and is
associated with the development and progression of tumors (13,14). For example, expression of CDC20 is
upregulated and associated with poor prognosis in breast cancer
(15). CDC20 may also be a
potential predictive indicator for prognosis of breast cancer with
co-expressed TPX2 gene, indicating that CDC20 inhibition may be of
value as a novel strategy for cancer treatment (16). However, whether REC8 affects the
progression of breast cancer by regulating CDC20 requires further
investigation.
In the present study, the expression of REC8 was
examined by reverse transcription-quantitative (RT-q)PCR analysis
in breast cancer cells. Furthermore, the role and underlying
mechanisms of REC8 on cell proliferation, migration and invasion
via targeting CDC20 were investigated by clone formation test,
wound healing assay and Transwell assay, respectively.
Materials and methods
Cell culture
The breast epithelial cell line MCF-10A and four
breast cancer cell lines (T47D, MCF-7, MDA-MB-231 and BT-549) were
purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. MCF-7 and T47d cell lines represent
luminal A condition [estrogen receptor (ER)+,
progesterone receptor (PR)+/− and human epidermal growth
factor receptor 2 (HER2−)], while MDA-MB-231 and BT-549
cell lines are triple-negative (ER−, PR− and
HER2−), All cells were cultured in DMEM/F12 Coon's
(DMEM/F12) containing 10% FBS (Hyclone; Cytiva), 100 U/ml
penicillin and 100 µg/ml streptomycin and maintained in a 5%
CO2 incubator at 37°C. The cells were passaged once
after 3 days. Cells in the logarithmic growth phase were used in
the subsequent experiments.
Cell transfection
For overexpression of REC8 and CDC20, pcDNA3.1
vector containing full-length REC8 (OV-REC8) and CDC20 (OV-CDC20)
and corresponding empty negative control vectors (OV-NC) were
designed and synthesized by Thermo Fisher Scientific, Inc.
Untransfected cells were used as the control.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to transfect plasmids into cells
(1×105 cells/well) at room temperature at 50 ng/ml
according to the manufacturer's instructions. After 12 h, the
medium was replaced with fresh DMEM and cells were routinely
cultured for 72 h at 37°C.
Bioinformatics
Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; string-db.org/) was used to predict
interactions between REC8 and CDC20. The expression of REC8 in
breast cancer and normal tissue was predicted by Gene Expression
Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn/).
Kaplan-Meier (KM) Plotter
(kmplot.com/analysis/index.php?p=background) was used to determine
the prognostic value of REC8 in breast cancer tissue.
RT-qPCR analysis
Total RNA was extracted from MCF-7 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
reverse transcribed to cDNA using a Reverse Transcription System
(Promega Corporation) according to the manufacturer's protocol. PCR
was performed using SYBR Green Supermix and the ABI 7500 PCR system
(both from Applied Biosystems; Thermo Fisher Scientific, Inc.). The
qPCR conditions were as follows: 94°C for 30 sec, 55°C for 30 sec
and 72°C for 30 sec (22 cycles). The relative expression of target
genes was calculated by the 2−ΔΔCq method (17) and normalized to the housekeeping
gene GAPDH. The sequences of PCR primers were as follows: REC8
forward, 5′-TACCTGCTCCTGGTGCTCTC-3′ and reverse,
5′-AGGTAAGCAGGACCCAGTGA-3′; CDC20 forward,
5′-GACCACTCCTAGCAAACCTGG-3′ and reverse, 5′-GGGCGTCTGGCTGTTTTCA-3′
and GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTA-3′. The primer sequences were obtained
using the PCR sequence design tool provided by National Center for
Biotechnology Information
(ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
Cell Counting Kit (CCK)-8 assay
Cell viability was detected by CCK-8 assay.
Transfected cells were incubated on 96-well plates at a density of
2×103 cells/well for 24, 48 and 72 h at 37°C. MCF-7
cells were incubated for 24, 48 and 72 h with the specific CDC20
inhibitor apcin (cat. no. 5747/10; R&D Systems, Inc.) dissolved
in PBS to concentrations of 25 and 50 µM. CCK-8 solution (10 µl;
cat. no. C0037; Beyotime Institute of Biotechnology) was added to
each well and incubated for an additional 2 h. The absorbance of
each well was measured at a wavelength of 450 nm using the Synergy
2 Multi-Mode Microplate Reader (BioTek Instruments, Inc.).
MTT viability assay
MTT was first prepared as a stock solution of 5
mg/ml in PBS (pH 7.2) and filtered. The cell suspension was added
to a 96-well plate with 5×103 cells/well, and then
treated with different concentrations of apcin (25 and 50 µM) for
24, 48 and 72 h, followed by the addition of 10 µl MTT solution to
each well. After incubation for 4 h at 37°C, 100 µl dimethyl
sulfoxide (Adamas-Beta, Ltd.) was added to each well. After
agitation, the 96-well plate was read by microplate reader at 570
nm for absorbance density values in order to determine the cell
viability. The viable cells produced a dark blue formazan product,
whereas no such staining was formed in the dead cells. The
percentage of the viable cells was calculated using the following
formula: [100× (sample absorbance)/(control absorbance)].
Colony formation assay
The MCF-7 cells were inoculated in 6-well plates at
a density of 4×102 cells/well for 14 days at 37°C.
Subsequently, the cells were fixed with 70% ethanol for 5 min and
then stained with 0.05% crystal violet solution for 20 min at room
temperature. The number of colonies formed (>50 cells) was
counted under an Olympus BX40 light microscope (Olympus
Corporation; magnification, ×100).
Wound healing assay
Cell migration was detected by wound healing assay.
MCF-7 cells were inoculated in 12-well plates at a density of
2×104 cells per well. Serum-free DMEM/F12 was used in
place of medium and the cells were incubated overnight at 37°C
under 5% CO2. When cell confluence reached 90%, a linear
wound was created in the cell monolayer using a 200-µl sterile
pipette tip. Images were captured at 0 and 48 h under an Olympus
BX40 light microscope (Olympus Corporation; magnification,
×100).
Transwell assay
Transwell assay was performed to assess cell
invasion. The 24-well Transwell plates (Corning, Inc.) with 8-µm
pore inserts were coated with Matrigel (BD Biosciences) at 37°C for
30 min. Cells in logarithmic growth phase (3×104) were
cultured in the top of Matrigel-coated invasion chambers (BD
Biosciences) filled with serum-free DMME/F12. DMEM/F12 supplemented
with 10% FBS was added to the lower chamber as a chemoattractant.
Following 24 h culture at 37°C with 5% CO2, cells that
had invaded to the bottom chamber were fixed with 80% ethanol for
30 min and stained with 0.1% crystal violet solution for 5 min at
room temperature. The stained cells were observed under a Leica DM
IL inverted light microscope (Leica Microsystems, Inc.;
magnification, ×100).
TUNEL assay
MCF-7 cells (2×105 cells/well) were
collected and washed three times with PBS. Following fixing with 4%
paraformaldehyde at room temperature for 20 min, cells were washed
twice with PBS. Subsequently, a small amount of DAPI staining
solution (cat. no. C1005; Beyotime Institute of Biotechnology) was
added to cover the cells and incubated at room temperature for 3–5
min. Triton X-100 (0.2%) was then added to cells at room
temperature for 5 min. Subsequently, 50 µl TUNEL assay solution
(Roche Diagnostics GmbH) was added to the cells and incubated at
37°C in the dark for 60 min. The detection solution was discarded
and cells were washed three times with PBS. Cells were sealed with
anti-fluorescence quenched sealing solution (cat. no. P0126;
Beyotime Institute of Biotechnology) and three fields of view were
randomly selected for observation under a fluorescence microscope
(Zeiss GmbH; magnification, ×200).
Western blot analysis
Total RNA was extracted from MCF-7 cells using RIPA
buffer (cat. no. P0013B; Beyotime Institute of Biotechnology). The
protein concentration was detected by BCA protein quantitative kit
(cat. no. P0012; Beyotime Institute of Biotechnology). Protein (20
µg/lane) was separated by 10% SDS-PAGE and transferred to PVDF
membranes (cat. no. FFP30; Beyotime Institute of Biotechnology).
The membranes were blocked with 5% skimmed milk at room temperature
for 30 min and incubated with primary antibodies (all 1:1,000; all
Abcam) against REC8 (cat. no. ab192241), CDC20 (cat. no. ab215908),
MMP-2 (cat. no. ab92536), MMP-9 (ab76003), Bcl-2 (cat. no.
ab182858), caspase-3 (cat. no. ab32351), cleaved caspase-3 (cat.
no. ab2302), cleaved poly(ADP-ribose) polymerase (PARP; cat. no.
ab32064) and GAPDH (cat. no. ab9485) overnight at 4°C.
Subsequently, membranes were incubated with goat anti-rabbit
horseradish peroxidase binding IgG (1:5,000; cat. no. ab6721;
Abcam) at 37°C for 2 h. The ECL Plus kit (cat. no. P0018; Beyotime
Institute of Biotechnology) was used to visualize the protein bands
and protein expression levels were semi-quantified using ImageJ
software (version 1.46; National Institutes of Health) with GAPDH
as the loading control.
Statistical analysis
Data were analyzed using GraphPad Prism 7 (GraphPad
Software, Inc.). One-way ANOVA followed by Tukey's post hoc test
was used to compare differences between multiple groups and
unpaired Student's t-test was used to analyze differences between
two groups. Data are presented as the mean ± standard deviation
from ≥3 independent experiments. P<0.05 was considered to
indicate a statistically significant difference.
Results
REC8 expression is downregulated in
breast cancer cells and tissue
Information on expression of REC8 in the serum of
patients with breast cancer was obtained via GEPIA (http://gepia.cancer-pku.cn/) and KM Plotter
(kmplot.com/analysis/index.php?p=service); low expression of REC8
was found to be positively associated with poor prognosis of
patients with breast cancer (Fig. 1A
and B). Therefore, the expression of REC8 in breast cancer
cells was measured by RT-qPCR and western blotting. REC8 was
downregulated in T47D, MCF-7, MDA-MB-231 and BT-549 breast cancer
cell lines compared with the MCF-10A breast epithelial cell line
(Fig. 1C and D). The expression
of REC8 was lowest in MCF-7 cells. Thus, MCF-7 cells were selected
for subsequent experiments. These results indicated that REC8 was
downregulated in breast cancer cells and tissue.
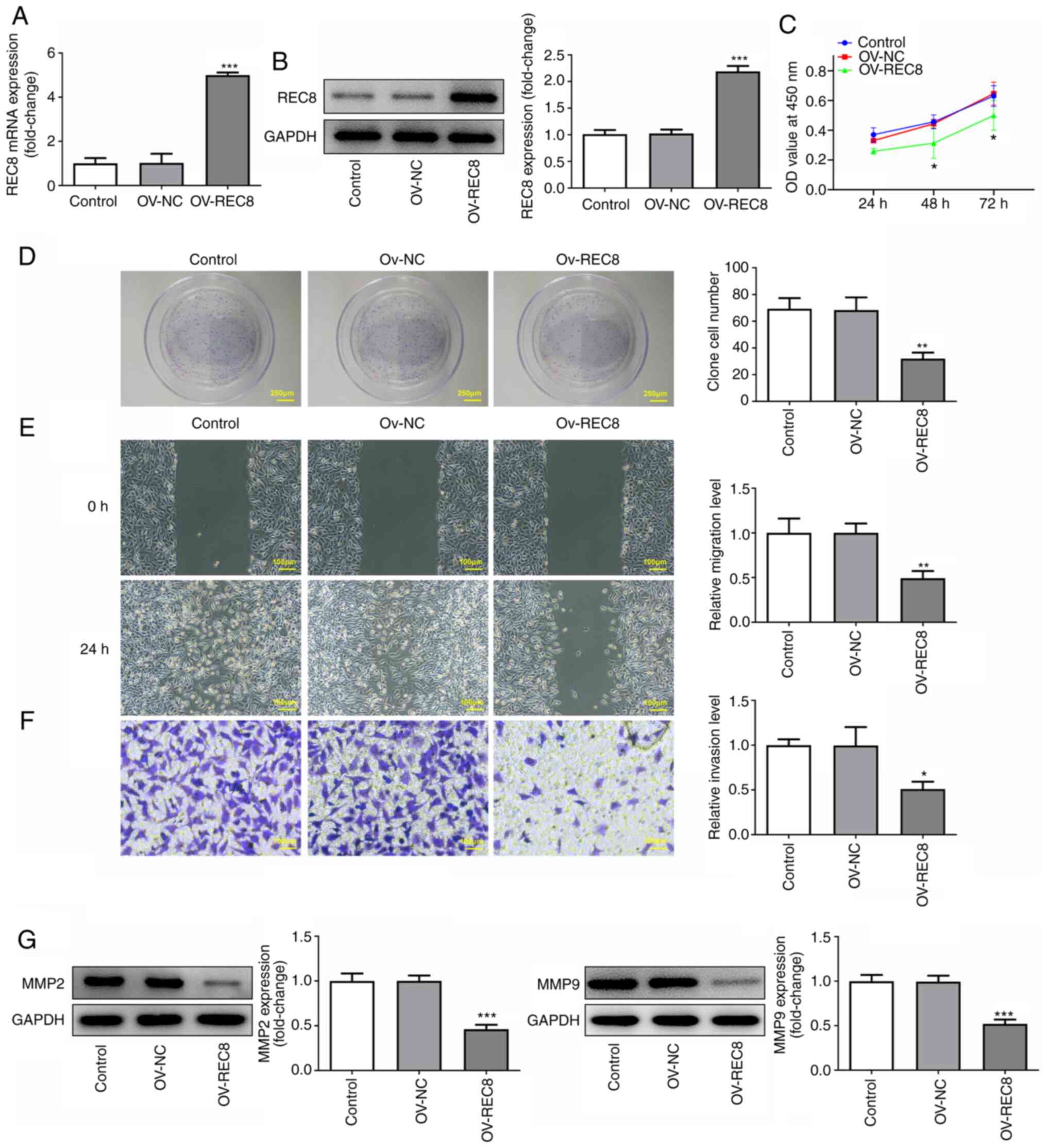
Overexpression of REC8 inhibits
proliferation, migration and invasion of MCF-7 cells
To study the specific role of REC8 in breast cancer,
REC8 was overexpressed in MCF-7 cells by transfection with
pcDNA-REC8. Protein and mRNA expression levels of REC8 in the
pcDNA-REC8 group were significantly higher compared with the
control (Fig. 2A and B). CCK-8
assay showed that overexpression of REC8 significantly inhibited
cell viability compared with the control at 48 and 72, but not 24,
h (Fig. 2C). Similar results were
obtained from the colony formation assay. Overexpression of REC8
significantly decreased the number of colonies compared with the
control (Fig. 2D). In addition,
cell migration and invasion were determined by wound healing and
Transwell assay, respectively. The results demonstrated that
overexpression of REC8 significantly decreased migration and
invasion of MCF-7 cells compared with the control (Fig. 2E and F). Furthermore, expression
levels of the invasion-associated factors MMP-2 and MMP-9 were
detected by western blotting. Overexpression of REC8 significantly
decreased expression of MMP-2 and MMP-9 compared with the control
(Fig. 2G). Taken together, these
findings indicated that overexpression of REC8 exerted an
inhibitory effect on proliferation, migration and invasion of MCF-7
cells.
Overexpression of REC8 induces
apoptosis in MCF-7 cells
The effect of REC8 on apoptosis of MCF-7 cells was
detected by TUNEL assay and western blotting. TUNEL staining assay
showed that overexpression of REC8 significantly enhanced apoptosis
of MCF-7 cells (Fig. 3A and B).
Compared with the control, overexpression of REC8 significantly
decreased the expression levels of the anti-apoptotic protein Bcl-2
but promoted expression of the pro-apoptotic proteins cleaved
caspase-3 and cleaved PARP (Fig.
3C). Taken together, these findings indicated that
overexpression of REC8 induced apoptosis in MCF-7 cells.
REC8 negatively regulates CDC20
expression
To determine the mechanism underlying the function
of REC8 in breast cancer, STRING was used to predict that CDC20 may
interact with REC8 (data not shown). Analysis of the association
between expression levels of REC8 and CDC20 in breast cancer cells
demonstrated that CDC20 was highly expressed in MCF-7 cells
(Fig. 4A and B) and
overexpression of REC8 significantly inhibited the expression of
CDC20 (Fig. 4C and D). Taken
together, these findings indicated that REC8 may negatively
regulate expression of CDC20 in MCF-7 cells.
REC8 suppresses proliferation,
migration and invasion and induces apoptosis of MCF-7 cells by
downregulating CDC20
To confirm whether REC8 serves an anticancer role by
inhibiting CDC20, an overexpression plasmid of CDC20 was
constructed; RT-qPCR and western blotting showed that expression
levels of CDC20 increased in MCF-7 cells (Fig. 5A and B). CCK-8, clone formation
assay, wound healing and Transwell experiments were performed to
test the effect of overexpressed CDC20 on proliferation, invasion
and migration of REC8-overexpressed cells. The results indicated
that overexpressed CDC20 significantly reversed the inhibitory
effect of overexpressed REC8 on cell proliferation, migration and
invasion (Fig. 5C-J).
As determined by TUNEL staining, the green
fluorescence of the OV-REC8 + OV-CDC20 group was significantly
reduced compared with that in the OV-REC8 group, indicating that
overexpression of CDC20 significantly reversed the pro-apoptotic
effects of OV-REC8 (Fig. 6A and
B). In addition, similar results were obtained when the
expression levels of apoptosis-related proteins (cleaved-PARP,
cleaved caspase-3 and Bcl-2) were further detected by western blot
analysis (Fig. 6C). These results
indicated that REC8 served an antitumor role by inhibiting
CDC20.
Apcin + OV-REC8 exerts an antitumor
effect
The effects of REC8 on proliferation, migration,
invasion and apoptosis of MCF-7 cells were detected following
treatment with CDC20 inhibitor apcin. The effect of apcin on cell
viability after treatment for 24, 48 and 72 h was detected by MTT
assay; the results showed that Apcin significantly inhibited the
viability of MCF-7 cells in a concentration-dependent manner
(Fig. 7A-C). Therefore, in
subsequent experiments, 25 µM apcin was selected to treat cells for
48 h. Compared with apcin alone, apcin + OV-REC8 showed a
significant inhibitory effect on MCF-7 cell viability (Fig. 7D). In addition, apcin + OV-REC8
effectively inhibited migration (Fig.
7E) and invasion (Fig. 7F) of
MCF-7 cells. Futhermore, western blot analysis showed that compared
with the Apcin 25 µM + OV-REC8 group, further overexpression of
REC8 decreased the expression of migration-related proteins MMP2
and MMP9. TUNEL staining (Fig. 8A and
B) and western blot (Fig. 8C)
detection of apoptosis-associated proteins (Bcl-2, cleaved PARP and
cleaved caspase-3) showed that apcin 25 µM + OV-REC8 significantly
promoted MCF-7 cell apoptosis compared with apcin 25 µM + OV-NC.
Taken together, these findings indicated that apcin + OV-REC8
exerted an antitumor effect.
Discussion
Breast cancer is the most common type of female
hormone-dependent malignancy. There are ~1.05 million new cases of
breast cancer annually, accounting for 23% of all female
malignancies; this incidence continues to rise at a rate of 3.1%
per year (18). Surgery,
chemotherapy, radiotherapy, endocrine therapy and targeted therapy
are the primary methods presently used in the clinical treatment of
breast cancer, but ~500,000 cases of breast cancer-associated
mortality are reported annually, accounting for 25% of all
malignant tumor mortality (19).
Therefore, the search for key molecules and genes involved in the
progression of breast cancer is crucial for its treatment.
REC8 is a component of mucin specifically expressed
during meiosis, which serves a key role in maintaining the
centromere and ensuring correct separation of chromosomes (6). There is a negative association
between the expression of REC8 and tumor occurrence, development,
deterioration and metastasis. Upregulation of REC8 is reported to
inhibit the proliferation and colony formation of thyroid cancer
cells (9). Among all types of
thyroid cancer, the greatest hypermethylation of REC8 occurs in the
most invasive anaplastic thyroid cancer, suggesting that REC8 may
be a novel tumor suppressor gene in thyroid cancer (9). REC8 is also hypermethylated in
Epstein-Barr virus-associated gastric cancer and overexpression of
REC8 significantly decreases cell viability, proliferation and
migration and induces early apoptosis to inhibit the occurrence and
development of gastric cancer (10,20). In addition, recent findings
demonstrated that REC8 inhibits proliferation and metastasis of
gastric cancer cells by downregulating EGR1 to inhibit
epithelial-to-mesenchymal transition in gastric cancer cells
(10). As a novel tumor
suppressor gene, REC8 has shown a good application prospect in
several types of tumors (9,21).
To the best of our knowledge, however, the role and mechanism of
action of REC8 in breast cancer has not been studied to date. In
the present study, REC8 expression was low in breast cancer cells
and overexpression of REC8 significantly inhibited the
proliferation, migration and invasion of MCF-7 cells and induced
apoptosis. These results demonstrated the inhibitory effect of REC8
on breast cancer.
To study the specific mechanism of REC8 in breast
cancer, STRING was used to show that CDC20 was associated with
REC8. CDC20 is a key cell cycle regulatory molecule in mitosis and
promotes mitosis by regulating ubiquitination and degradation of
isolated inhibitor protein and cyclin B (22). In addition, CDC20 is upregulated
in pancreatic cancer (23), lung
cancer (24) and hepatocellular
carcinoma (25), and predicts a
poor prognosis. CDC20 not only affects proliferation of tumor
cells, but expression of CDC20 is also positively associated with
the invasive ability of breast cancer cells (15). To clarify the effect of the
interaction between CDC20 and REC8 on the progression of breast
cancer, the effect of CDC20 overexpression on proliferation,
migration, invasion and apoptosis of MCF-7 cells overexpressing
REC8 was assessed. The results showed that overexpression of CDC20
significantly reversed the inhibitory effect of REC8 overexpression
on proliferation, migration and invasion of MCF-7 cells and
inhibited apoptosis of MCF-7 cells, indicating that REC8 exerted an
antitumor effect by inhibiting CDC20. Furthermore, the present
study demonstrated that the combination of apcin and OV-REC8
effectively inhibited the viability, proliferation, migration and
invasion of MCF-7 cells and promoted cell apoptosis.
In conclusion, the present study demonstrated that
REC8 inhibited proliferation, migration and invasion and induced
apoptosis of breast cancer cells by downregulating the expression
of CDC20. To the best of our knowledge, the present study is the
first to propose a mechanism by which REC inhibits breast cancer by
regulating CDC20. However, the present study did not determine the
molecular mechanism underlying REC8 inhibition of CDC20 expression.
It is also necessary to verify the present results using other
breast cancer cell lines as well as in vivo animal models.
In addition, the role of apcin in REC8 knockdown cells will be
investigated in future.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZHC and SDH conceptualized and designed the current
study. DPL, ZHC and SDH acquired, analyzed and interpreted data.
ZHC and SDH drafted the manuscript and revised it critically for
important intellectual content. All authors agreed to be held
accountable for the current study in ensuring questions related to
the integrity of any part of the work are appropriately
investigated and resolved. ZHC and SDH confirm the authenticity of
all the raw data. All authors read approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ullah MF: Breast cancer: Current
perspectives on the disease status. Adv Exp Med Biol. 1152:51–64.
2019. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Alterio C, Scala S, Sozzi G, Roz L and
Bertolini G: Paradoxical effects of chemotherapy on tumor relapse
and metastasis promotion. Semin Cancer Biol. 60:351–361. 2020.
View Article : Google Scholar
|
|
5
|
Keklikoglou I, Cianciaruso C, Güç E,
Squadrito ML, Spring LM, Tazzyman S, Lambein L, Poissonnier A,
Ferraro GB, Baer C, et al: Chemotherapy elicits pro-metastatic
extracellular vesicles in breast cancer models. Nat Cell Biol.
21:190–202. 2019. View Article : Google Scholar
|
|
6
|
Han J, Bai Y, Wang J, Xie XL, Li AD, Ding
Q, Cui ZJ, Yin J, Jiang XY and Jiang HQ: REC8 promotes tumor
migration, invasion and angiogenesis by targeting the PKA pathway
in hepatocellular carcinoma. Clin Exp Med. 21:479–492. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Sohaily S, Biankin A, Leong R, Corish
MK and Warusavitarne J: Molecular pathways in colorectal cancer. J
Gastroenterol Hepatol. 27:1423–1431. 2012. View Article : Google Scholar
|
|
8
|
Ly P, Brunner SF, Shoshani O, Kim DH, Lan
W, Pyntikova T, Flanagan AM, Behjati S, Page DC, Campbell PJ and
Cleveland DW: Chromosome segregation errors generate a diverse
spectrum of simple and complex genomic rearrangements. Nat Genet.
51:705–715. 2019. View Article : Google Scholar
|
|
9
|
Liu D, Shen X, Zhu G and Xing M: REC8 is a
novel tumor suppressor gene epigenetically robustly targeted by the
PI3K pathway in thyroid cancer. Oncotarget. 6:39211–39224. 2015.
View Article : Google Scholar
|
|
10
|
Zhao J, Geng L, Duan G, Xu W, Cheng Y,
Huang Z, Zhou Z and Gong S: REC8 inhibits EMT by downregulating
EGR1 in gastric cancer cells. Oncol Rep. 39:1583–1590.
2018.PubMed/NCBI
|
|
11
|
Gu Q, Li F, Ge S, Zhang F, Jia R and Fan
X: CDC20 Knockdown and acidic microenvironment collaboratively
promote tumorigenesis through inhibiting autophagy and apoptosis.
Mol Ther Oncolytics. 30:94–106. 2020. View Article : Google Scholar
|
|
12
|
Guo C, Kong F, Lv Y, Gao N, Xiu X and Sun
X: CDC20 inhibitor apcin inhibits embryo implantation in vivo and
in vitro. Cell Biochem Funct. 6:810–816. 2020. View Article : Google Scholar
|
|
13
|
Zhang Q, Huang H, Liu A, Li J, Liu C, Sun
B, Chen L, Gao L, Xu D and Su C: Cell division cycle 20 (CDC20)
drives prostate cancer progression via stabilization of β-catenin
in cancer stem-like cells. EBioMedicine. 42:397–407. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karra H, Repo H, Ahonen I, Löyttyniemi E,
Pitkänen R, Lintunen M, Kuopio T, Söderström M and Kronqvist P:
Cdc20 and securin overexpression predict short-term breast cancer
survival. Br J Cancer. 12:2905–2913. 2014. View Article : Google Scholar
|
|
15
|
Alfarsi LH, Ansari RE, Craze ML, Toss MS,
Masisi B, Ellis LO, Rakha EA and Green AR: CDC20 expression in
oestrogen receptor positive breast cancer predicts poor prognosis
and lack of response to endocrine therapy. Breast Cancer Res Treat.
178:535–544. 2019. View Article : Google Scholar
|
|
16
|
Cheng L, Huang YZ, Chen WX, Shi L, Li Z,
Zhang X, Dai XY, Wei JF and Ding Q: Cell division cycle
proteinising prognostic biomarker of breast cancer. Biosci Rep.
40:BSR201912272020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guerrero-Preston R, Hadar T, Ostrow KL,
Soudry E, Echenique M, Ili-Gangas CL, Pérez G, Perez J, Mieville
PB, Deschamps .et al: Differential promoter methylation of kinesin
family member 1a in plasma is associated with breast cancer and DNA
repair capacity. Oncol Rep. 32:505–512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu J, Liang Q, Wang J, Gao J, Zhang J,
Zeng Y, Chiu PWY, Ng EKW and Sung JJY: REC8 functions as a tumor
suppressor and is epigenetically downregulated in gastric cancer,
especially in EBV-positive subtype. Oncogene. 36:182–193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu M, Xu W, Su M and Fan P: REC8
suppresses tumor angiogenesis by inhibition of NF-κB-mediated
vascular endothelial growth factor expression in gastric cancer
cells. Biol Res. 53:412020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kapanidou M, Curtis NL and Bolanos-Garcia
VM: Cdc20: At the crossroads between chromosome segregation and
mitotic exit. Trends Biochem Sci. 42:193–205. 2017. View Article : Google Scholar
|
|
23
|
Li D, Zhu J, Firozi PF, Abbruzzese JL,
Evans DB, Cleary K, Friess H and Sen S: Overexpression of oncogenic
STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin Cancer
Res. 9:991–997. 2003.PubMed/NCBI
|
|
24
|
Zhang W, Gong W, Ai H, Tang J and Shen C:
Gene expression analysis of lung adenocarcinoma and matched
adjacent non-tumor lung tissue. Tumori. 100:338–345. 2014.
|
|
25
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar
|