Introduction
Bladder cancer is the fourth leading cause of
cancer-related mortality worldwide. In 2012, the economic burden
associated with bladder cancer was 4.9 billion in the European
Union, accounting for 5% of the total health care costs for cancer
(1). In China in 2015, it was
estimated that there were 66,8000 new cases of bladder cancer,
which led to a considerable loss in productivity and a heavy
economic burden, also due to the fact that the nature of cancer
requires lifetime surveillance (2). The diagnosis, treatment and 5-year
survival rates of patients with bladder cancer have greatly
improved over the past 30 years (3). However, patient outcomes,
particularly those of patients who are not responsive or intolerant
to chemo-immunotherapy remain poor. Therefore, there is an urgent
need for the development of novel markers which can be used to
identify the most effective treatment regimens for patients.
Genomic instability is a trademark of cancer
(4,5). Pathological studies show that 70–75%
of newly diagnosed bladder cancer cases are non-muscle-invasive
bladder cancers (NMIBCs) and the remainder are classified as
muscle-invasive bladder cancers (MIBCs). NMIBCs are easily
manageable; however, ~15% of high-grade cases progress to MIBC
(6). Studies demonstrate that
non-homologous end joining (NHEJ) is involved in double-strand
break repair in MIBCs; in MIBCs, the failure to repair stalled
replication forks is observed (7,8).
As proteins directly participate in these molecular alterations and
mediate cancer cell biology mechanisms, proteins may be more
accurate and specific markers for cancer (9).
The human RecQ helicases are a highly conserved
protein family which serve vital roles in DNA metabolism and
genetic stability. There are five human RecQ helicases and they
contain a deeply conserved helicase domain that can uncoil double
strand structure in an ATP-dependent and 3′-to-5′ manner (10). Bloom syndrome protein (BLM), also
known as RECQL2, belongs to this family. The mutation of BLM leads
to Bloom syndrome, which is mostly characterized by a
predisposition to developing various types of cancers, including
bladder cancer (11,12). BLM interacts with proteins
involved in replication fork migration and the NHEJ pathway. Due to
the lack of BLM, human epithelial cells exhibit hyper-recombination
and mice or yeast exhibit a cancer-prone phenotype (13,14).
The loss or inactivation of BLM has been shown to
lead to cancer development via structural changes in oncogenes or
tumor suppressor genes (15,16). It has been demonstrated that BLM
may serve important roles in the progress of oncogenesis (17). Furthermore, previous studies have
found that the nonsense mutation of BLM increases the risk of
developing breast or ovarian cancer (18,19). RecQ helicase expression has also
been used to predict whether cancers are sensitive to DNA-damaging
chemotherapeutic agents (20,21). However, to date, to the best of
our knowledge, there are no reports on the mechanisms of action of
BLM in bladder cancer. Thus, in order to elucidate the role of BLM
in bladder cancer, the present study analyzed BLM expression
patterns in bladder cancer and matched adjacent healthy tissues. In
addition, in order to further determine the functions of BLM in
bladder cancer, the cell cycle, proliferation and apoptosis, as
well as sensitivity to chemotherapeutic drugs was analyzed in an
in vitro model.
Materials and methods
Tissue microarray (TMA) and
immunohistochemical (IHC) analysis
TMA chips that covered 68 bladder cancer tissue
specimens and 54 adjacent normal bladder cancer specimens were
purchased from Shanghai Outdo Biotech Co., Ltd. The present study
was approved by the ethics committee of Beijing Chao-Yang Hospital,
Capital Medical University (approval number 2017-ke-47) and was
conducted in accordance with the Declaration of Helsinki. Patients
gave written consent for their information to be stored on the
hospital database and to be used in research. Immunohistochemical
staining was performed according to the instructions of the S-P kit
(OriGene Technologies, Inc.). The TMAs were firstly fixed with 4%
paraformaldehyde and blocked with 10% goat serum for 30 min at room
temperature. Then, the TMAs were stained with BLM primary antibody
(cat. no. NBP1-89929; Novus Biologicals, Ltd.) at a dilution of
1:100 overnight at room temperature. Negative and positive controls
(by omission of the primary antibody and IgG-matched serum) were
included in each test.
Evaluation of immunohistochemistry
score
The BLM protein staining scores were evaluated by
two experienced pathologists and the concordance between the two
pathologists was excellent. The whole field of the section was
scored and intensities of staining were grouped as follows: 0 = no
staining, 1 = weak staining, 2 = moderate staining, 3 = strong
staining. The cut-off value used to separate patients into high and
low BLM expression groups was ≥2.
Cell lines and culture
Human bladder cancer cell lines (J82 and 5637) were
obtained from China Infrastructure of Cell Line Resources. Cells
were cultured routinely in RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) in a standard culture conditions
(5% CO2 at 37°C).
Transient siRNA transfection
Cells were transfected with 20 nM negative control
[short interfering (si)RNA-CON] or BLM siRNA (siRNA-BLM) (sense:
5′-CCCACUACUUUGCAAGUAATT-3′, antisense:
5′-UUACUUGCAAAGUAGUGGGAA-3′) according to the manufacturer's
instructions (Ambion; Thermo Fisher Scientific, Inc.). Briefly,
15×104 cells were seeded and cultured in 6-well plates
and medium was changed to Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) at ~70% confluence. Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) and siRNA-CON or siRNA-BLM
was diluted into Opti-MEM separately and incubated for 5 min at
room temperature then mixed and incubated for 20 min to allow the
formation of lipid-siRNA complex. After 6 h incubation, the medium
was changed to RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.).
After 48 to 72 h, the cells were harvested and used for
experiments.
Cell viability assay
Cell viability was assessed by the Cell Counting
Kit-8 (CCK-8) (Beyotime Institute of Biotechnology) for three times
independently. Briefly, the cells was plated into a 96 well plate
and treated with corresponding agents for the indicating time.
CCK-8 reagent (10 µl/well) was added to each well and incubated for
2 h. Then the cell viability was determined by using a micro-plate
reader at a 450 nm wavelength (Multiskan; Thermo Fisher Scientific,
Inc.).
Cell proliferation assay
A total of 4×104 cells were cultured in
96-well plates and exposed to 50 µM EdU solution (Guangzhou RiboBio
Co., Ltd.) for 4 h at 37°C and then fixed in 4% formaldehyde
overnight at 4°C and permeabilized in 0.5% Triton X-100 for 10 min.
Cells were incubated with 100 µl Apollo reaction cocktail for 30
min at room temperature in the dark and DNA was stained with
Hoechst 33342 (100 µl/well) for 30 min at room temperature in the
dark. The stained cells were analyzed using a fluorescent
microscope (6 fields were selected randomly; magnification,
×40).
Cell cycle assay
A total of 1×106 cells were fixed with
pre-cooled 70% ethanol and treated with 10 µg/ml RNase
(Sigma-Aldrich; Merck KGaA), then the cells incubated at 37°C for
30 min before staining with 50 µg/ml propidium iodide (PI) for 30
min at 4°C (Invitrogen; Thermo Fisher Scientific, Inc.). Cell cycle
distribution was analyzed using a Gallios flow cytometer (Beckman
Coulter, Inc.) and ModFit software version 3.2 (Verity Software
House) was utilized for cell cycle analysis.
Cell apoptosis assay
The experiment was performed following the protocols
described by the kit manufacturer (BD Biosciences). A total of
5×105 cells were harvested and re-suspended in binding
buffer at a concentration of 1×106 cells/ml. 100 µl of
the single cells suspension was mixed with 5 µl of Annexin V-FITC
and 5 µl of PI and further incubated for 15 min at room temperature
in the dark. Finally, 400 µl of binding buffer was added to the
mixture and the cells were analyzed by a Gallios flow cytometer
(Beckman Coulter, Inc.). The FlowJo software (BD Biosciences;
Version 7.6) was used to analyze the early apoptosis rate, late
apoptosis rate and total apoptosis rate respectively. The cells
undergoing early apoptosis were defined as Annexin
V-FITC-positive/PI-negative, and the late apoptotic cells were
defined as Annexin V-FITC-positive/PI-positive. The total apoptosis
rate was the percentage of early + late apoptotic cells.
IC50 determination
The transfected cells were seeded into a 96-well
plate at a density of 5×104 cells per well. Then the
cells were treated with different concentrations of cisplatin (0,
2, 4 and 8 µmol/l) for different periods of time (12, 24 and 48 h).
The IC50 value of cisplatin in the J82 and 5637 was
calculated at 24 h by probit regression. Cisplatin was obtained
from Qilu Pharmaceutical Co., Ltd.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from 5×105 cells
using TRIzol reagent (Thermo Fisher Scientific, Inc.), in
accordance with the manufacturer's instructions. RT-qPCR was
performed according to the manufacturer's instructions (Taq
Plantinum PCR MasterMix, Tiangen Biotech Co., Ltd.). The primers
were designed using the software Primer Premier (BLM: forward
5′-GGATCCTGGTTCCGTCCGC-3′, reverse 5′-CCTCAGTCAAATCTATTTGCTCG-3′,
β-actin: forward: 5′-TGACGTGGACATCCGCAAAG-3′, reverse:
5′-TCTTCATTGTGCTGGGTGCC-3′). The PCR program included a cycle of
95°C for 15 min, followed by 40 cycles of denaturation at 95°C for
10 sec, annealing/extension at 60°C for 32 sec. All samples were
normalized against the internal control (β-actin) and analyzed
using the 2−ΔΔCq method (22).
Western blot analysis
Cells were lysed with RIPA buffer (10 mM Tris-HCl,
pH 7.5; 150 mM NaCl; 1% NP-40; 0.25% sodium deoxycholate) and
quantified using the BCA protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts (40 µg) protein were separated on a
10% SDS-polyacrylamide gel and electrotransferred to PVDF membranes
(MilliporeSigma). Membranes were immersed in blocking buffer with
10% fat free milk for 1 h at room temperature. The blocked PVDF
membrane was incubated with antibodies against BLM (1:1,000; cat.
no. 2742; Cell Signaling Technology, Inc.) for 3 h at room
temperature. Following incubation with the primary antibodies, the
membrane was incubated with horseradish peroxidase-conjugated goat
anti-rabbit or anti-mouse secondary antibodies for 1 h (1:5,000;
cat. nos. 31466 and PA174421; Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature. Protein bands were detected by using
enhanced chemiluminescence. β-actin (1:1,000; cat. no. 3700; Cell
Signaling Technology, Inc.) was used as the loading control.
Proteins bands were visualized using an ECL reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), and the immunoblots were
quantified using ImageJ software (version 3.0; National Institutes
of Health).
Statistical analysis
Two groups were compared using Mann Whitney test if
data were non-parametric or using unpaired Student's t-test if data
were parametric. Multiple groups were compared using one-way
analysis of variance, followed by Sidak's or Tukey's multiple
comparisons test. Survival curves were estimated using the
Kaplan-Meier analysis and were compared using the log-rank test. A
Cox regression model was used to identify prognostic factors for
survival of patients with bladder cancer. The contingency table
presented in Table I was analyzed
using a chi-squared test or Fisher's exact test when the expected
count in <20% of the cells of the analyzed contingency table is
5 or fewer (i.e. age, gender, differentiation, muscle invasion and
lymph node metastasis). P<0.05 was considered to indicate a
statistically significant difference.
 | Table I.Association between BLM expression
and clinicopathological characteristics in bladder cancer
patients. |
Table I.
Association between BLM expression
and clinicopathological characteristics in bladder cancer
patients.
|
|
| BLM expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Number | High (n=19) | Low (n=49) | P-value |
|---|
| Age (years) |
|
|
|
|
|
<60 | 17 | 4 | 13 | 0.761 |
|
≥60 | 51 | 15 | 36 |
|
| Sex |
|
|
|
|
|
Male | 57 | 17 | 40 | 0.715 |
|
Female | 11 | 2 | 9 |
|
| Pathology
grade |
|
|
|
|
|
I–II | 22 | 6 | 16 | 1.000 |
|
>II | 46 | 13 | 33 |
|
|
Differentiation |
|
|
|
|
| Well,
moderate | 65 | 18 | 47 | 1.000 |
|
Poor | 3 | 1 | 2 |
|
| Muscle
invasion |
|
|
|
|
| No | 11 | 3 | 8 | 1.000 |
|
Yes | 57 | 16 | 41 |
|
| Tumor invasion
depth |
|
|
|
|
| Tis,
T1, T2 | 39 | 13 | 26 | 0.287 |
| T3,
T4 | 29 | 6 | 23 |
|
| Lymph node
metastasis |
|
|
|
|
| No | 59 | 17 | 42 | 1.000 |
|
Yes | 9 | 2 | 7 |
|
| Distant
metastasis |
|
|
|
|
| M0 | 68 | 19 | 49 | - |
| M1 | 0 | 0 | 0 |
|
| AJCC stage |
|
|
|
|
|
I–II | 41 | 15 | 26 | 0.059 |
|
>II | 27 | 4 | 23 |
|
Results
Differential expression of BLM in
bladder cancerous and non-cancerous tissues
A total of 68 bladder cancer and 54 adjacent healthy
bladder tissues were analyzed in the present study. The patient
demographics are summarized in Table
I. The expression of BLM in cancer tissues was assessed using
immunohistochemistry. Bladder cancer tissues expressed a high level
of BLM, while the adjacent healthy tissue expressed a low level of
BLM (Fig. 1). When the
immunostaining scores of BLM were further compared between
cancerous tissue and adjacent healthy tissue, a significantly
higher expression level of BLM was found in the cancerous tissues
in all patients (P<0.0001) or in patients with bladder cancer of
grade II or higher (P<0.001; Fig.
1B and C). These results suggested that the expression of BLM
was significantly higher in bladder cancer tissues than in adjacent
normal tissues.
Survival analysis was then performed for the
patients with bladder cancer. In the low BLM expression group
(non-staining group and weak staining group), 33 of the 49 (67.35%)
patients survived, while in the high BLM expression group (moderate
staining group and strong staining group), 8 of the 19 (42.10%)
patients survived. There was a significant difference in the
survival rate between the high and low BLM expression groups
(P=0.013; Fig. 1D).
In Cox regression analysis, two prognostic factors
for the overall survival of patients with bladder cancer were
identified using univariate analysis: The American Joint Committee
on Cancer (AJCC) stage (I–II vs. >II, P=0.008) and TNM stage
(I/II vs. III/V, P=0.001). However, the BLM expression level (low
vs. high) was not a statistically significant prognostic factor
(P=0.981). Furthermore, multivariate analysis revealed that the
AJCC (P=0.030) and TNM stage (P=0.008) were significant independent
predictors of the poor survival of patients with bladder cancer
(Table II). Additionally, a
nomogram of the median survival time was drawn, containing BLM
expression, age and sex (data not shown).
 | Table II.Risk factors associated with overall
survival of bladder cancer patients. |
Table II.
Risk factors associated with overall
survival of bladder cancer patients.
| Variables | HR (95% CI) | P-value |
|---|
| Age | 1.004
(0.972-1.036) | 0.802 |
| Gender | 0.854
(0.389-1.876) | 0.694 |
|
Differentiation | 0.496
(0.237-1.040) | 0.063 |
| Neural
invasion | 1.787
(0.858-3.723) | 0.121 |
| TNM stage | 2.345
(1.087-5.057) | 0.030a |
| AJCC | 3.040
(1.432-6.457) | 0.008a |
| BLM expression | 2.439
(0.732-5.255) | 0.981 |
Expression and silencing of BLM in
bladder cancer lines
To investigate the role of BLM in bladder cancer,
the expression of BLM was first assessed in two bladder cancer
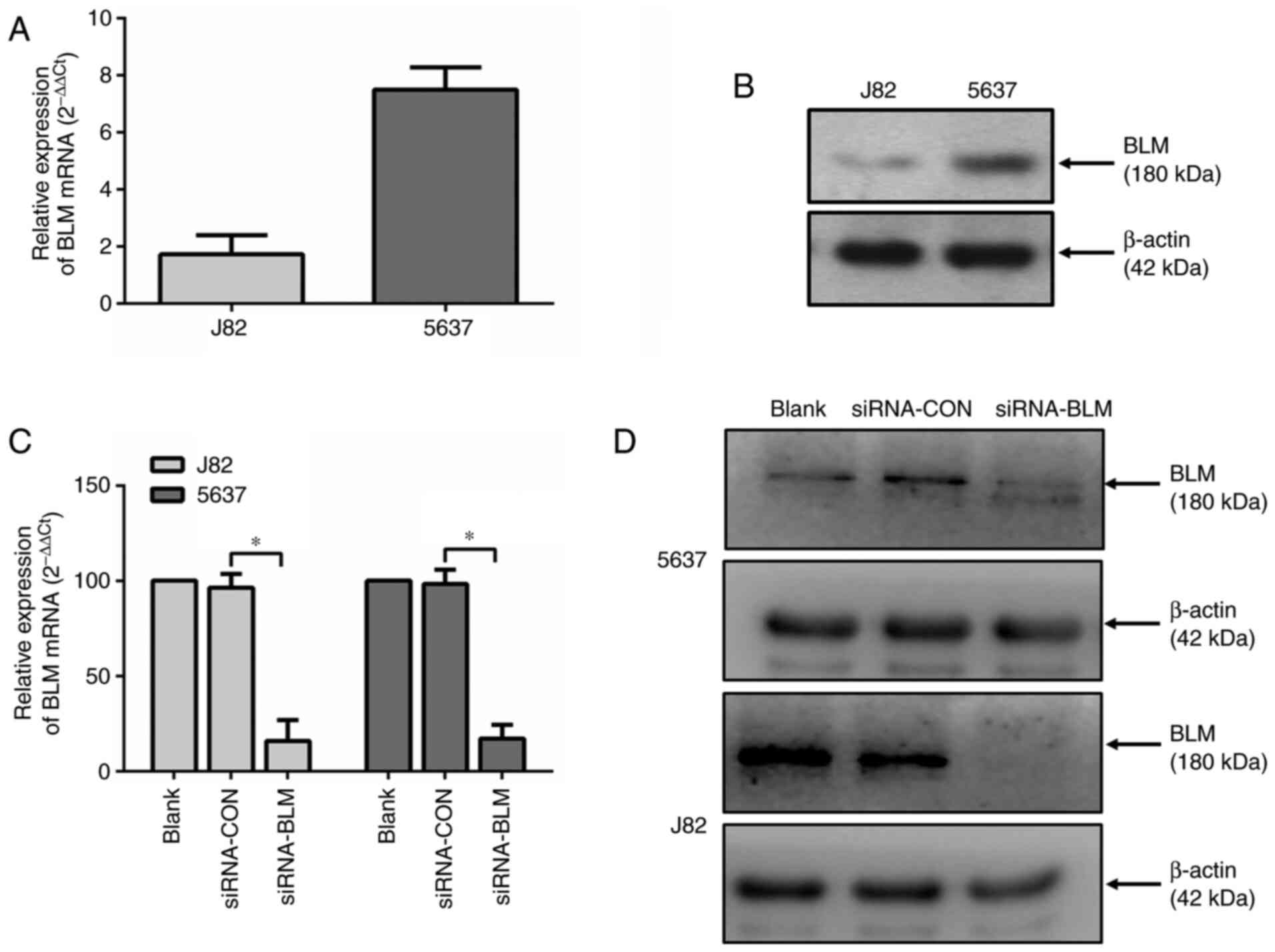
lines (J82 and 5637). As shown in Fig. 2A and B, both cell lines expressed
BLM. The mRNA and protein expression level of BLM in the J82 cells
was lower compared with that in the 5637 cells.
Subsequently, both bladder cancer cell lines were
transfected with a control siRNA (siRNA-CON) or a BLM-specific
siRNA (siRNA-BLM). As shown in Fig.
2C and D, both the mRNA and protein expression levels of BLM
were markedly decreased in these two cell lines (P<0.0001)
following transfection with siRNA-BLM.
Silencing of BLM inhibits the
viability and proliferation and promotes the apoptosis of bladder
cancer cells in vitro
The cell cycle was analyzed to identify whether cell
cycle perturbation occurs after the silencing of BLM expression.
The results of cell cycle assay (Fig.
3) revealed that both cell lines transfected with siRNA-BLM
were arrested in the G0G1 phase (P<0.001).
In the siRNA-BLM group, the J82 cells in the S phase were
significantly decreased compared with the siRNA-CON group
(P<0.0001) and the 5637 cells in the G2M phase were
significantly decreased compared with the siRNA-CON group
(P<0.05). These results indicated that the silencing of BLM
expression led to G1 arrest in the bladder cancer
cells.
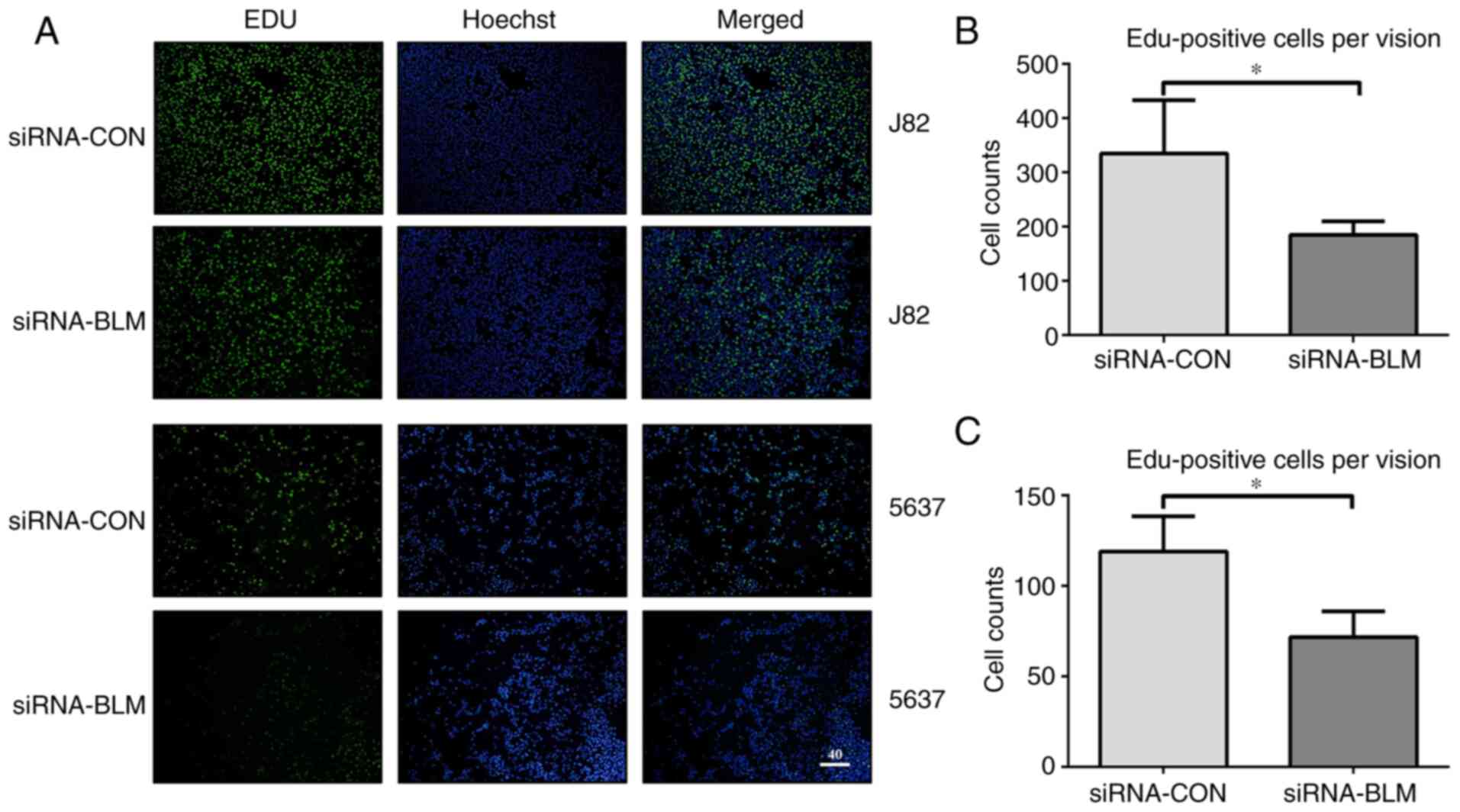
It was hypothesized that cell proliferation was
inhibited as there were fewer in the S phase. To verify this
hypothesis, a cell proliferation assay was performed. The numbers
of EdU-labeled J82 and 5637 bladder cancer cells transfected with
siRNA-CON were 335.25±97.79 and 118.71±19.20 in each field,
respectively. In addition, the numbers of EdU-labeled J82 and 5637
bladder cancer cells transfected with siRNA-BLM were 185±24.90 and
76.34±16.45, respectively (P<0.05). Therefore, the silencing of
BLM significantly decreased bladder cancer cell proliferation
(Fig. 4).
To examine cell functions in response to BLM
knockdown in the J82 and 5637 cells, cell apoptosis assay was
performed. The results revealed that cell apoptosis was
significantly increased following transfection with siRNA-BLM
compared with the siRNA-CON group (P<0.05, Fig. 5).
Silencing of BLM sensitizes bladder
cancer cell lines to cisplatin
Previous studies have demonstrated that the RecQ
helicase level is negatively associated with genomic instability
induced by DNA damaging agents in various type of cells (21,23). As platinum-containing anticancer
drugs are widely used in intravesical instillation, cisplatin was
selected to induce DNA damage in the present study. To determine
the cytotoxicity of cisplatin in bladder cancer cell lines, the J82
and 5637 cells were first treated with various concentrations of
cisplatin (0, 2, 4 and 8 µmol/l) for different periods of time (12,
24 and 48 h). The results revealed that cisplatin significantly
increased the death of the J82 and 5637 cells in a concentration-
and time-dependent manner (Fig. 6A
and B). The IC50 of cisplatin to J82 and 5637 at 24
h were (10.44±1.00) µmol/l and (6.91±0.32) µmol/l respectively.
Subsequently, the siRNA-BLM-siRNA-CON-transfected J82 and 5637
cells were treated with cisplatin at the IC50
concentration for 24 h. As shown in Fig. 6C and D, the silencing of BLM
significantly sensitized the J82 and 5637 cells to cisplatin; the
death rate of the siRNA-BLM-transfected cells was significantly
higher than that of the control group (P<0.05).
Discussion
BLM monitors genomic integrity and functions as a
genome ‘caretaker’; however, to the best of our knowledge, the
mechanisms of action of BLM in bladder cancer have not yet been
reported. The present study first confirmed that BLM was highly
expressed in human bladder cancer tissues when compared with
adjacent healthy tissues. These results were consistent with those
of other studies which found that BLM mRNA expression is
significantly deregulated in breast (24) and colorectal cancer (25). It was hypothesized this may due to
the varied expression of BLM throughout the cell cycle, with its
highest expression being found during the S phase (26). Even minimal changes in BLM
expression can disrupt genomic integrity and function. However, the
molecular mechanisms which lead to the upregulation of BLM are not
yet fully understood. In addition, no association between BLM
expression and the survival rate of patients with bladder cancer of
all grades or of a high grade was found. Therefore, the present
findings suggested that further studies on BLM are required to
determine whether it can be used as a predictive biomarker for
bladder cancer.
The present study examined the function of BLM in
bladder cancer progression in vitro using the J82 and 5637
bladder cancer cells. Silencing of BLM significantly led to cell
cycle arrest in the G0G1 phase and inhibited
cell proliferation while it promoted cell apoptosis. As is known,
homologous recombination occurs during the S and G2/M phase of the
cell cycle and NHEJ is pivotal for the repair of DNA double-strand
breaks, particularly during the G0 and G1
phase of the cell cycle (27).
Therefore, without BLM, NHEJ cannot be processed smoothly and the
cell cycle is arrested in the G0 and G1
phase. With the accumulation of cells in the G1 phase,
cell death eventually occurs. Similar with the findings of the
present study, Mao et al (20) found that the silencing of BLM in
the GM639 and U-2 OS cells significantly suppresses cell
proliferation. Chen et al (28) also demonstrated that BLM knockdown
leads to a reduction in prostate cancer cell proliferation. On the
other hand, BLM can resolve DNA double-strand breaks, a type of
cell DNA damage followed by apoptosis. Therefore, cells lacking BLM
eventually undergo apoptosis (29). These results suggested that the
discovery of specific BLM inhibitors may greatly enhance the
therapeutic effect in a variety of cancers.
In the present study, in light of the biological
behavioral changes, a drug sensitivity test in vitro was
conducted and the results revealed that the silencing of BLM
expression sensitized bladder cancer cells to cisplatin. Although
immense efforts have been made to explore genetic therapeutics over
the past few years, the cisplatin-based chemotherapeutic regimen
remains the first-line therapy for MIBCs. However, resistance to
cisplatin is a major obstacle to successful treatment. Therefore, a
better understanding of the mechanisms of cisplatin resistance may
provide crucial information which would be of clinical
significance. DNA repair pathway alterations are known to drive
cancer behavior and therapeutic efficacy (30). In the present study, the
inhibition of BLM expression could enhance cell sensitivity to
cisplatin. Over the past few years, the role of BLM as a DNA sensor
protein that recognizes DNA damage has been noted (31). As an important component of the
DNA damage response, the response of BLM to DNA damage signals may
occur through secondary nucleic acid structures. The dysregulated
expression of BLM in cancer cells is observed in the cytoplasm in
response to these lesions (32).
Accordingly, the findings of the present study suggest that the
inhibition of BLM in bladder cancer using small molecules or
inhibitors may prove to be an effective therapy, similar to the
study by Zhang et al (33), which found that a small molecule
termed HJNO inhibited breast cancer cell expansion by targeting BLM
helicase.
However, there are some limitations to the present
study. Although it confirmed that the silencing of BLM
significantly sensitized J82 and 5637 cells to cisplatin, whether
the alterations of survival rate could be recognized by the
difference of the BLM expression in MIBC patients receiving
cisplatin treatment remains to be elucidated. Therefore, the
influence of BLM differential expression on the survival rate of
MIBC patients receiving cisplatin treatment and its related
molecular mechanisms will be explored in future work.
In conclusion, the findings of the present study
suggested that BLM served an oncogenic role in bladder cancer. The
results provided preliminary evidence that BLM may be a predictive
biomarker and a promising therapeutic target in bladder cancer.
However, further studies are required to determine the precise
regulatory mechanisms of BLM in bladder cancer.
Acknowledgements
Not applicable.
Funding
This work was supported by Beijing Natural Science Foundation
(grant no. 7182058).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SF was responsible for study design, data
collection, statistical analysis, data interpretation and
literature search. XQ was responsible for study design, statistical
analysis, data interpretation, literature search and drafting the
manuscript. DF was responsible for data collection, statistical
analysis and data interpretation. XZ was responsible for data
interpretation, literature search, critical revision of the
manuscript for scientific and factual content and sourcing funds.
SF and XQ confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
Beijing Chao-Yang Hospital, Capital Medical University (approval
number 2017-ke-47) and was conducted in accordance with the
Declaration of Helsinki. Patients gave written informed consent for
their information to be used in the research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Leal J, Luengo-Fernandez R, Sullivan R and
Witjes JA: Economic burden of bladder cancer across the European
union. Eur Urol. 69:438–447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berdik C: Unlocking bladder cancer.
Nature. 551:S34–S35. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sameni S and Hande MP: Plumbagin triggers
DNA damage response, telomere dysfunction and genome instability of
human breast cancer cells. Biomed Pharmacother. 82:256–268. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marzec P, Armenise C, Pérot G, Roumelioti
FM, Basyuk E, Gagos S, Chibon F and Déjardin J:
Nuclear-receptor-mediated telomere insertion leads to genome
instability in ALT cancers. Cell. 160:913–927. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujii Y: Prediction models for progression
of non-muscle-invasive bladder cancer: A review. Int J Urol.
25:212–218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Morrison CD, Liu P, Woloszynska-Read A,
Zhang J, Luo W, Qin M, Bshara W, Conroy JM, Sabatini L, Vedell P,
et al: Whole-genome sequencing identifies genomic heterogeneity at
a nucleotide and chromosomal level in bladder cancer. Proc Natl
Acad Sci USA. 111:E672–E681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagata M, Muto S and Horie S: Molecular
biomarkers in bladder cancer: Novel potential indicators of
prognosis and treatment outcomes. Dis Markers. 2016:82058362016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Newman JA and Gileadi O: RecQ helicases in
DNA repair and cancer targets. Essays Biochem. 64:819–830. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Larsen NB and Hickson ID: RecQ helicases:
Conserved guardians of genomic integrity. In: DNA Helicases and DNA
Motor Proteins. Advances in Experimental Medicine and Biology. Vol.
767. Spies M: Springer; New York, NY: pp. 161–184. 2013, View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Croteau DL, Popuri V, Opresko PL and Bohr
VA: Human RecQ helicases in DNA repair, recombination, and
replication. Annu Rev Biochem. 83:519–552. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krogh BO and Symington LS: Recombination
proteins in yeast. Annu Rev Genet. 38:233–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Traverso G, Bettegowda C, Kraus J,
Speicher MR, Kinzler KW, Vogelstein B and Lengauer C:
Hyper-recombination and genetic instability in BLM-deficient
epithelial cells. Cancer Res. 63:8578–8581. 2003.PubMed/NCBI
|
|
15
|
Hickson ID: RecQ helicases: Caretakers of
the genome. Nat Rev Cancer. 3:169–178. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chandra S, Priyadarshini R, Madhavan V,
Tikoo S, Hussain M, Mudgal R, Modi P, Srivastava V and Sengupta S:
Enhancement of c-Myc degradation by BLM helicase leads to delayed
tumor initiation. J Cell Sci. 126:3782–3795. 2013.PubMed/NCBI
|
|
17
|
Turley H, Wu L, Canamero M, Gatter KC and
Hickson ID: The distribution and expression of the Bloom's syndrome
gene product in normal and neoplastic human cells. Br J Cancer.
85:261–265. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anisimenko MS, Kozyakov AE, Paul GA and
Kovalenko SP: The frequency of the BLM p.Q548X (c.1642C>T)
mutation in breast cancer patients from Russia is no higher than in
the general population. Breast Cancer Res Treat. 148:689–690. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Prokofyeva D, Bogdanova N, Dubrowinskaja
N, Bermisheva M, Takhirova Z, Antonenkova N, Turmanov N, Datsyuk I,
Gantsev S, Christiansen H, et al: Nonsense mutation p.Q548X in BLM,
the gene mutated in Bloom's syndrome, is associated with breast
cancer in Slavic populations. Breast Cancer Res Treat. 137:533–539.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mao FJ, Sidorova JM, Lauper JM, Emond MJ
and Monnat RJ: The human WRN and BLM RecQ helicases differentially
regulate cell proliferation and survival after chemotherapeutic DNA
damage. Cancer Res. 70:6548–6555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Lu X, Zhou G, Lou H and Luo G:
RECQL5 is an important determinant for camptothecin tolerance in
human colorectal cancer cells. Biosci Rep. 31:363–369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kumari A, Owen N, Juarez E and McCullough
AK: BLM protein mitigates formaldehyde-induced genomic instability.
DNA Repair (Amst). 28:73–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arora A, Abdel-Fatah TM, Agarwal D,
Doherty R, Moseley PM, Aleskandarany MA, Green AR, Ball G,
Alshareeda AT, Rakha EA, et al: Transcriptomic and protein
expression analysis reveals clinicopathological significance of
bloom syndrome helicase (BLM) in breast cancer. Mol Cancer Ther.
14:1057–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lao VV, Welcsh P, Luo Y, Carter KT,
Dzieciatkowski S, Dintzis S, Meza J, Sarvetnick NE, Monnat RJ Jr,
Loeb LA and Grady WM: Altered RECQ helicase expression in sporadic
primary colorectal cancers. Transl Oncol. 6:458–469. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Singh DK, Popuri V, Kulikowicz T, Shevelev
I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL and
Bohr VA: The human RecQ helicases BLM and RECQL4 cooperate to
preserve genome stability. Nucleic Acids Res. 40:6632–6648. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saleh-Gohari N and Helleday T:
Conservative homologous recombination preferentially repairs DNA
double-strand breaks in the S phase of the cell cycle in human
cells. Nucleic Acids Res. 32:3683–3688. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen K, Xu H and Zhao J: Bloom syndrome
protein activates AKT and PRAS40 in prostate cancer cells. Oxid Med
Cell Longev. 2019.2019:3685817, 2019. View Article : Google Scholar
|
|
29
|
Roos WP and Kaina B: DNA damage-induced
cell death by apoptosis. Trends Mol Med. 12:440–450. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma J, Setton J, Lee NY, Riaz N and Powell
SN: The therapeutic significance of mutational signatures from DNA
repair deficiency in cancer. Nat Commun. 9:32922018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tikoo S and Sengupta S: Time to bloom.
Genome Integr. 4:142010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Böhm S and Bernstein KA: The role of
post-translational modifications in fine-tuning BLM helicase
function during DNA repair. DNA Repair (Amst). 22:123–132. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang W, Yang S and Liu J, Bao L, Lu H, Li
H, Pan W, Jiao Y, He Z and Liu J: Screening antiproliferative drug
for breast cancer from bisbenzylisoquinoline alkaloid tetrandrine
and fangchinoline derivatives by targeting BLM helicase. BMC
Cancer. 19:10092019. View Article : Google Scholar : PubMed/NCBI
|