Introduction
Long non-coding (lnc)RNAs are defined as RNAs
>200 nucleotides that are not translated into functional
proteins. It is well documented that a number of lncRNAs have
important cellular functions and roles in different mechanisms of
gene regulation. lncRNAs control the expression of nearby genes by
influencing gene transcription. They can also interact with
proteins by building complex secondary structures (1,2).
lncRNAs can recruit chromatin remodeling complex and place them in
nuclei at specific positions, thus regulating other
chromosome-specific genes (3).
The function of the encoded genes may involve structure and/or
regulation, covering all stages of gene transcription and
translation, including splicing, flipping and translation away from
its genetic basis (4).
lncRNAs effect some physiological cell functions,
while the changes in their expression are inherent in many
diseases, such as ischemic stroke (IS) and cancer. In patients with
ischemic injury or ischemic injury animal models, abnormal
expression of lncRNAs have been reported using techniques such as
microarray or RNA-sequencing (RNA-seq) (5). Specifically, uncoded antonym RNA has
been identified in lncRNA gene bank entries for inhibitors of CDK4
(INK4), antisense non-coding RNA in the INK4 locus (ANRIL), H19,
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),
maternally expressed gene 3 and N1LR, which affect cell
inflammation, apoptosis, angiogenesis and other physiological and
pathological processes (6). ANRIL
is present in more than eight splice variants that are ~3.9 kb in
length (7), located on chromosome
9p21, which is association with myocardial infarction and IS
(8). The regulation of gene
expression by ANRIL is mediated by combining multiple fusion
inhibitors of compound 1 or 2 (Polycomb repressive complex) and
gene silencing is directed to the position INK4b-ARF-INK4a.
Together, these findings indicate that ANRIL has a direct
effect on the pathobiology of atherosclerosis. Therefore,
ANRIL is considered a good candidate for atherosclerotic
disease risk, such as coronary artery disease (CAD) and IS
(9,10).
Inflammatory responses are involved in endothelial
dysfunction, propagation of atherosclerosis and ischemic brain
injury (11,12). Cerebral ischemia and
ischemia-reperfusion injury cause an inflammatory cascade reaction,
including inflammatory cell infiltration, release of toxic
inflammatory mediators and oxidative stress and excitotoxicity,
thus leading to further nerve tissue damage and cell death
(13). Over recent years,
multiple studies have reported that ANRIL regulates the
inflammatory response and proliferation of cells through different
signaling pathways in pathological aspects of ischemic stroke
(14–16). For example, Zhao reported that
lncRNA ANRIL knockdown improves cerebral infarction-induced
neurological deficits and decreases the number of apoptotic neurons
in an animal model. The possible underlying mechanism may be
related to the effect of inhibition of lncRNA ANRIL knockdown on
the NF-κB signaling pathway (17). Previous studies have also reported
that in animal models of diabetes with cerebral ischemia, the
increase of vascular endothelial growth factor (VEGF) is caused by
overexpression of ANRIL and angiogenesis is achieved by activating
the NF-κB signaling pathway (17,18). However, it is unclear how ANRIL
affects the expression of potential downstream genes that regulate
endothelial dysfunction and inflammatory response.
To study the potential function of ANRIL in
regulating expression and alternative splicing, ANRIL-regulated
transcriptomes were assessed in human umbilical vein endothelial
cells (HUVECs) using RNA-seq. The present study provided an
important database and platform to evaluate the role of ANRIL in
mediating the mechanisms of inflammatory response.
Materials and methods
Cell culture
HUVECs (cat. no. 8000; ScienCell Research
Laboratories, Inc.) were cultured in extracellular matrix (ECM,
1001, Sciencell) at 37°C (with 5% CO2 Atmosphere)
containing 5% fetal bovine serum (1001, Sciencell) supplemented
with 100 U/ml penicillin, 100 g/ml streptomycin and 1% growth
factor (1001, Sciencell). The cultured cells were processed
according to the manufacturer's protocols as follows: Original
culture solution was aspirated, cells were washed using PBS, 0.05%
trypsin (183, ScienCell) was used for digestion (37°C) for 2 min
and Complete medium containing 10% fetal bovine serum was added to
stop digestion. Cultured wells were blown into single cells using a
pipette gun and a 15 µl aliquot of cell suspension was counted, the
remaining cell suspension was centrifuged at 143 × g for 5 min
(37°C), resuspended with culture medium and were seeded
(1×105/well) into 24-well plates. Each group had 3
replicates and was cultured overnight at 37°C under 5%
CO2.
Plasmid transfection
After cells were plated and cultured overnight at
37°C, plasmid transfection was performed. HUVEC cells was performed
using Lipofectamine 2000 (11668019, Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
ANRIL gene was constructed into pcDNA3.1 vector synthesized from
Youbio Biotech and transfected to HUVEC cells with 500 ng plasmid.
After incubated at 37°C for 24 h, they were used for RT-qPCR
analysis. Following transfection, medium was replaced with fresh
complete medium and incubated under 5% CO2 at 37°C for
24 h and imaging was performed using a Optical light microscope.
Samples were collected for subsequent experiments
Assessment of gene overexpression
GAPDH was used as an internal control gene to
evaluate overexpression of ANRIL. Complementary DNA (cDNA) was
synthesized as follows: The collected HUVEC cells were subjected to
total RNA extraction using TRIZOL (15596-018, Ambion). The RNA was
further purified with two phenol-chloroform treatments and then
treated with RQ1 DNase (M6101, Promega) to remove DNA. The quality
and quantity of the purified RNA were determined by measuring the
absorbance at 260/280 nm (A260/A280) using Smartspec Plus
(Bio-Rad). The integrity of RNA was further verified by 1.5%
agarose gel electrophoresis. Reverse transcription at 37°C for 15
min. Then for pre degeneration at 95°C for 10 min, 40 cycles of
degeneration at 95°C for 15 sec, annealing and extension at 60°C
for 1 min. Bio-Rad S1000 thermocycler was used for reverse
transcription (RT)-PCR using SYBR-Green Bestar RT-PCR Master Mix
(DBI Bioscience). The concentration of each transcript was
standardized to the GAPDH mRNA expression level using the
2−ΔΔCq method (19).
The thermocycling conditions were as follows: Denaturing at 95°C
for 10 min, followed by 40 cycles of denaturing at 95°C for 15 sec
and annealing and extension at 60°C for 1 min. PCR amplification
was performed in triplicate for each sample. Primers for
quantitative (q)PCR analysis are presented in Table SI.
RNA extraction and seq
Total RNA was extracted using TRIzol®
(Ambion; Thermo Fisher Scientific, Inc.) and treated with RQ1
RNase-Free DNase (Promega Corporation) to remove DNA. The integrity
of RNA was assessed using 1.5% agarose gel electrophoresis. The
mRNA was purified using VAHTS mRNA capture beads (cat. no. N401-01,
Vazyme, China)For each sample, 1 µg of total RNA was used for
RNA-seq library preparation. The purified RNA was treated with RQ1
DNase (Promega) to remove DNA before used for directional RNA-seq
library preparation by KAPA Stranded mRNA-Seq Kit for
Illumina® Platforms (cat. no. KK8544, Roche).
Polyadenylated mRNAs were purified and fragmented. Fragmented mRNAs
were converted into double strand cDNA. Following end repair and A
tailing, the DNAs were ligated to Diluted Roche Adaptor (KK8726,
Roche). After purification of ligation product and size fractioning
to 300–500 bps, the ligated products were amplified and purified,
quantified and stored at −80°C before sequencing. The strand marked
with dUTP (the 2nd cDNA strand) is not amplified, allowing
strand-specific sequencing.
For high-throughput sequencing, the libraries were
prepared following the manufacturer's instructions and applied to
Illumina Novaseq 6000 system for 150 nt paired-end sequencing.
RNA-seq raw data cleaning and
alignment
Original reads containing more than 2 uncertain
bases were discarded. The adapter and low-quality bases were
trimmed from the original sequencing reads using Fasxtoolkit
(version 0.0.13; hannonlab.cshl.edu/fastx_toolkit/index.html).
Subsequently, the cleaned reads were aligned to the GRch38 genome
using TopHat2 (20) which allowed
a maximum of four misread errors. A single mapped survey was used
for counting genetic surveys and calculating fragments per kilobase
of transcript per million fragments mapped (FPKM) values (21).
Differentially expressed genes (DEGs)
and alternative splicing (AS) analysis
Principal component analysis (PCA, performed by R
package factoextra (https://cloud.r-project.org/package=factoextra)).
demonstrated a separation between Control and ANRIL overexpression
samples The edgeR Bioconductor package (version no. 3.32.1)
(22) was used to assess DEGs.
The fold-change (FC) of the DEGs, log2FC and false discovery rate
(FDR) were obtained following testing. The cutoff thresholds were
set to a FC of ≥2 or ≤-2 and FDR <0.05. To clean up the feature
categories in DEGs, the KOBAS 2.0 server was used to determine the
Gene Ontology (GO) terminology and Kyoto Encyclopedia of Genes and
Genomes (KEGG) path (23).
AS events (ASEs) and regulated ASEs (RASEs) between
the samples were defined and quantified using the ABLas pipeline
(24,25). Briefly, the ABLas pipeline was
used to assess ten types of ASE based on the splice junction reads,
including exon skipping (ES), mutually exclusive exons (MXE),
alternative 5’ splice site (A5SS), intron retention (IR),
alternative 3′ splice site (A3SS), mutually exclusive 3′ UTRs
(3pMXE), mutually exclusive 5′ UTRs (5pMXE), cassette exon, A3SS +
ES and A5SS + ES. To evaluate the validity of the RNA-seq data,
RT-qPCR was performed as aforementioned for selected DEGs ((CEBPB,
CXCL11, PTGS2, THEMIS2, IL6, HMOX1, NLRC5, SECTM1, TNFSF18 and
TRIM21) and ASEs (CTNNB1, IL23, ATP13A2, SIGIRR, HDAC7, and
normalized using the reference gene GAPDH. The same RNA samples for
RNA-seq were used for RT-qPCR and are available from the Gene
Expression Omnibus website using accession number GSE197115
(ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197115).
Statistical analysis
An unpaired two-tailed t-test (between two groups)
was performed for cell and RT-qPCR data. P<0.05 was considered
to indicate a statistically significant difference. The data are
presented as the mean ± standard deviation. GraphPad Prism (version
no. 8.0; GraphPad Software, Inc.) was used to perform unpaired
Student's t-test. Each experiment was performed for at least three
biological replicates, except RNA-seq.
Results
RNA-seq analysis of ANRIL-regulated
transcriptome profile in HUVECs
RNA-seq was used to explore the molecular mechanisms
of ANRIL-mediated transcriptional regulation. ANRIL was
overexpressed in HUVECs following transfection using a vector
expressing ANRIL (ANRIL-OE) compared with empty negative control
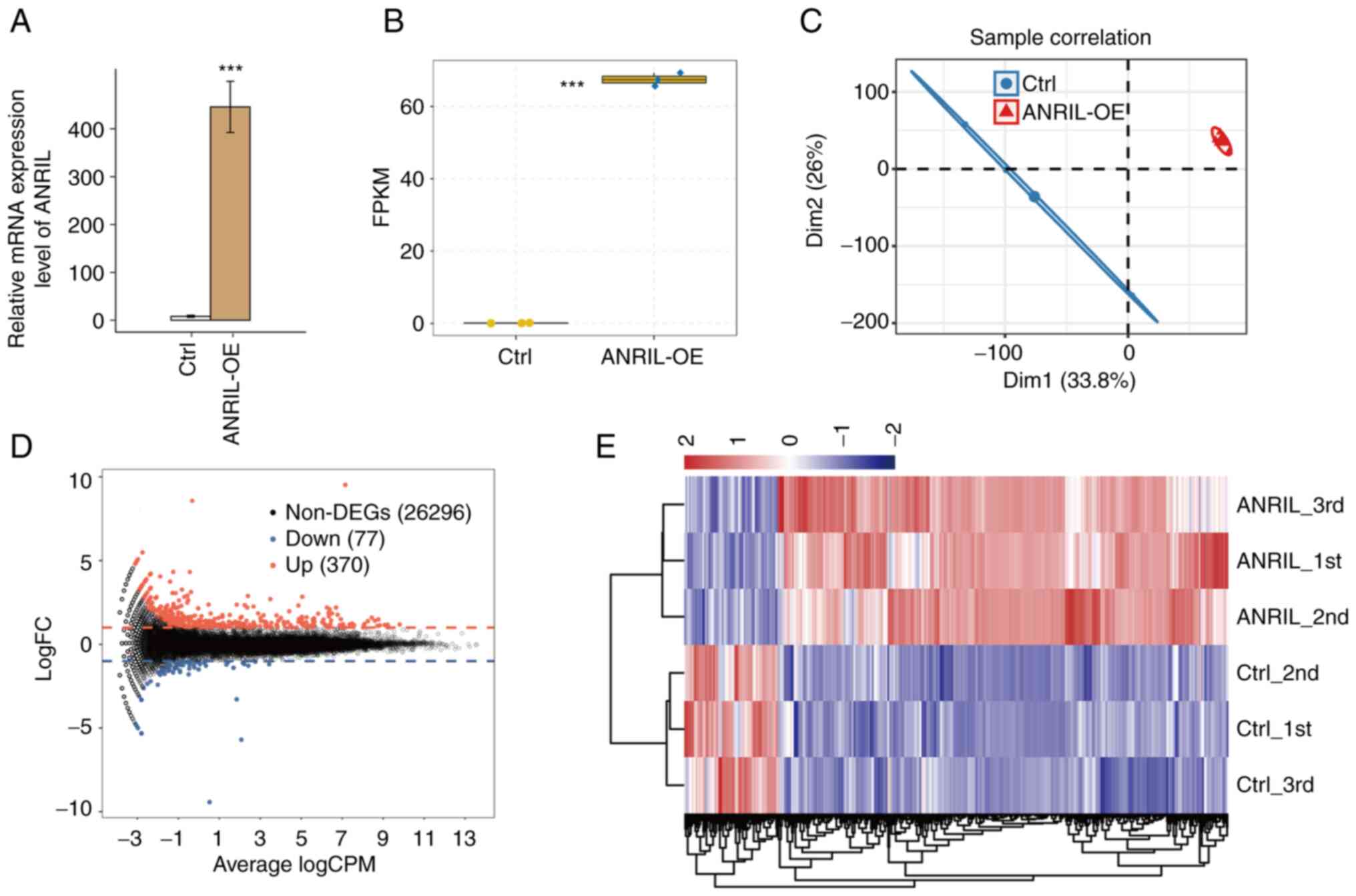
vector (NC). The results of RT-qPCR demonstrated that ANRIL mRNA
expression levels were significantly increased in HUVECs with
ANRIL-OE (Fig. 1A and B). A total
of six RNA-seq libraries were constructed and sequenced for the
ANRIL-OE and NC group with each group having three biological
repeats (ANRIL_1st, ANRIL_2nd, ANRIL_3rd, Ctrl_1st, Ctrl_2nd and
Ctrl_3rd). For high-throughput sequencing, the library was prepared
according to the manufacturer's protocols and used the Illumina
NovaSeq 6000 system for terminal sequencing to produce 150
nucleotide paired-end reads. A mean total of 74.18±6.15 million
original reads were produced for each sample. After the adapter and
polluted sequences were removed, each sample had a mean total of
72.56±6.03 million clean reads. The average number of read
pairs/sample was 69.82±2.79 million, which were aligned with the
human genome (Table I).
 | Table I.Summary of RNA-seq reads used in
analysis. |
Table I.
Summary of RNA-seq reads used in
analysis.
| Sample | ANRIL_1st | ANRIL_2nd | ANRIL_3rd | Ctrl_1st | Ctrl_2nd | Ctrl_3rd | Mean ± SD |
|---|
| Raw | 71189488 | 68752622 | 82908788 | 71254178 | 81115056 | 69881880 |
74183668±6159878 |
| Clean (%) | 69634309 | 67136142 | 81023158 | 69873252 | 79428266 | 68291552 |
72564447±6036790 |
|
| (97.82) | (97.65) | (97.73) | (98.06) | (97.92) | (97.72) |
|
| Total
mappeda (%) | 66924893 | 64433515 | 77763393 | 67444711 | 76559923 | 65803658 |
69821682±5791250 |
|
| (97.62) | (97.6) | (97.6) | (97.92) | (97.89) | (97.88) |
|
| Total uniquely | 61071119 | 58348374 | 67865079 | 62954592 | 70991974 | 59635640 |
63477796±4956815 |
| mappedb (%) | (91.25) | (90.56) | (87.27) | (93.34) | (92.73) | (90.63) |
|
| Splice
readsc (%) | 24821085 | 24093961 | 27782615 | 25306098 | 29117397 | 25457522 |
26096446±1930734 |
|
| (40.64) | (41.29) | (40.94) | (40.2) | (41.02) | (42.69) |
|
FPKM values were calculated to represent levels of
gene expression. RNA-seq yielded robust expression results for
60,498 genes (Table II). A
correlation matrix was calculated based on the FPKM value of the
expression gene in all samples. There was a high Pearson's
correlation value between ANRIL-OE and the control (>0.99),
which indicated similar expression of most genes. Furthermore,
significant segregation between the sample groups was demonstrated
in the FPKM uniform numerical sequence for DEGs, indicating
agreement between the composite datasets (Fig. 1C and E). These results indicated
that expression of lncRNA-ANRIL altered the replication level of
genes. To further compare the gene expression profile, edgeR was
performed to identify the DEGs between the ANRIL-OE and control
cells, with a cut-of as fold change ≥2 or ≤0.5 and a 5% false
discovery rate (FDR) (22). A
total of 370 up- and 77 downregulated genes were identified between
ANRIL-OE and NC cells. Indicating that ANRIL overexpression
extensively regulates gene expression in HUVECs (Fig. 1D).
 | Table II.Mapped normalized reads on expressed
genes in reference genome (60,498 total genes in genome). |
Table II.
Mapped normalized reads on expressed
genes in reference genome (60,498 total genes in genome).
| Sample | Expressed genes
with FPKM >0 (%) | Expressed genes
with FPKM ≥1 (%) |
|---|
| ANRIL_1st | 21066 (34.82) | 11245 (53.38) |
| ANRIL_2nd | 20920 (34.58) | 11238 (53.72) |
| ANRIL_3rd | 21319 (35.24) | 11254 (52.79) |
| Ctrl_1st | 20980 (34.68) | 11392 (54.30) |
| Ctrl_2nd | 20943 (34.62) | 11264 (53.78) |
| Ctrl_3rd | 20696 (34.21) | 11101 (53.64) |
ANRIL regulates expression of genes
involved in the inflammatory response and IκB/NF-κB signaling
pathway in HUVECs
To reveal the potential roles of DEGs, GO functional
analysis was performed and 26,743 DEGs were annotated. The results
revealed 370 upregulated and 77 downregulated genes annotated with
GO categories biological process terms, respectively (Fig. 2A and B; Tables SII and SIII). Upregulated genes were
significantly enriched in inflammatory response and NF-κB signaling
pathway, including Mullerian-inhibiting factor (AMH), Cystathionine
gammalyase (CTH), Heme oxygenase 1 (HMOX1), Kruppel-like factor 4
(KLF4), Protein NLRC5, Secreted and transmembrane protein 1
(SECTM1), Tumor necrosis factor ligand superfamily member 18
(TNFSF18), E3 ubiquitin-protein ligase Tripartite-motif protein 21
(TRIM21), CCAAT/enhancer-binding protein beta (CEBPB), C-X-C motif
chemokine 10 (CXCL10), C-X-C motif chemokine 11 (CXCL11), IL-1
alpha (IL1A), Interleukin-6 (IL6), Prostaglandin G/H synthase 2
(PTGS2), Protein THEMIS2, Toll-like receptor 3 (TLR3), Tumor
necrosis factor alpha-induced protein 3 (TNFAIP3); Fig. 2C and D). These results indicated a
key role of ANRIL in expression of inflammatory factors.
 | Figure 2.Functional analysis of
ANRIL-regulated DEGs in HUVECs. (A) Top 10 GO biological processes
of upregulated genes. (B) Top GO biological processes of
downregulated genes. (C) ANRIL-upregulated genes were enriched in
NF-κB-associated pathways. (D) Hierarchical clustering of the
expression levels of inflammatory response and NF-κB-associated
genes effected by ANRIL overexpression in HUVECs. (E) Gene
expression levels of ANRIL-regulated inflammatory response genes
assessed using RNA-seq data. (F) Relative expression levels of DEGs
identified using RNA-seq were assessed using RT-qPCR. For RT-qPCR,
GAPDH was used as the reference gene. Student's t test was
performed to compare ANRIL-OE and control cells. ***P<0.001 vs.
Ctrl. HUVECs, human umbilical vein endothelial cells; ANRIL,
lncRNA-antisense non-coding RNA in the INK4 locus; DEGs,
differentially expressed genes; GO, Gene Ontology; Ctrl, control;
CEBPB, CCAAT/enhancer binding protein (C/EBP) beta; CXCL11,
chemokine (C-X-C motif) ligand 11; PTGS2,
prostaglandin-endoperoxide synthase 2; THEMIS2, thymocyte selection
associated family member 2; IL6, interleukin 6; HMOX1, heme
oxygenase 1; TRIM21, tripartite motif containing 21; SECTM1,
secreted and transmembrane 1; TNFSF18, tumor necrosis factor
(ligand) superfamily, member 18; NLRC5, Protein NLRC5. |
To verify the effect of ANRIL-OE on the expression
of these DEGs, qPCR was conducted to quantify the changes in mRNA
levels of these genes. Ten upregulated DEGs were randomly selected
for qPCR analysis, including CEBPB, CXCL11, PTGS2, THEMIS2, IL6,
HMOX1, NLRC5, SECTM1, TNFSF18 and TRIM21 These results demonstrated
that mRNA expression levels of all 10 selected DEGs were
significantly increased, except IL6, which was consistent with RNA
sequence analysis (Fig. 2E and
F).
When the corrected P-value for the KEGG pathways was
set at P<0.05, numerous cancer-associated pathways including
‘cytokine-cytokine receptor interaction’, ‘NF-κB signaling
pathway’, ‘Toll-like receptor signaling pathway’, ‘TNF signaling
pathway’ and ‘inflammatory mediator regulation of TRP channels’
were demonstrated to be enriched (Figs. S1 and S2; Tables SIV and SV).
ANRIL overexpression regulates AS of
transcription factors in HUVECs
To the best of our knowledge, there have been few
previous reports of studies of regulation of AS of ANRIL in HUVECs.
The purpose of the present study was to elucidate the role of ANRIL
in the regulation of AS. It was hypothesized that by regulating the
expression of key functional genes, ANRIL may be involved in
regulating inflammatory responses. Therefore, whether ANRIL
affected expression of DEGs via AS regulation was evaluated. The
splice reads from ANRIL-OE and NC HUVECs were mapped to the
reference genome and 203,974 exons annotated (55.53% of total
annotated ones). Next, a total of 134,447 known splice sites and
51,606 novel splice junctions were identified using TopHat2. AS
events in the spliced joints were identified using the ABLas
pipeline (25,16), which resulted in 58,543 known
alternative splicing events (ASEs) and 40,880 novel ASEs (Tables II and III).
 | Table III.Known or novel splicing events
detected using the ABLas pipeline. |
Table III.
Known or novel splicing events
detected using the ABLas pipeline.
| A, Known splicing
events |
|---|
|
|---|
| Sample | 3pMXE | 5pMXE | A3SS | A3SS + ES | A5SS | A5SS + ES | ES | IR | MXE | Cassette exon | Total | Detected
junctions |
|---|
| ANRIL_1st | 963 | 1443 | 6030 | 673 | 6045 | 763 | 3268 | 4514 | 357 | 1787 | 25843 | 179431 |
| ANRIL_2nd | 980 | 1529 | 5915 | 640 | 6113 | 768 | 3288 | 4441 | 378 | 1775 | 25827 | 178905 |
| ANRIL_3rd | 999 | 1632 | 6288 | 681 | 6357 | 801 | 3380 | 4696 | 386 | 1896 | 27116 | 186053 |
| Ctrl_1st | 922 | 1489 | 5799 | 582 | 6078 | 735 | 3126 | 4439 | 343 | 1756 | 25269 | 178976 |
| Ctrl_2nd | 940 | 1555 | 5916 | 594 | 6142 | 800 | 3176 | 4388 | 373 | 1845 | 25729 | 180273 |
| Ctrl_3rd | 918 | 1453 | 5600 | 591 | 5780 | 738 | 3036 | 4186 | 317 | 1701 | 24320 | 173350 |
| Total | 2390 | 3959 | 14507 | 1699 | 14629 | 1856 | 6596 | 8584 | 842 | 3481 | 58543 | 300041 |
|
| B, Novel
splicing events |
|
| Sample | 3pMXE | 5pMXE | A3SS |
A3SS&ES | A5SS |
A5SS&ES | ES | IR | MXE | Cassette
exon | Total | Detected
junctions |
|
| ANRIL_1st | 657 | 1005 | 3468 | 401 | 3258 | 383 | 1320 | 3249 | 131 | 477 | 14349 | 47618 |
| ANRIL_2nd | 672 | 1032 | 3404 | 371 | 3235 | 387 | 1320 | 3173 | 134 | 459 | 14187 | 46874 |
| ANRIL_3rd | 688 | 1149 | 3641 | 391 | 3415 | 428 | 1352 | 3391 | 143 | 505 | 15103 | 51606 |
| Ctrl_1st | 614 | 1010 | 3145 | 304 | 3160 | 359 | 1142 | 3135 | 126 | 409 | 13404 | 42558 |
| Ctrl_2nd | 628 | 1036 | 3290 | 306 | 3136 | 407 | 1135 | 3083 | 136 | 422 | 13579 | 44500 |
| Ctrl_3rd | 628 | 966 | 3095 | 310 | 2962 | 380 | 1100 | 2934 | 104 | 366 | 12845 | 40754 |
| Total | 1912 | 3185 | 10649 | 1267 | 10156 | 1284 | 3652 | 6951 | 424 | 1400 | 40880 | 140131 |
A strict cut-off value P≤0.05 and modified T-value
≥0 were used to identify RASEs with high confidence adjustment. A
total of 1,714 RASEs was identified. ASEs regulated by ANRIL
included 330 ES, 296 A5SS, 286 A3SS and 191 cassette exons Fig. 3A). To analyze whether the increase
in ASEs was due to altered transcription, we overlapped the genes
whose level of expression and alternative splicing were both
regulated by ANRIL, and identified twenty one such genes:
LINC01002, NPIPB4, TXNRD1, OAS1, SLC3A2, NBPF14, STX1A, LINC01063,
DDX60L, CTB-55O6.8, RP11-875O11.1, DDIT3, FXYD2, MEF2B, MX1, WARS,
IFITM1, NBPF26, PI4KAP1, SLC22A20 and PSAT1 (Fig. 3B). These results suggested that
transcriptional regulation and AS may be linked.
 | Figure 3.AS pattern analysis following ANRIL
overexpression in HUVECs. (A) Frequency distribution of types of
ANRIL-regulated ASEs. (B) Overlap analysis between DEGs and
ANRIL-RASGs. Top 10 (C) GO biological processes and (D) KEGG
pathways identified for RASGs. Integrative Genomics Viewer-sashimi
plots of ANRIL-regulated (E) A3SS of CTNNB1 and (F) ES of IL32
demonstrate AS changes in ANRIL overexpression and control cells;
transcripts for the gene presented below. The schematic diagrams
depicts the structures of ASEs, The exon sequences were denoted by
boxes and intron sequences by horizontal lines. RNA-seq
quantification and RT-qPCR validation of ASEs were presented in the
histogram. The altered ratio of AS events in RNA-seq data was
calculated as follows: AS1 junction reads/(AS1 junction reads + AS2
junction reads). The altered ratio of AS events in RT-qPCR data was
calculated as follows: AS1 transcript level/AS2 transcript level.
Student's t test was performed to compare ANRIL-OE and Ctrl cells.
**P<0.01 and ***P<0.001 vs. Ctrl. DEGs, differentially
expressed genes; RASGs, regulated alternative splicing genes;
HUVECs, human umbilical vein endothelial cells; ANRIL,
lncRNA-antisense non-coding RNA in the INK4 locus; GO, Gene
Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; A3SS,
alternative 3′ splice site; ES, exon skipping; ASEs, alternative
splicing events; RNA-seq, RNA-sequencing; RT-qPCR, reverse
transcription-quantitative PCR; Ctrl, control. |
GO analysis was performed to assess the potential
features of regulated AS genes(RASGs). The results demonstrated
that RASGs were enriched in GO biological processes, such as ‘gene
expression’, ‘translation’, ‘DNA repair’, ‘RNA processing’ and
‘positive regulation of the IκB kinase/NF-κB signaling pathway’
(Fig. 3C). Enriched KEGG pathways
(P<0.05) included ‘RNA transport’, ‘Notch signaling pathway’,
‘axon guidance’, ‘apoptosis’ and ‘VEGF signaling pathway’ (Fig. 3D). These results demonstrated that
ANRIL overexpression affected expression of DEGs and including
inflammatory response by regulating AS of transcription factors and
co-activators. To evaluate ASEs identified using the RNA-Seq data,
five possible ASEs were assessed using RT-qPCR. The five events
validated by RT-qPCR were consistent with RNA-Seq results. The five
genes which contained verified splicing events were as follows:
Catenin Beta 1(CTNNB1), IL23, ATPase type 13A2 (ATP13A2), Single
Immunoglobulin and Toll-interleukin 1 Receptor domain (SIGIRR) and
Histone Deacetylase 7A (HDAC7)(Fig.
3E and F; Fig. S3A-C).
Discussion
In the immune system, lncRNAs serve a key role in
regulating cell survival, inflammation and angiogenesis (4,9,10).
The effect of ANRIL on the occurrence of IS has been previously
reported by numerous studies (9,10).
However, the specific mechanism remains unknown. The inflammatory
response may be an important regulatory pathway. Here, ANRIL
overexpression regulated the expression of genes associated with
inflammatory responses, activated the NF-κB signaling pathway and
affected the AS of certain genes (CTNNB1, IL23) with key functions
in the inflammatory response in HUVECs.
In the occurrence and development of IS,
inflammation serves a key role. Numerous studies (26,27) have reported that ANRIL promotes
the inflammatory response, leading to angiogenesis and vascular
endothelial injury, which is similar to the results of GO analysis
following high-throughput sequencing in the present study. But
These results were inconsistent with respect to the inflammatory
reaction and mRNA expression levels of ANRIL in cells and tissue,
which suggested that ANRIL acted as an anti-inflammatory gene
participating in acute IS progression (28,29). This evidence indicated that ANRIL
may serve a bidirectional role in the pathological process of IS,
which can alleviate nerve injury by activating vascular endothelial
growth factors and promote inflammatory progress of IS via the
NF-κB signaling pathway. It has been reported that a number of
lncRNAs are abnormally expressed in inflammatory disease and
participate in activating numerous inflammation-associated genes
and signaling pathways (30). It
has also been indicated that lncRNA changes in IS are present and
provide a unique view of multiple adjustment genes in ANRIL-OE
cells, which are significantly enriched in inflammatory reactions
and the NF-κB pathway. Certain of these gene (NLRC5, CEBPB, PTGS2)
had not been reported to be associated with ANRIL regulation in
previous studies. Among these genes, lncRNAs promotes inflammation
by regulating genes such as lncRNA MALAT1, which regulates PTGS2 by
targeting microRNA-26b to aggravated inflammatory reactions in
myocardial ischemia-reperfusion injury (31). Moreover, Liu et al
(32) reported that
downregulation of NLRC5 decreases cell apoptosis and intracellular
oxidative stress in an acute myocardial infarction model rat, which
was regulated by lncRNA. Certain studies have also reported that
CEBPB and IL6 genes were regulated by lncRNAs and involved in the
inflammatory response (33,34). Furthermore, Liu et al
(35) reported that ANRIL
regulates HUVEC dysfunction by regulating the let-7b/TGF-βR1
signaling pathway to mediate the development of atherosclerosis.
Knockdown of ANRIL significantly promoted cell proliferation and
tubule formation and inhibited inflammatory activation and
apoptosis of HUVEC. Other previous studies reported that
lncRNA-ANRIL knockdown inhibits proliferation and migration of
oxidized low-density lipoprotein-induced HUVECs by sponging
microRNA-339-5p and inactivating the RAS/RAF/ERK signaling pathway
(36,37). To the best of our knowledge,
however, there is a lack of research on the regulation of other
genes by lncRNAs. The results of present study suggested that
lncRNA-ANRIL regulated these genes (NLRC5, CEBPB) to participate in
the inflammatory response. These findings extend understanding of
the role of ANRIL in the inflammatory response. Furthermore, the
identification of downstream genes provides additional possible
research directions.
NF-κB, a classical nuclear transcription factor, has
been reported to be involved in inflammation and cell survival
(4). Many of the upregulated
genes in ANRIL-OE cells were enriched in ‘NF-κB pathway’. These
genes may affect the NF-κB signaling pathway as downstream
regulatory genes of ANRIL. The study on the correlation between
upregulated genes and lncRNAs have reported that lncRNAs regulate
the NF-κB signaling pathway via certain genes, such as caspase
recruitment domain family member 8 (CARD8) which is the target of
ANRIL and that ANRIL is an inhibitor of the NF-κB signaling pathway
(38). Activation of ANRIL
inhibits the inflammatory process by inhibiting NF-κB signaling
pathway via activation of CARD8. The present study identified
potential controlled by lncRNA ANRIL that regulate the NF-κB
signaling pathway. Certain genes have been confirmed to exist in
the lncRNA/NF-κB regulatory pathway, including KLF4 and NLRC5. KLF4
decrease transcription activity of NF-κB by establishing a positive
feedback loop of NF-κB/KLF4, which is controlled by
NF-κB-interacting lncRNA (39),
and lncRNA ARID 2-IR, which was located within the intron region
between the fourth and fifth exons of Arid2 on the chromosome 15 of
the mouse genome, mediates renal inflammation driven by NF-κB via a
NLRC5-dependent mechanism (40).
To the best of our knowledge, however, the role of genes in the
ANRIL/NF-κB regulatory pathway remains unknown. GO and KEGG
analysis in the present study demonstrated the identity of
candidate genes, which need to be assessed by further tests.
AS is a process in which exons are included or
excluded from the final mRNA transcripts, thus allowing genes to
produce several isoforms with cell- and tissue-specific functions.
AS and its role in transcript diversity is critical to the
susceptibility and severity of disease and different types of AS
have previously been reported following stroke (41). In previous study, only a few
lncRNAs were reported to have an important role in RNA splicing
(42). In the present study,
ANRIL was a multifunctional lncRNA that regulated both AS and
translation and ASEs occurred in genes involved in the inflammatory
response, including CTNNB1, IL32, ATP13A2, SIGIRR and HDAC7.
SIGIRR, a negative regulator of the TLR signaling pathway, has been
reported to attenuate colonic tissue inflammation via inhibition of
TLR4/NF-κB overactivation by downregulating TLR4, MyD88 and NF-κB
p65 expression in a mouse model of colitis (43). HDACs drive innate immune
cell-mediated inflammation and serve as key molecular links between
TLR-inducible aerobic glycolysis and macrophage inflammatory
responses (44). CTNNB1, which is
a central mediator in the canonical Wnt signaling pathway, has been
extensively reported to serve an oncogenic role in numerous types
of cancer and stimulate proliferation of microvascular endothelial
cells (45,46). Moreover, it has been reported that
lncRNAs facilitate tumor progression by targeting CTNNB1 to
activate the Wnt/β-catenin signaling pathway (47,48). The present study identified CTNNB1
as an ANRIL-regulated AS gene, which indicated that
CTNNB1-dependent transcriptional regulation may be involved in the
inflammatory response. The findings of the present study regarding
AS of multiple genes mediated by ANRIL suggested a more complex
network between these inflammatory response genes.
The present study had certain limitations. First, it
focused on the effects of lncRNA ANRIL on inflammatory-associated
pathways in HUVECs but further studies are required to elucidate
the underlying molecular mechanism of the effects in HUVECs and
other types of cell in detail. Second, there was a lack of research
on protein expression levels and additional experiments are needed
to demonstrate the effect on protein expression levels.
Furthermore, future studies using animal models and patient tissue
samples should be conducted to elucidate the mechanism underlying
ANRIL-mediated effects in vivo.
In the present study, it was demonstrated that ANRIL
was involved in the inflammatory response and genetic transcription
of NF-κB. These findings may provide a new direction for the
understanding of vascular disease and tumorigenesis, which need to
be further evaluated by future studies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Weili Quan and Mr
Ning Li (both Center for Genome Analysis, ABLife Inc., Wuhan,
Hubei, China) for helpful discussions.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
AW and JM conceived and designed the experiments.
AW, XL, LD, JL, MS and DFH performed the experiments and
constructed the table and figures. XL, LD, MS and JL provided the
reagents, materials and analysis tools. AW and JM wrote the paper.
JL, DFH, MS and JM revised the manuscript. All authors read and
approved the final version of the manuscript. AW, XL, LD, JL, MS,
DH and JM confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ren W and Yang X: Pathophysiology of long
non-coding RNAs in ischemic stroke. Front Mol Neurosci. 11:962018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grammatikakis I and Lal A: Significance of
lncRNA abundance to function. Mamm Genome. 33:271–280. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lorenzen JM and Thum T: Long noncoding
RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol.
12:360–373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen S, Wu Y, Qin X, Wen P, Liu J and Yang
M: Global gene expression analysis using RNA-seq reveals the new
roles of panax notoginseng saponins in ischemic cardiomyocytes. J
Ethnopharmacol. 268:1136392021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS
and Feng ZP: Long non-coding RNAs in ischemic stroke. Cell Death
Dis. 9:2812018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Folkersen L, Kyriakou T, Goel A, Peden J,
Mälarstig A, Paulsson-Berne G, Hamsten A, Watkins H,
Franco-Cereceda A, Gabrielsen A, et al: Relationship between CAD
risk genotype in the chromosome 9p21 locus and gene expression.
Identification of eight new ANRIL splice variants. PLoS One.
4:e76772009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen W, Bharati S, Yi L, Benowitz L, Chen
Q, Zhang Z, Patel NJ, Aziz-Sultan AM, Chiocca AE and Wang X:
Monogenic, polygenic, and microRNA markers for ischemic stroke. Mol
Neurobiol. 56:1330–1343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amouyel P: From genes to stroke subtypes.
Lancet Neurol. 11:931–933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bai N, Liu W, Xiang T, Zhou Q, Pu J, Zhao
J, Luo D, Liu X and Liu H: Genetic association of ANRIL with
susceptibility to Ischemic stroke: A comprehensive meta-analysis.
PLoS One. 17:e02634592022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun R, Wang L, Guan C, Cao W and Tian B:
Carotid atherosclerotic plaque features in patients with acute
ischemic stroke. World Neurosurg. 112:e223–e228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Luo X, Chen F, Yuan W, Xiao X,
Zhang X, Dong Y, Zhang Y and Liu Y: LncRNA SNHG1 regulates
cerebrovascular pathologies as a competing endogenous RNA through
HIF-1α/VEGF signaling in ischemic stroke. J Cell Biochem.
119:5460–5472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mo Y, Sun YY and Liu KY: Autophagy and
inflammation in ischemic stroke. Neural Regen Res. 15:1388–1396.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhong W, Li YC, Huang QY and Tang XQ:
lncRNA ANRIL ameliorates oxygen and glucose deprivation (OGD)
induced injury in neuron cells via miR-199a-5p/CAV-1 axis.
Neurochem Res. 45:772–782. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang X, Hu J and Zhou H: Knock-down of
long non-coding RNA ANRIL suppresses mouse mesangial cell
proliferation, fibrosis, inflammation via regulating Wnt/β-Catenin
and MEK/ERK pathways in diabetic nephropathy. Exp Clin Endocrinol
Diabetes. 130:30–36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu B, Cao W and Xue J: LncRNA ANRIL
protects against oxygen and glucose deprivation (OGD)-induced
injury in PC-12 cells: Potential role in ischaemic stroke. Artif
Cells Nanomed Biotechnol. 47:1384–1395. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao JH, Wang B, Wang XH, Wang JR and Xu
CW: Influence of lncRNA ANRIL on neuronal apoptosis in rats with
cerebral infarction by regulating the NF-κB signaling pathway. Eur
Rev Med Pharmacol Sci. 23:10092–10100. 2019.PubMed/NCBI
|
|
18
|
Zhang B, Wang D, Ji TF, Shi L and Yu JL:
Overexpression of lncRNA ANRIL up-regulates VEGF expression and
promotes angiogenesis of diabetes mellitus combined with cerebral
infarction by activating NF-κB signaling pathway in a rat model.
Oncotarget. 8:17347–17359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim D, Pertea C, Trapnell H, Pimentel R,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trapnell CBA, Williams G, Pertea A,
Mortazavi G, Kwan MJ, van Baren SL, Salzberg SL, Wold BJ and
Pachter L: Transcript assembly and quantification by RNA-Seq
reveals unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie CX, Mao J, Huang Y, Ding J, Wu S, Dong
L, Kong G, Gao C, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:(Web Server issue). W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y
and Zhang L: CELF1 preferentially binds to exon-intron boundary and
regulates alternative splicing in HeLa cells. Biochim Biophys Acta
Gene Regul Mech. 1860:911–921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin L, Li G, Yu D, Huang W, Cheng C, Liao
S, Wu Q and Zhang Y: Transcriptome analysis reveals the complexity
of alternative splicing regulation in the fungus Verticillium
dahliae. BMC Genomics. 18:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu HW, Hu ZL, Li H, Tan QF, Tong J and
Zhang YQ: Knockdown of lncRNA ANRIL suppresses the production of
inflammatory cytokines and mucin 5AC in nasal epithelial cells via
the miR-15a-5p/JAK2 axis. Mol Med Rep. 23:1452021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou B, Li L, Qiu X, Wu J, Xu L and Shao
W: Long non-coding RNA ANRIL knockdown suppresses apoptosis and
pro-inflammatory cytokines while enhancing neurite outgrowth via
binding microRNA-125a in a cellular model of Alzheimer's disease.
Mol Med Rep. 22:1489–1497. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng L, Guo J and Ai F: Circulating long
noncoding RNA ANRIL downregulation correlates with increased risk,
higher disease severity and elevated pro-inflammatory cytokines in
patients with acute ischemic stroke. J Clin Lab Anal.
33:e226292019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Tang X, Liu K, Hamblin MH and Yin
KJ: Long noncoding RNA Malat1 regulates cerebrovascular pathologies
in ischemic stroke. J Neurosci. 37:1797–1806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruan Z, Wang S, Yu W and Deng F: LncRNA
MALAT1 aggravates inflammation response through regulating PTGS2 by
targeting miR-26b in myocardial ischemia-reperfusion injury. Int J
Cardiol. 288:1222019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Z, Liu J, Wei Y, Xu J, Wang Z, Wang P,
Sun H, Song Z and Liu Q: LncRNA MALAT1 prevents the protective
effects of miR125b5p against acute myocardial infarction through
positive regulation of NLRC5. Exp Ther Med. 19:990–998.
2020.PubMed/NCBI
|
|
33
|
Wu H, Liu B, Chen Z, Li G and Zhang Z:
MSC-induced lncRNA HCP5 drove fatty acid oxidation through
miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and
chemo-resistance of gastric cancer. Cell Death Dis. 11:2332020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang L and Li J: lncRNA GMDS-AS1
upregulates IL-6, TNF-α and IL-1β, and induces apoptosis in human
monocytic THP-1 cells via miR-96-5p/caspase 2 signaling. Mol Med
Rep. 25:672022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu X, Li S, Yang Y, Sun Y, Yang Q, Gu N,
Li J, Huang T, Liu Y, Dong H, et al: The lncRNA ANRIL regulates
endothelial dysfunction by targeting the let/TGF-βR1signalling
pathway. J Cell Physiol. 236:2058–2069. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang T, Zhao HY, Zhang XB, Gao XL, Peng
WP, Zhou Y, Zhao WH and Yang HF: LncRNA ANRIL regulates cell
proliferation and migration via sponging miR-339-5p and regulating
FRS2 expression in atherosclerosis. J Eur Rev Med Pharmacol Sci.
24:1956–1969. 2020.PubMed/NCBI
|
|
37
|
Ying Liu X, Li S, Yang Y, Sun Y, Yang Q,
Gu N, Li J, Huang T, Liu Y, Dong H, et al: The lncRNA ANRIL
regulates endothelial dysfunction by targeting the let-7b/TGF-βR1
signalling pathway. J Cell Physiol. 236:2058–2069. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bai Y, Nie S, Jiang G, Zhou Y, Zhou M,
Zhao Y, Li S, Wang F, Lv Q, Huang Y, et al: Regulation of CARD8
expression by ANRIL and association of CARD8 single nucleotide
polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke.
45:383–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu X, Du J, Yu J, Guo R, Feng Y, Qiao L,
Xu Z, Yang F, Zhong G, Liu F, et al: LncRNA NKILA regulates
endothelium inflammation by controlling a NF-κB/KLF4 positive
feedback loop. J Mol Cell Cardiol. 126:60–69. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang P, Yu C, Yu J, Yu J, Li Z, Lan HY
and Zhou Q: Arid2-IR promotes NF-κB-mediated renal inflammation by
targeting NLRC5 transcription. Cell Mol Life Sci. 78:2387–2404.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Suñé-Pou M, Prieto-Sánchez S,
Boyero-Corral S, Moreno-Castro C, El Yousfifi Y, Suñé-Negre JM,
Hernández-Munain C and Suñé C: Targeting splicing in the treatment
of human disease. Genes (Basel). 8:872017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang JG, Sachdeva M, Xu E, Robinson TJ,
Luo L, Ma Y, Williams NT, Lopez O, Cervia LD, Yuan F, et al: The
long noncoding RNA NEAT1 promotes sarcoma metastasis by regulating
RNA splicing pathways. Mol Cancer Res. 18:1534–1544. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu J, Chen Y, Liu D, Liu W, Hu S, Zhou N
and Xie Y: Ectopic expression of SIGIRR in the colon ameliorates
colitis in mice by downregulating TLR4/NF-κB overactivation.
Immunol Lett. 183:52–61. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gupta KD, Shakespear MR, Curson JEB,
Murthy AMV, Iyer A, Hodson MP, Ramnath D, Tillu VA, von Pein JB,
Reid RC, et al: Class IIa histone deacetylases drive toll-like
receptor-inducible glycolysis and macrophage inflammatory responses
via pyruvate kinase M2. Cell Rep. 30:2712–2728. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Moon-Sung L, Hee-Jung B, Lee J, Jeoung DI,
Kim YM and Lee H: Tetraspanin CD82 represses Sp1-mediated Snail
expression and the resultant E-cadherin expression interrupts
nuclear signaling of β-catenin by increasing its membrane
localization. J Cell Signal. 52:83–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chai W, Liu R, Li F, Zhang Z and Lei B:
Long noncoding RNA TSLNC8 enhances pancreatic cancer aggressiveness
by regulating CTNNB1 expression via association with HuR. Hum Cell.
34:165–176. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pan Z, Ding J, Yang Z, Li H, Ding H and
Chen Q: LncRNA FLVCR1-AS1 promotes proliferation, migration and
activates Wnt/β-catenin pathway through miR-381-3p/CTNNB1 axis in
breast cancer. Cancer Cell Int. 20:2142020. View Article : Google Scholar : PubMed/NCBI
|