Introduction
As a type of gastrointestinal cancer, colorectal
cancer (CRC) can occur anywhere from the cecum to the rectum
(1). Epidemiological studies have
suggested that the incidence of colon cancer is higher in men
compared with women (2). The
incidence and mortality rates of colon cancer both increase
progressively with age (3). The
commonest treatment approaches for CRC include chemical and
surgical therapy, and radiotherapy, along with targeted therapy
(4). Surgical treatment is
appropriate for patients with early tumors without invasion or
metastasis. The majority of patients can be cured following
surgical resection of the primary lesion, which is considered as
the most effective treatment strategy for colon cancer (5). However, when the tumor reaches an
intermediate and advanced stage, the effectiveness of surgical
treatment is low, the risk of recurrence is greater and the quality
of life of patients is also relatively poor (6). Therefore, the development of more
effective treatment approaches and the identification of novel
therapeutic targets for patients with CRC are of great
significance.
Being located on chromosome 5 (5q12.1), DEP domain
protein 1B (DEPDC1B) gene encodes a protein encompassing two
conserved domains, namely DEP and RhoGAP (7). DEP, as a globular domain, can
interact with G protein-coupled receptors (GPCRs) to regulate GPCR
signaling. The RhoGAP domain is associated with Rho-GTPase signal
transduction, which in turn is closely associated with cell
proliferation, migration and invasion, cell cycle progression as
well as cytoskeletal recombination (8). It has been reported that DEPDC1B is
upregulated in several types of cancer, such as prostate cancer
(PCa), soft tissue sarcoma, cervical cancer and malignant melanoma
(9–12). A previous study indicated that
DEPDC1B could promote the proliferation of oral cancer cells by
regulating the Ras-related C3 botulinum toxin substrate 1
(Rac1)/extracellular signal-regulated protein kinase 1/2 signaling
pathway (13). Another study
revealed that DEPDC1B, which could act as an oncogene in non-small
cell lung cancer (NSCLC), was negatively associated with patient
survival, while it could enhance the migration and invasion
abilities of NSCLC cells by activating the Wnt/β-catenin pathway
(14), but the role of DEPDC1B in
CRC and its underlying mechanism has not yet reported. It has been
reported that NUP37, as a component of the nuclear pore complex,
serves a crucial role in the regulation of gene expression and
heterochromatin function, as well as in the formation of the
nuclear envelope (15). Huang
et al (16) demonstrated
that NUP37 deficiency could attenuate cell proliferation and induce
G1 phase cell cycle arrest and apoptosis in NSCLC cells.
Furthermore, NUP37 could enhance the proliferation, migration and
invasion abilities of gastric cancer cells via triggering the
PI3K/AKT/mammalian target of rapamycin signaling pathway (17). However, the role of NUP37 in CRC
and its association with DEPDC1B remain to be elucidated.
Therefore, the current study aimed to explore the expression of
DEPDC1B in CRC cells and uncover the effect of DEPDC1B on
regulating CRC cell proliferation, metastasis, cell cycle and
apoptosis.
Materials and methods
Bioinformatics analysis
The Coexpedia database (http://www.coexpedia.org/) was used to analyze the
co-expression between DEPDC1B and nucleoporin 37 (NUP37). PPA_pred
database (https://www.iitm.ac.in/bioinfo/PPA_Pred/prediction.html#)
was used to predict the binding between DEPDC1B and NUP37.
Cell culture and treatment
The human CRC cell lines SW620, HCT8 and HCT116 were
purchased from Procell Life Science & Technology Co., Ltd.,
while the human HIEC intestinal epithelial cell line was provided
by Ningbo Mingzhou Biotechnology Co., Ltd. Cells were cultured in
DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS and 1% penicillin/streptomycin solution at 37°C in a
humidified atmosphere with 5% CO2.
Cell transfection
NUP37-specific pcDNA overexpression plasmid
(Ov-NUP37) and the corresponding negative control (Ov-NC), the
specific small interfering (si)RNA targeting DEPDC1B
(si-DEPDC1B-1:5′-GAUUCUAAGUCAAAUGCAAAU-3′;
si-DEPDC1B-2:5′-GAAGAGAUAUGGAAGUCUAUG-3′; 50 nM), the specific
siRNA targeting NUP37 (si-NUP37-1: 5′-GAGUUGCUGUAAAGAUUAAAU-3′;
si-NUP37-2: 5′-GUGUGUGUAUAUAUAUAUAUU-3′; 50 nM) and the siRNA
control (si-NC: 5′-UUCUCCGAACGUGUCACGU-3′; 50 nM) were obtained
from Shanghai GenePharma Co., Ltd. HCT8 cells were transfected with
the above recombinants using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37°C,
according to the manufacturer's instructions. Subsequent
experiments were performed 48 h post-transfection.
Cell Counting Kit-8 (CCK-8) assay
Following inoculation of untreated or transfected
HCT8 cells into 96-well plates at a density of 5×104
cells/ml, cells were cultured in DMEM with 10% FBS for 24, 48 and
72 h at 37°C. Subsequently, cells were treated with 10 µl WST-8
(Beyotime Institute of Biotechnology) at 37°C followed by
incubation for an additional 2 h. The optical density value in each
well was measured at a wavelength of 450 nm using a microplate
reader.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Following seeding into six-well plates
(4×105 cells/well), HCT8 cells were cultured overnight
at 37°C. Subsequently, HCT8 cells were fixed with 4%
polyformaldehyde for 15 min at room temperature and 0.5% Triton
X-100 at room temperature for 15 min. Cells were then stained using
the Cell-Light™ EdU Cell Proliferation Detection Assay
(Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature
for 30 min followed by counterstaining with DAPI at room
temperature for 10 min. Finally, five areas were randomly selected
at ×200 magnification under a fluorescent microscope (Nikon
Corporation).
Wound healing assay
Following inoculation into a six-well plate, the
transfected cells were cultured at 37°C until they reached 80–90%
confluency. Subsequently, a wound was created on the cell monolayer
using a 20-µl pipette tip and cells were then cultured at 37°C in
serum-free medium. Following incubation for 24 h, the area occupied
by the migrated cells in the wound was determined. The migration
rate was calculated using the following formula: (Wound width at 0
h-wound width at 24 h)/wound width at 0 h ×100%. Cells were
analyzed at five randomly selected fields under a light microscope
(magnification, ×100).
Transwell assay
Initially, the Transwell chambers (Corning, Inc.)
were pre-treated with 0.1 ml Matrigel (Becton-Dickinson) at 37°C
for 30 min. The collected cells (density, 2×105
cells/ml) were cultured in DMEM supplemented with 1% FBS at 37°C
for 48 h. Cell suspensions were seeded into the top chamber, while
the lower chamber was supplemented with medium containing 10% FBS
at 37°C for 48 h. Following incubation, a cotton swab was employed
to remove cells on the upper surface of the Transwell membrane.
Subsequently, invaded cells on the lower surface were fixed with
100% methanol for 10 min at room temperature, followed by staining
with 0.1% crystal violet for 10 min at room temperature. Finally,
the invaded cells were counted at five randomly selected fields
under a light microscope (magnification, ×100).
Co-immunoprecipitation (Co-IP)
assay
Total proteins were extracted from cells
(4×107) using an IP lysis buffer (20 mM Tris-HCl, 150 mM
NaCl, 1% Triton X-100, pH 7.5). The supernatant was centrifuged at
14,000 × g at 4°C for 10 min to obtain whole-cell extracts. A part
of the cell lysate was isolated as input to correct for
non-specific binding and 250 µl cell lysates were then cultivated
with rabbit-IgG, followed by incubation with DEPDC1B (dilution,
1:100; cat no. 8283; ProSci, Inc.) or NUP37 antibody (dilution,
1:100; cat no. PAB088Hu01; Wuhan USCN Business Co., Ltd.) with
Protein A/G PLUS-Agarose beads (MilliporeSigma) at 4°C overnight.
Subsequently, the precipitate was washed in lysis buffer and then
centrifuged (700 × g at 4°C for 5 min). After discarding the
supernatant, 25 µl 2C loading buffer was added, and then samples
were heated in boiling water for 7 min to elute proteins, followed
by western blotting as mentioned below.
Flow cytometry
The FITC Annexin V/PI Apoptosis Detection kit I
(Guangzhou RiboBio Co., Ltd.) was used to assess cell apoptosis.
The PBS-rinsed HCT8 cells were re-suspended in binding buffer and
were then supplemented with 5 µl Annexin V-FITC and 10 µl propidium
iodide (PI; 10 mg/ml) and incubated for 15 min in the dark at room
temperature. Cell apoptosis was analyzed using FlowJo software
(FlowJo LLC). Cell apoptosis rate (the percentage of early + late
apoptotic cells) was obtained from three different replications.
Furthermore, cell cycle was also assessed by flow cytometry.
Briefly, cells that were harvested using trypsinization, were fixed
with 70% ethanol at 4°C overnight, followed by staining with a
solution containing 50 µg/ml PI and 100 µg/ml RNase I at room
temperature for 1 h in PBS. Cell cycle was then analyzed at the
Flow Cytometry Core Facility of University of Colorado Denver (UCD)
with a FACScan flow cytometer (BD Biosciences).
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA isolated from 1×104 HCT8 cells
utilizing TRIzol® reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols was reverse
transcribed into cDNA according to the manufacturer's protocols
using the PrimeScript RT Master Mix kit (Perfect Real Time; Takara
Bio, Inc.), according to the manufacturer's protocol. cDNA was
amplified by qPCR using the SYBR Premix Ex Taq™ II kit
(Takara Bio Inc.) according to the manufacturer's instructions. The
thermocycling conditions were as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 58°C
for 60 sec. The following primer pairs were used for qPCR: DEPDC1B
forward, 5′-AGACCGTGGAGCTTTTTCGTG-3′ and reverse,
5′-TTCAGGGCCGAAGTTTTGACT-3′; NUP37 forward,
5′-AAGAAGCAGACGTTGAAGGCA-3′ and reverse,
5′-CTCTGGGCTCCAAGCTATGC-3′; and GAPDH forward,
5′-AATGGGCAGCCGTTAGGAAA-3′ and reverse, 5′-GCGCCCAATACGACCAAATC-3′.
The relative mRNA levels were determined using the
2−ΔΔCq method (18).
GAPDH mRNA level was used for normalization. RT-qPCR was performed
in triplicate.
Xenograft experiments
All animal experiments were approved by The First
Affiliated Hospital of Nanchang University (approval no.
SD-2021-011) and strictly followed the Guidelines for the Care and
Use of Laboratory Animals by the National Institute of Health
(19). A total of 15 male BALB/c
nude mice (Beijing Vital River Laboratory Animal Technology Co,
Ltd.) were housed in a temperature-controlled room (22±1°C) at
40–70% humidity with a 12-h light/dark cycle. The animals had free
access to standard food pellets and water. All nude mice (aged, 4–5
weeks; weight, 18–20 g) were randomly divided into three groups
(n=5/group). Α total of 5×106 cells transfected with
si-DEPDC1B with or without Ov-NUP37, were subcutaneously injected
into the right flank of each nude mouse. Tumor volume and body
weight were measured every three days. Tumor volume was calculated
using the following formula: Volume=(length × width2)/2.
The allowed maximum tumor diameter and volume were 1.5 cm and 2,000
mm3, respectively. Any difficulties in eating and water
intake, any symptoms of discomfort (self-harm, abnormal posture,
respiratory distress, crying), the long-term appearance of
abnormalities with no signs of recovery (diarrhea, bleeding, faeces
on genital area), if the animal's distress was judged to be
intolerable, such as heavy weight loss (≥20% within a few days) or
the transplanted cancer cells grew to a size of ≥1.5 cm, were used
as the humanitarian endpoints of the present study and the
experiment was immediately terminated and the animal was
sacrificed. After three weeks, mice were sacrificed by
CO2 (50% vol/min) asphyxiation followed by cervical
dislocation. The tumor tissues were then weighed and subjected to
immunohistochemical and western blot analyses.
Immunohistochemistry assay
Cancer tissues from mice were fixed with 4%
paraformaldehyde for 24 h at room temperature, embedded in paraffin
and cut into 5-µm sections. The sections were then re-hydrated and
deparaffinized by immersion in xylene (100% ×2) followed by
immersion in a graded alcohol series (100% ethanol for 1 min twice;
95% ethanol for 1 min and 70% ethanol for 1 min twice). For antigen
retrieval, the tissues were microwaved in citrate-buffered solution
(pH 6.0) for 3 min. Following blocking with 10% goat serum for 30
min at room temperature, the sections were first probed with Ki-67
antibody (dilution, 1:300; cat no. ab15580; Abcam) at 4°C overnight
and then with HRP-labelled goat anti-rabbit secondary antibody
(cat. no. ab6721; dilution, 1:1,000; Abcam) for 30 min at room
temperature. Finally, the sections were first stained with DAB and
then with hematoxylin for 3 min at room temperature. Images of the
tissue sections in five randomly selected fields were captured
under a light microscope (magnification, ×100).
Western blotting
Total proteins were extracted from tissues and cells
utilizing a RIPA buffer (Auragene Bioscience Co.) and were then
quantified using a BCA Protein Assay kit (Beijing Dingguo
Changsheng Biotechnology Co., Ltd.), according to the standard
protocol. Following separation by 10% SDS-PAGE (Bio-Rad
Laboratories, Inc.), the proteins (50 µg) were transferred onto
PVDF membranes (MilliporeSigma). The membranes, which were first
blocked in 5% non-fat milk for 1 h at 37°C in 0.1% tris-buffered
saline with 0.1% Tween-20 (TBST), were incubated with specific
antibodies targeting DEPDC1B (1:1,000; cat. no. LS-C674693;
LifeSpan Biosciences), MMP-2 (1:1,000; cat. no. ab92536; Abcam),
MMP-9 (1:1,000; cat. no. ab76003; Abcam), Bcl-2 (1:1,000; cat. no.
ab692; Abcam), Bax (1:1,000; cat. no. ab32503; Abcam), Cyclin D1
(1:1,000; cat. no. ab16663; Abcam), cyclin B1 (1:1,000; cat. no.
ab32053; Abcam), NUP37 (1:1,000; cat. no. LS-C135188; LifeSpan
Biosciences), phosphorylated (p-)PI3K antibody (1:1,000; cat. no.
ab182651; Abcam), PI3K antibody (1:1,000; cat. no. ab86714; Abcam),
p-Akt antibody (1:1,000; cat. no. ab38449; Abcam), Akt antibody
(1:500, ab8805; Abcam) and GAPDH (1:2,500, ab9485; Abcam) at 4°C
overnight. Subsequently, the PBST-rinsed membranes were probed with
the HRP-conjugated goat anti-rabbit or mouse secondary antibodies
(cat. nos. sc-2004 or sc-2005; 1:5,000; Santa Cruz Biotechnology,
Inc.) for 1 h at room temperature. Finally, the protein bands were
visualized utilizing an ECL detection system (Beyotime Institute of
Biotechnology), according to the manufacturer's protocol. Finally,
the protein bands were quantified by means of densitometry
(QuantityOne 4.5.0 software; Bio-Rad Laboratories, Inc.).
Statistical analysis
All data are expressed as the mean ± SD. The results
were analyzed using SPSS software (version 18.0; SPSS, Inc.).
Differences among multiple groups were analyzed using one-way ANOVA
with a post hoc Bonferroni multiple comparison test. P<0.05 was
considered to indicate a statistically significant difference.
Results
DEPDC1B is upregulated in CRC
cells
To investigate the role of DEPDC1B in CRC
progression, the expression levels of DEPDC1B were detected in CRC
cells. As shown in Fig. 1A and B,
the mRNA and protein expression levels of DEPDC1B were notably
increased in CRC cells compared with those in HIEC cells. Since
HCT8 cells showed the highest DEPDC1B expression levels, these
cells were used for the following experiments.
DEPDC1B silencing inhibits the
proliferation, invasion and migration of CRC cells
To determine the biological role of DEPDC1B in CRC
cells, a specific siRNA clone targeting DEPDC1B was transfected
into HCT8 cells. The results demonstrated that the mRNA and protein
expressions levels of DEPDC1B were conspicuously reduced following
cell transfection with si-DEPDC1B (Fig. 2A and B). The si-DEPDC1B-1 clone
exhibited the most potent silencing ability. Therefore,
si-DEPDC1B-1 was chosen for the subsequent assays. Furthermore,
CCK-8 assays showed that DEPDC1B silencing markedly suppressed cell
viability (Fig. 2C). In addition,
EdU assays demonstrated that the colony formation ability of
DEPDC1B-depleted CRC cells was decreased compared with cells
transfected with si-NC (Fig. 2D).
Additionally, DEPDC1B knockdown attenuated the migration ability of
HCT8 cells compared with the control group (Fig. 2E and F). Transwell assays also
indicated that the invasion ability of HCT8 cells was restrained by
DEPDC1B knockdown (Fig. 2G and H).
Finally, western blotting showed that DEPDC1B silencing
specifically decreased the protein expression levels of migration
and invasion-related proteins MMP-2 and MMP-9 (Fig. 2I).
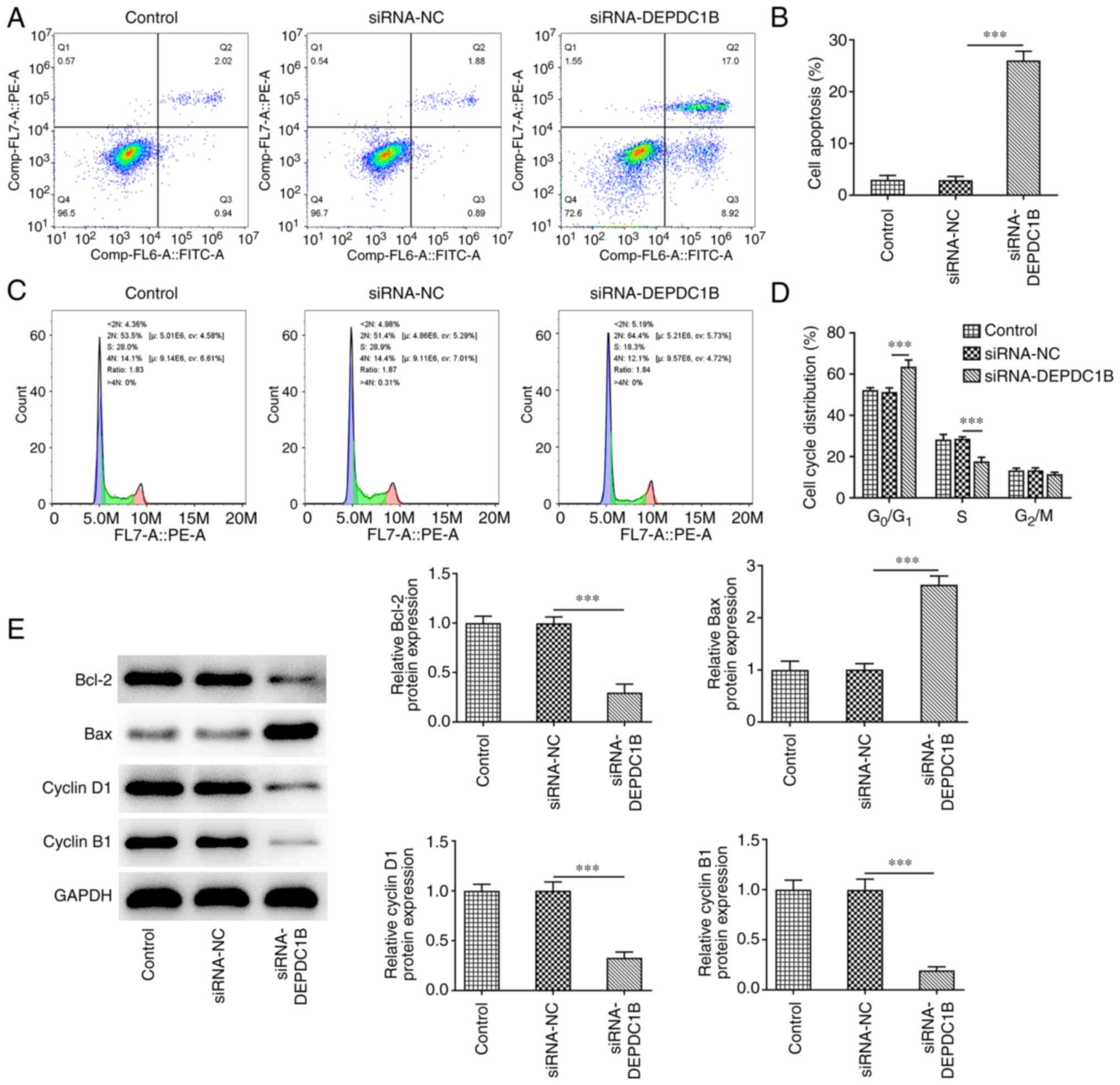
DEPDC1B silencing promotes CRC cell
apoptosis and cell cycle arrest
Subsequently, the effect of DEPDC1B silencing on CRC
cell apoptosis and cell cycle arrest was explored. As shown in
Fig. 3A and B, the apoptosis rate
of HCT8 cells transfected with si-DEPDC1B was notably elevated. In
addition, the percentage of cells in the
G0/G1 phase of the cell cycle was markedly
increased, while that in the S phase was diminished in
DEPDC1B-depleted cells, indicating the cell cycle was arrested
after DEPDC1B silencing (Fig.
3C,D). Additionally, DEPDC1B knockdown in HCT8 cells
upregulated Bax and downregulated Bcl-2, cyclin D1 and cyclin B1
(Fig. 3E).
DEPDC1B directly binds to NUP37
As shown in Fig. 4A and
B, the mRNA and protein expression levels of NUP37 were
significantly increased in CRC cells compared with the control
group. The protein expression levels of NUP37 were notably reduced
following DEPDC1B knockdown (Fig.
4C). Bioinformatics analysis using the Coexpedia database
predicted that DEPDC1B was co-expressed with NUP37 and the score
was 2.927 (Fig. 4D). Additionally,
the Co-IP assay results verified the interaction between DEPDC1B
and NUP37 (Fig. 4E).
NUP37 knockdown restrains the
proliferation, invasion and migration of CRC cells
To investigate the effect of NUP37 on
DEPDC1B-mediated CRC progression, NUP37 was silenced in HCT8 cells.
The results revealed that NUP37 knockdown downregulated NUP37 in
HCT8 cells (Fig. 5A and B).
Evidently, si-NUP37-1 had an improved transfection efficiency
compared with si-NUP37-1. Therefore, si-NUP37-1 was selected for
the subsequent experiments. Additionally, the CCK-8 assay results
showed that NUP37 silencing significantly attenuated the
proliferation capability of HCT8 cells (Fig. 5C). Furthermore, NUP37 depletion
reduced the number of positive-stained cells compared with the
control group (Fig. 5D). In
addition, wound healing and Transwell assays demonstrated that the
migration and invasion abilities of HCT8 cells were markedly
attenuated following NUP37 knockdown (Fig. 5E-H). Finally, western blotting
demonstrated that MMP-2 and MMP-9 were both downregulated in
NUP37-depleted cells (Fig.
5I).
 | Figure 5.NUP37 knockdown restrains the
proliferation, invasion and migration of CRC cells. The (A) mRNA
and (B) protein levels of NUP37 were detected after transfection
with siRNA-NUP37-1/2 by reverse transcription-quantitative PCR and
western blotting. Cell proliferation was evaluated by (C) CCK-8 and
(D) EdU assays. Original magnification, ×200. (E and F) Cell
migration was evaluated by wound healing assay. Original
magnification, ×100. (G and H) Cell invasion was evaluated by
Transwell assay. Original magnification, ×100. (I) Levels of MMP2
and MMP9 were measured by western blotting. Data are expressed as
mean ± SD. **P<0.01, ***P<0.001. NUP37, nucleoporin 37; CRC,
colorectal cancer; si, small interfering; EdU,
5-Ethynyl-2′-deoxyuridine; NC, negative control. |
NUP37 silencing accelerates the
apoptosis and cycle arrest of CRC cells
As shown in Fig. 6A and
B, NUP37 silencing obviously enhanced cell apoptosis compared
with NC cells. The results obtained from flow cytometric assays
demonstrated that the percentage of cells in the
G0/G1 phase of the cell cycle was markedly
increased, while the percentage of cells in the S phase was reduced
in NUP37-depleted cells (Fig. 6C and
D). Finally, NUP37 deficiency decreased the levels of Bcl-2,
cyclin D1 and cyclin B1, and increased those of Bax in HCT8 cells
(Fig. 6E).
Overexpression of NUP37 reverses the
effects of DEPDC1B knockdown on HCT8 cell proliferation,
metastasis, apoptosis and cycle arrest
To further explore the biological function of NUP37
in HCT8 cells, NUP37 was overexpressed. The transfection efficiency
is presented in Fig. 7A and B. As
shown in Fig. 7C, NUP37
overexpression markedly strengthened the proliferation of
DEPDC1B-depleted HCT8 cells. Additionally, NUP37 overexpression
greatly increased the number of positive-stained cells (Fig. 7D). Wound healing and Transwell
assays revealed that the cell migration and invasion rates were
increased in NUP37-overexpressing cells (Fig. 7E-H). In addition, the western
blotting results demonstrated that the protein levels of MMP2 and
MMP9, the two protein that were associated with cell migration and
invasion capacity, were enhanced in cells co-transfected with
si-DEPDC1B and Ov-NUP37 (Fig. 7I).
Additionally, the apoptosis rate of cells co-transfected with
si-DEPDC1B and Ov-NUP37 was obviously decreased compared with cells
transfected with si-DEPDC1B only (Fig.
8A and B). Furthermore, NUP37 overexpression reduced the number
of cells in the G0/G1 phase of the cell cycle
and increased those in the S phase in si-DEPDC1B-transfected cells
(Fig. 8C and D). Finally, the
levels of Bcl-2 and Bax were detected to assess the apoptotic
levels, and the levels of cyclin D1 and cyclinB1 were detected to
measure the cell cycle. The results showed that the protein
expression levels of Bcl-2, cyclin D1 and cyclinB1 were increased
and the level of Bax were reduced in NUP37 overexpressing cells
(Fig. 8E).
 | Figure 7.DEPDC1B knockdown inhibits the
proliferation and metastasis of HCT8 cells through NUP37. The (A)
mRNA and (B) protein levels of NUP37 were detected after the
transfection with Ov-NUP37 by reverse transcription-quantitative
PCR and western blotting. Cell proliferation was evaluated by (C)
CCK-8 and (D) EdU assays. Original magnification, ×200. (E and F)
Cell migration was evaluated by wound healing assay. Original
magnification, ×100. (G and H) Cell invasion was evaluated by
Transwell assay. Original magnification, ×100. (I) Levels of MMP2
and MMP9 were measured by western blotting. Data are expressed as
mean ± SD. *P<0.05, **P<0.01, ***P<0.001. DEPDC1B, DEP
domain protein 1B; NUP37, nucleoporin 37; Ov, overexpression; si,
small interfering; NC, negative control. |
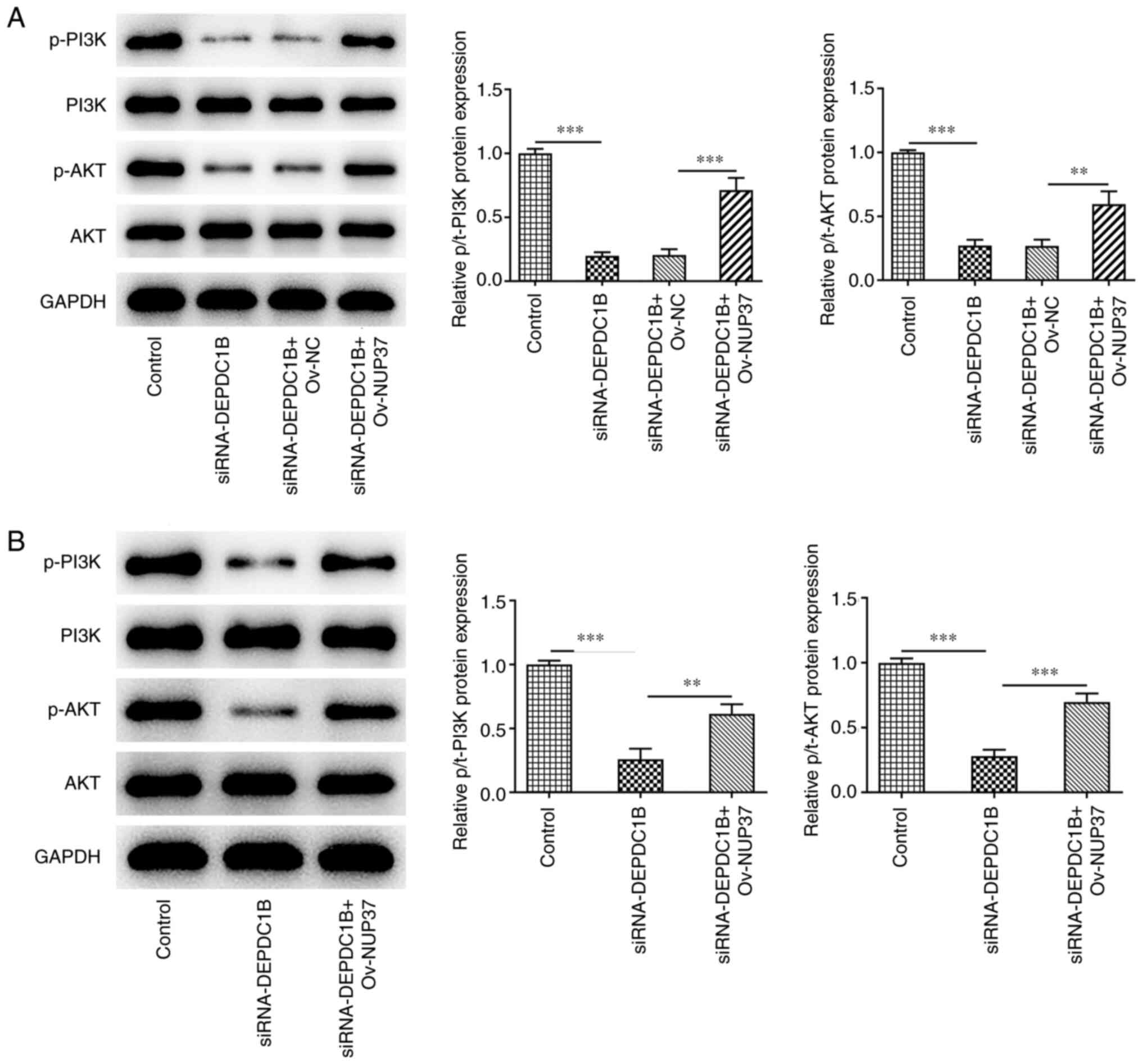
DEPDC1B silencing inhibits the growth
of CRC and the PI3K/AKT pathway in vivo via regulating NUP37
To further investigate the biological role of the
DEPDC1B/NUP37 axis in CRC in vivo, HCT8 cells transfected
with si-DEPDC1B or Ov-NUP37 were separately subcutaneously injected
into the flanks of nude mice. As shown in Fig. 9A-C, DEPDC1B knockdown notably
decreased tumor weight and volume compared with the control group.
However, NUP37 overexpression reversed the effect of DEPDC1B
silencing on tumor weight and volume. In addition,
immunohistochemistry assays showed that the expression levels of
Ki-67 were reduced in the tumor tissues of nude mice treated with
si-DEPDC1B. However, NUP37 overexpression upregulated Ki-67
(Fig. 9D). To assess the extent of
activation of the Akt-PI3K signaling pathway, the protein levels of
p-PI3K, PI3k, p-AKT and AKT were detected. The western blotting
results revealed that si-DEPDC1B reduced the phosphorylation of
PI3K and AKT, while NUP37 overexpression reversed the effect of
DEPDC1B knockdown on the expression levels of p-PI3K and p-AKT in
HCT8 cells (Fig. 10A) and murine
tissues (Fig. 10B).
Discussion
CRC is associated with 700,000 deaths every year,
exceeded only by lung, liver and stomach cancers (20). Surgical resection is the mainstay
of treatment, while systemic chemotherapy and local pelvic
radiotherapy are critical adjuvant treatment modalities (21). However, the prognosis of CRC is far
from satisfactory, particularly for patients suffering from
metastatic lesions (22). Targeted
therapy is a novel effective treatment approach, which can
successfully prolong the overall survival of patients with CRC
(23). The current study aimed to
evaluate the therapeutic potential of DEPDC1B in CRC cell
migration, apoptosis and cycle arrest and to uncover its underlying
molecular mechanisms.
DEPDC1B is a recently identified gene located on
human chromosome 5q12.1. It has been reported that DEPDC1B is
involved in cell proliferation, apoptosis and cell cycle
distribution (24). A previous
study also demonstrated that DEPDC1B could critically regulate the
progression of several types of cancer, such as hepatocellular and
NSCLC (25). Li et al
(26) showed that DEPDC1B is
significantly upregulated in patients with PCa and is positively
associated with a high Gleason score and poor prognosis. DEPDC1B
increases the levels of Rac1-GTP to enhance the activation of the
Rac1/p21activated kinase 1 signaling pathway, thus inducing
epithelial-mesenchymal transition and promoting PCa metastasis and
progression. Additionally, Lai et al (27) showed that DEPDC1B deficiency
suppresses the proliferation and migration and promotes the
apoptosis of bladder cancer cells both in vitro and in
vivo. Nevertheless, the role of DEPDC1B in colon cancer remains
to be elucidated. The results of the present study revealed that
DEPDC1B was upregulated in CRC cell lines as well as in tissues
derived from a CRC mouse model. In addition, DEPDC1B depletion
attenuated the proliferation, migration and invasion abilities of
CRC cells, while it promoted CRC cell apoptosis and cell cycle
arrest by inhibiting Bcl-2 and promoting Bax level, as well as
repressing cyclin D1 and cyclin B1.
To further reveal the mechanism underlying the
effect of DEPDC1B on regulating colorectal adenocarcinoma, the
Coexpedia database was used to predict the co-expression between
DEPDC1B and NUP37. NUP37 was upregulated in CRC cell lines and its
protein expression levels were significantly decreased following
DEPDC1B knockdown. Co-IP assays also verified the interaction
between DEPDC1B and NUP37. In addition, NUP37 knockdown attenuated
the proliferation, migration and invasion capabilities of CRC
cells, while it accelerated cell apoptosis and cell cycle arrest,
which were then reversed by NUP37 overexpression.
Previous studies show that both DEPDC1B and NUP37
can promote the expression of PI3K/AKT signaling-related proteins
(28,29). Therefore, the current study aimed
to investigate whether DEPDC1B could be involved in the regulation
of PI3K/AKT signaling via targeting NUP37. Functional experiments
indicated that DEPDC1B deficiency could conspicuously inhibit the
PI3K/AKT signaling pathway. However, NUP37 overexpression reversed
the effects of DEPDC1B silencing on PI3K/AKT signaling both in
vitro and in vivo.
There are several limitations to the present study.
It did not explore the roles of DEPDC1B overexpression in HCT8
cells. In addition, CRC clinical specimens were not involved in
this study. Moreover, the present study only performed assays in
one cell line HCT8; using several cell lines may improve the
results and will be considered in a further study.
In conclusion, the results of the present study
suggested that DEPDC1B silencing could inhibit the proliferation,
migration and invasion abilities and enhance the apoptosis and cell
cycle arrest of CRC cells via NUP37. The above findings could
provide a novel insight into prospective strategies for treating
CRC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HX and YL designed the study, drafted and revised
the manuscript. HX and ML analyzed the data and searched the
literature. All authors performed the experiments and all authors
read and approved the final manuscript. HX and YL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All of the experimental protocols were approved by
The First Affiliated Hospital of Nanchang University (approval no.
SD-2021-011) and strictly followed the Guidelines for the Care and
Use of Laboratory Animals by National Institute of Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li J, Ma X, Chakravarti D, Shalapour S and
DePinho RA: Genetic and biological hallmarks of colorectal cancer.
Genes Dev. 35:787–820. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh
Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M and Saad A:
Colorectal cancer epidemiology: Recent trends and impact on
outcomes. Curr Drug Targets. 22:998–1009. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heinimann K: Hereditary colorectal cancer:
Clinics, diagnostics and management. Ther Umsch. 75:601–606.
2018.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zielińska A, Włodarczyk M, Makaro A,
Sałaga M and Fichna J: Management of pain in colorectal cancer
patients. Crit Rev Oncol Hematol. 157:1031222021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sinha R: Colorectal cancer. Clin Radiol.
76:8702021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang L, Tang L, Xu R, Ma J, Tian K, Liu Y,
Lu Y, Wu Z and Zhu X: DEPDC1B regulates the progression of human
chordoma through UBE2T-mediated ubiquitination of BIRC5. Cell Death
Dis. 12:7532021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Figeac N, Pruller J, Hofer I, Fortier M,
Ortuste Quiroga HP, Banerji CRS and Zammit PS: DEPDC1B is a key
regulator of myoblast proliferation in mouse and man. Cell Prolif.
53:e127172020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai S, Chen T, Du T, Chen X, Lai Y, Ma X,
Wu W, Lin C, Liu L and Huang H: High levels of DEPDC1B predict
shorter biochemical recurrence-free survival of patients with
prostate cancer. Oncol Lett. 14:6801–6808. 2017.PubMed/NCBI
|
|
10
|
Pollino S, Benassi MS, Pazzaglia L, Conti
A, Bertani N, Righi A, Piccinni-Leopardi M, Picci P and Perris R:
Prognostic role of XTP1/DEPDC1B and SDP35/DEPDC1A in high grade
soft-tissue sarcomas. Histol Histopathol. 33:597–608.
2018.PubMed/NCBI
|
|
11
|
Zhang F and Zhou Q: Knockdown of BRCC3
exerts an anti-tumor effect on cervical cancer in vitro. Mol
Med Rep. 18:4886–4894. 2018.PubMed/NCBI
|
|
12
|
Xu Y, Sun W, Zheng B, Liu X, Luo Z, Kong
Y, Xu M and Chen Y: DEPDC1B knockdown inhibits the development of
malignant melanoma through suppressing cell proliferation and
inducing cell apoptosis. Exp Cell Res. 379:48–54. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su YF, Liang CY, Huang CY, Peng CY, Chen
CC, Lin MC, Lin RK, Lin WW, Chou MY, Liao PH and Yang JJ: A
putative novel protein, DEPDC1B, is overexpressed in oral cancer
patients, and enhanced anchorage-independent growth in oral cancer
cells that is mediated by Rac1 and ERK. J Biomed Sci. 21:672014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Y, Liu L, Cai J, Wu J, Guan H, Zhu X,
Yuan J and Li M: DEPDC1B enhances migration and invasion of
non-small cell lung cancer cells via activating Wnt/β-catenin
signaling. Biochem Biophys Res Commun. 450:899–905. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walther TC, Alves A, Pickersgill H,
Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen
T, et al: The conserved Nup107-160 complex is critical for nuclear
pore complex assembly. Cell. 113:195–206. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Wang T, Wang F, Hu X, Zhan G, Jin
X, Zhang L and Li Y: NUP37 silencing induces inhibition of cell
proliferation, G1 phase cell cycle arrest and apoptosis in
non-small cell lung cancer cells. Pathol Res Pract. 216:1528362020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang J, Lv W, Liu Y, Fu W, Chen B, Ma Q,
Gao X and Cui X: Nucleoporin 37 promotes the cell proliferation,
migration, and invasion of gastric cancer through activating the
PI3K/AKT/mTOR signaling pathway. In Vitro Cell Dev Biol Anim.
57:987–997. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
National Research Council (US), .
Committee for the update of the guide for the care and use of
laboratory animals: The national academies collection: Reports
funded by national institutes of health. Guide for the care and use
of laboratory animals. 8th edition. National Academies Press (US),
National Academy of Sciences; Washington, DC: 2011, PubMed/NCBI
|
|
20
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal Carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:2017. View Article : Google Scholar
|
|
21
|
Johdi NA and Sukor NF: Colorectal cancer
immunotherapy: Options and strategies. Front Immunol. 11:16242020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5:222020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marchesi S, Montani F, Deflorian G,
D'Antuono R, Cuomo A, Bologna S, Mazzoccoli C, Bonaldi T, Di Fiore
PP and Nicassio F: DEPDC1B coordinates de-adhesion events and
cell-cycle progression at mitosis. Dev Cell. 31:420–433. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun Y and Zhang Z: In silico
identification of crucial genes and specific pathways in
hepatocellular cancer. Genet Test Mol Biomarkers. 24:296–308. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Wang Q, Peng S, Yao K, Chen J, Tao
Y, Gao Z, Wang F, Li H, Cai W, et al: The metastatic promoter
DEPDC1B induces epithelial-mesenchymal transition and promotes
prostate cancer cell proliferation via Rac1-PAK1 signaling. Clin
Transl Med. 10:e1912020. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lai CH, Xu K, Zhou J, Wang M, Zhang W, Liu
X, Xiong J, Wang T, Wang Q, Wang H, et al: DEPDC1B is a tumor
promotor in development of bladder cancer through targeting SHC1.
Cell Death Dis. 11:9862020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Li T, Huang X, Wu W, Li J, Wei L,
Qian Y, Xu H, Wang Q and Wang L: DEPDC1B promotes migration and
invasion in pancreatic ductal adenocarcinoma by activating the
Akt/GSK3β/Snail pathway. Oncol Lett. 20:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan Y, Ping W, Zhang R, Hao Z and Zhang
N: DEPDC1B collaborates with GABRD to regulate ESCC progression.
Cancer Cell Int. 22:2142022. View Article : Google Scholar : PubMed/NCBI
|