Introduction
Obesity is an independent risk factor for asthma
(1). American health survey data
show that the incidence of asthma in individuals with obesity is
~11.1% (2). Individuals with
obesity have a 1.9-fold higher risk of asthma susceptibility than
those without obesity (1).
Moreover, >50% of patients with severe asthma are also obese
(3). Obesity increases the
deposition of collagen fibers, hyperplasia of airway elastin fibers
(4) and other ultrastructural
airways changes, such as mucous cell hyperplasia (5–9).
According to the 2014 Global Asthma Prevention and Treatment
Initiative guidelines (10),
asthma associated with obesity is a new phenotype of asthma that is
associated with poorer asthma control, reduced response to oral
corticosteroids and notable decline in lung function. Bronchial
epithelial cells (BECs) serve as the first barrier in the airway,
protecting against harmful substances and pathogens (11). Cell damage to BECs serves a key
role in promoting airway remodeling (12). However, the changes of airway
epithelial cells in asthma associated with obesity are not
clear.
Pyroptosis is a type of programmed cell death
characterized by the involvement of the gasdermin (GSDM) family and
is accompanied by the release of proinflammatory cytokines such as
interleukin-1β (IL-1β) and IL-18. Research has shown that
pyroptosis serves a crucial role in respiratory illnesses (13). Exposure to environmental allergens
such as Dermatophagoides farina 1 or toluene diisocyanate activates
the NOD-like receptor thermal protein domain associated protein 3
(NLRP3)/caspase-1/GSDMD signaling pathway, inducing pyroptosis and
resulting in airway inflammation (14,15).
A previous study demonstrated that schisandrin B effectively
inhibits activation of the NLRP3 inflammasome and decreases
pyroptosis via the microRNA-135a-5p/Transient receptor potential
canonical type 1(TRPC1)/STAT3/NF-κB axis (16). This mechanism mitigates airway
inflammation and remodeling in asthma. Furthermore, there are
reports indicating that cigarette smoke extract activates the
NLRP3/caspase-1/GSDMD signaling pathway, resulting in pyroptosis in
BECs, leading to impaired BEC barrier function and severe lung
injury (17,18). Obesity increases airway damage in
asthma; however, it is unclear whether pyroptosis serves a role or
if there is an upstream initiating factor.
In 2007, Moffatt et al (19) published genome-wide association
research results of asthma for the first time in Nature magazine
and revealed that ORMDL3 was associated with asthma and other
diseases, as well as autophagy and apoptosis (19). A previous study confirmed that
ORMDL3 participates in airway remodeling by regulating the
ERK/MMP-9/VEGF pathway (20).
ORMDL3 belongs to the Orm protein (serum mucoid) family, encodes a
transmembrane protein located on the endoplasmic reticulum membrane
and regulates sphingomyelin metabolism. High ORMDL3 expression
in vivo and in vitro causes inflammation and promotes
ceramide production (21–23). The contribution of ORMDL3 to
induction of pyroptosis remains uncertain. The present study aimed
to investigate the effect of ORMDL3 on BECs and on the airway
remodeling in asthma associated with obesity, as well as to
elucidate its underlying mechanisms.
Materials and methods
Animals
A total of 30 female BALB/c mice (age, 4 weeks;
weight, 11±1 g) were obtained from the Experimental Animal Center
of Shandong University (Jinan, China). Mice were housed in a
controlled environment with a 12/12-h light/dark cycle, maintained
at 20–26°C and humidity level of 60–70%. They were provided with ad
libitum access to chow and water. Importantly, no mice were
prematurely sacrificed during the study. All mice were randomly
assigned to three groups: i) Normal [non-sensitized lean (NSL)];
ii) asthmatic (SL) and iii) obese mice with asthma [sensitized
obese (SO group)]. NSL and SL groups were fed a standard chow for
14 weeks, while the mice in the SO group received high-fat diet
(HFD) containing 60 kcal% for 14 weeks. After 14 weeks of feeding,
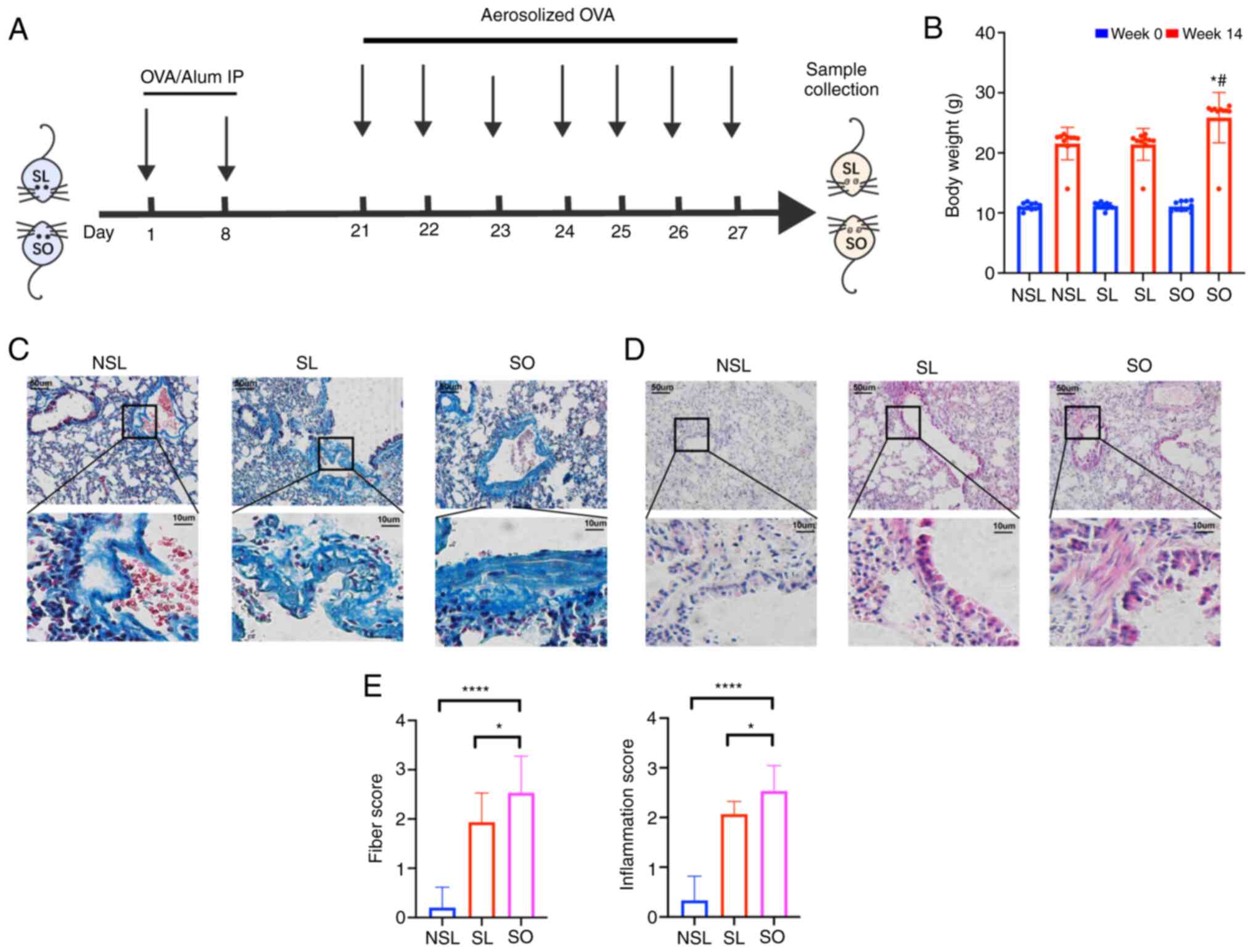
the SL and SO groups were formed using ovalbumin (OVA) (Fig. 1A), where the mice were sensitized
by intraperitoneal (i.p.) injection on days 1 and 8 with 20 µg OVA
(Sigma-Aldrich, Merck KGaA) and 2 mg aluminum hydroxide
(Sigma-Aldrich, Merck KGaA) in 200 µl phosphate-buffered saline
(PBS) (19). On days 21-27, mice
in SL and SO group were challenged with 1% OVA through ultrasonic
atomization for 30 min every day, as previously described (24). Mice in the NSL group received 200
µl PBS instead of OVA. The study received ethical approval from the
Animal Ethics Committee of Shandong First Medical University
(approval no. 2020-1328).
Tissue collection
On day 28 after asthma modelling, mice were
euthanized by cervical dislocation following anesthesia with i.p.
injection of sodium pentobarbital (35 mg/kg). A total of 50% of the
mouse lung tissue was dissected and fresh-frozen in liquid nitrogen
before being stored at −80°C until further processing. The rest of
the mouse lung tissue was fully fixed using 4% paraformaldehyde
solution for 48 h at 4°C for subsequent experiments.
Hematoxylin and eosin (H&E) and
Masson staining
The lung tissue was fixed using 4% paraformaldehyde
(48 h; 4°C). The lung tissues were embedded in paraffin and cut
into 5-µm thick sections. Subsequently, the paraffin-embedded
sections were stained with hematoxylin and eosin (H&E) staining
for 5 min and Masson staining for 8 min, both at 37°C. The
pathological changes were observed using a light microscope
(magnification, ×20). IM50 Image Manager software Version 1.20
(Leica Microsystems, Inc.) was used to measure lung pathology
changes, including inflammatory situation and collagen deposition.
H&E staining was used to investigate inflammatory situation,
while Masson staining was used to evaluate collagen deposition in
lung tissue. To assess the severity of fibrosis, Szapiel's method
(25) was applied using the
following scoring system: i) 0, normal tissues without alveolitis
or fibrosis; ii) 1, mild alveolitis or fibrosis with ≤20% lung
lesions; iii) 2, moderate alveolitis or fibrosis with 20–50% lung
lesions and iv) 3, severe alveolitis or fibrosis with ≥50% lung
lesions. The severity of fibrosis were calculated by reviewing five
high-powered fields in every specimen.
Protein extraction and trypsin
digestion
Tissue samples were removed storage and an
appropriate amount (30 mg) was weighed. Next, samples were
subjected to ultrasonic lysis with 300 µl of SDS lysis solution
(Beyotime, China) at a frequency of 1.0 sec on and 1.0 sec off, for
a total duration of 3 min on ice at 0°C. The remaining debris was
eliminated by centrifugation at 12,000 × g at 4°C for 10 min. The
supernatant was collected and the protein concentration was
determined using a BCA kit. Trypsin (Beijing Hualishi Tech. Ltd)
was added to the protein supernatant for complete enzymolysis once
protein concentration was measured. The reaction of enzymolysis was
carried out at 37°C for 12 h. Then, peptides were lyophilized. The
lyophilized samples were resuspended in 30 µl 100 mM TEAB and
Labeling reaction in a 1.5 ml Eppendorf (EP) tube. 20 µl
acetonitrile were added to TMT reagent and mixed for
centrifugation. Then 10 µl TMT label reagent was added to each
sample and incubated for 1 h. Finally, the labeling peptides
solutions were lyophilized.
Liquid chromatography-mass
spectrometry analysis (LC-MS/MS) and data search
Samples were loaded onto a pre-column Acclaim™
PepMap™ C18 (100 µm × 2 cm, Thermo Fisher Scientific, Inc.) at a
flow rate of 300 nl/min, followed by separation on an analytical
column Acclaim™ PepMap™ RSLC (75 µm × 15 cm, Thermo Fisher
Scientific, Inc.). The nitrogen gas temperature was 220°C while the
nebulizer pressure was adjusted to 2200 psi. Mass spectrum scanning
was conducted at a full scanning mass nucleus ratio in the m/z
range of 350-1,500, with MS/MS scanning performed on the 10 highest
peaks. All MS/MS spectra were collected under data-dependent
positive ionization mode using high-energy collision dissociation,
with collision energy set at 30. The resolution of MS was set to
15,000, with automatic gain control set and a maximum injection
time of 40 msec. The dynamic exclusion time was 60 sec. The raw
data were searched using Proteome Discoverer™ 2.2 (Thermo Fisher
Scientific, Inc.)software. The database used for the search was
Uniprot Mus musculus database (26). False positive rates for peptide
identification were controlled at ≤1%. Specific database search
parameter settings are shown in Table
I.
 | Table I.Database search parameter
settings. |
Table I.
Database search parameter
settings.
| Parameter | Setting |
|---|
| Peptide label | Tandem Mass Tag
6-plex |
| Cysteine
alkylation | Iodoacetamide |
| Digestion | Trypsin |
| Instrument | Thermo Scientific™
Q Exactive™ HF |
| Database | Mus musculus
fasta |
Bioinformatics analysis
Principal component analysis (PCA) was performed to
assess the differences between the SO and NSL groups based on the
expression levels of all proteins (27). Volcano plots were used to show
distribution of DEPs between different samples (28). Peptide length distribution analysis
was performed to evaluate the conformity of peptide lengths
identified by mass spectrometry based on enzymatic hydrolysis and
mass spectrometry fragmentation mode (29). Moreover, heatmap was generated to
visualize the protein expression patterns in the proteomics study.
The heatmap displayed the relative abundance of proteins across
different samples, providing a clear overview of the protein
expression levels (30). The
clusterProfiler package (version 4.2.2) (31) was used to perform Gene Ontology
(GO) (32) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) (33)
pathway enrichment analyses for differentially expressed proteins
(DEPs). To indicate a statistically significant difference, a
threshold of fold Change) ≥1.30 or FC ≤0.77, along with a
significance level of P<0.05, was considered. Proteins
associated with cell pyroptosis were downloaded from the Gene Set
Enrichment Analysis (GSEA; gsea-msigdb.org/gsea/index.jsp;
R-MMU-5620971) database. For upregulated proteins in lysosomal
pathway, protein-protein interaction (PPI) network was constructed
using the Search Tool for the Retrieval of Interacting Genes
(STRING; string-db.org/) database with a physical score >0.132.
Molecular Complex Detection (MCODE) algorithm10 was applied to
identify densely connected network components. GSEA was conducted
to analyze pyroptosis-associated genes. Marker genes corresponding
to each cell type in mouse lung tissue were obtained from the
CellMarker database (xteam.xbio.top/CellMarker/) (34).
Immunohistochemical and
immunofluorescent staining
The lung tissues were fixed in 4% formalin at room
temperature for 48 h, followed by paraffin embedding. Serial
sections of 5 µm thickness were cut and used for staining. For
immunohistochemical staining, each sections were incubated
overnight at 4°C with GSDMD primary antibody (1:1,000; cat. no.
ab219800; Abcam). The stained sections were observed under a light
microscope (Olympus Corporation, Tokyo, Japan) at a magnification
of ×100. Five random images per section were collected for
analysis.
For immunofluorescent staining, primary antibodies
used were ORMDL3 (1:1,000; cat. no. ab211522; Abcam), NLRP3 (1:100;
cat. no. ab263899; Abcam). A secondary antibody, Alexa Fluor
488-conjugated goat anti-rabbit/mouse IgM antibody (1:1,000; Life
Technologies, Inc.), was used. The sections were then incubated
with a 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)
solution (1:1,000; Dojindo Laboratories, Inc.) for nuclear
staining. Digital section images were captured using an Olympus
BX51 imaging system (Olympus Corporation) and quantified with
Image-Pro Plus (version 6.0; Media Cybernetics).
Human bronchial epithelial (HBE) cell
infected by ORMDL3-overexpressing lentivirus
HBE cells (BEAS-2B) purchased from Shanghai
EK-Bioscience Biotechnology Co., Ltd (Shanghai, China). To package
ORMDL3-overexpressing lentivirus, ORMDL3 (NM_139280) sequence,
GV492 vector, pHelper1.0, and pHelper2.0 (all GeneChem Corporation
(Shanghai, China) to synthesize the Ubi-MCS-gcGFP construct. Then,
the Ubi-MCS-gcGFP (20 µg), pHelper1.0 (15 µg) and pHelper2.0 (10
µg) were mixed and transfected into 293T cells using GeneChem
Transfection kit (GeneChem, Shanghai, China) according to the
manufacturer's instructions. After transfection for 48 h at 37°C,
the viral supernatants were collected. Subsequently, BEAS-2B were
seeded into a six-well dish at 2×105 cells/well and then
infected with lentiviral vectors loaded with ORMDL3 at an optimum
multiplicity of infection (MOI). Following 16 h of infection, the
regular culture medium was replaced with fresh medium before
subsequent experiments. After 72 h culture at 37°C, expression of
the reporter gene was assessed. Cells were collected for downstream
experiments when the rate of positive cell infection reached 85% or
higher. The group infected with lentivirus overexpressing ORMDL3
can be named as the overexpression (OE) group, while the group
infected with empty vector serves as the negative control (NC)
group.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from infected HBE using the
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RT
was performed using M-MLV reverse transcriptase Kit (Promega,
Fitchburg). RT was performed at 42°C for 1 h, followed by
incubation at 70°C for 10 min in a water bath. The expression
levels of ORMDL3, GSDMD, NLRP3 and cathepsin D (CTSD) were assessed
via RT-qPCR using SYBR Master Mixture (Takara, Canada)
according to the manufacturer's protocol. Amplification was
performed in following procedure: 95°C for 30 sec, 95°C for 5 sec,
and 60°C for 30 sec, for 40 cycles. The threshold cycle of each
sample was recorded, and data were analyzed by normalization to
GAPDH values using the 2−ΔΔCq method (35). The primer sequences were as
follows: ORMDL3, forward 5′-CCTCACCAACCTCATTCACAAC-3′ and reverse
5′-TACAGCACGATGGGTGTGATG-3′; GAPDH, forward
5′-TGACTTCAACAGCGACACCCA-3′ and reverse
5′-CACCCTGTTGCTGTAGCCAAA-3′; NLRP3, forward
5′-ATGCCCAAGGAGGAAGAG-3′ and antisense 5′ CCAACCACAATCTCCGAAT 3′;
CTSD, sense 5′-AGGCCCCGTCTCAAAGTA-3′ and reverse 5′
ATGCCAATCTCCCCGTAG 3′; GSDMD, forward, 5′-GTGGTTAGGAAGCCCTCAAG-3′
and reverse, 5′ CATGGCATCGTAGAAGTGGA 3′; and CTSD forward
5′-AGGCCCCGTCTCAAAGTA-3′ and reverse 5′ ATGCCAATCTCCCCGTAG 3′.
Statistical analysis
Data analysis was performed using GraphPad Prism
(version 9.0; Dotmatics) and SPSS (version 27.0; IBM Corp). Data
are presented as the mean ± standard deviation (mean ± SD) for at
least three independent experiments. An Unpaired t-test was used
for two-group comparisons. One-way ANOVA was used to assess
differences between multiple groups, followed by Dunnett's T3 (when
equal variances not assumed) or the Bonferroni's post hoc test
(when equal variance was assumed) for pairwise comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Airway remodeling is aggravated in
asthma associated with obesity
After 14 weeks, weight of the SO group was 20%
higher than that of the NSL and SL groups (Fig. 1B). The pathological changes in
mouse lung tissue were detected, including inflammatory
infiltration and collagen deposition. Masson staining was used to
observe collagen deposition. Collagen deposition in the SL and SO
groups was significantly higher than that in the NSL group
(Fig. 1C and E). The infiltration
of monocytes, neutrophils, as well as the shedding of airway
epithelial cells, was observed by H&E staining (Fig. 1D). The inflammatory score in the
lung tissue of the SO group was increased compared with that of the
NSL and SL groups (Fig. 1E).
H&E staining can reflect inflammation changes in the lungs,
while Masson staining can indicate the presence of collagen
deposition. Once lung collagen deposition occurs, it represents
irreversible damage, indicating a more severe pathological
condition in the lungs. Therefore, Masson staining is of
significant importance in assessing lung pathology changes. These
results showed that the airway remodeling in obese mice with asthma
was the most serious.
Data validation and DEP identification
by proteomics
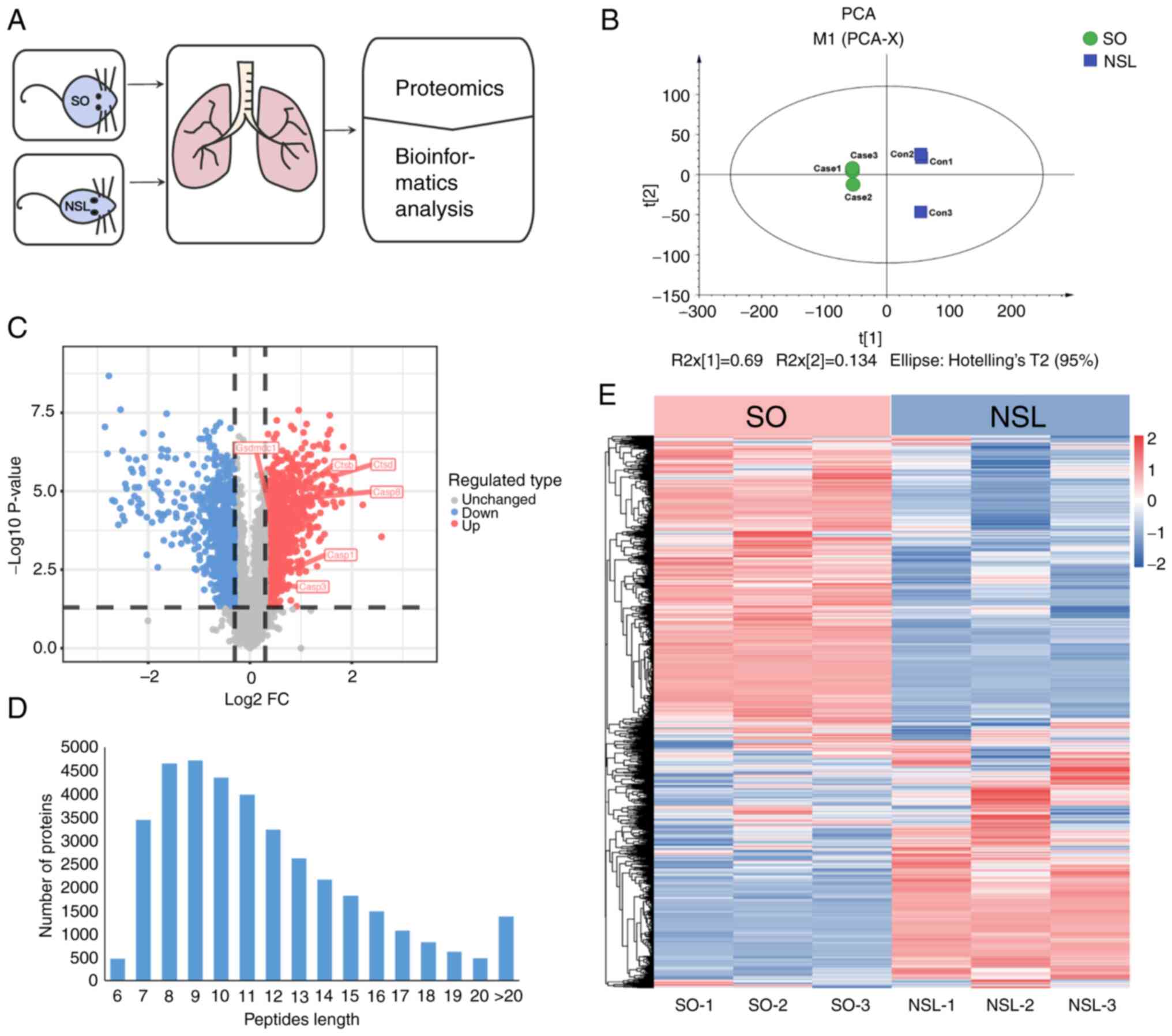
To determine how asthma associated with obesity
affected lung tissue, SO and NSL mouse lung tissues were collected
for proteomics (Fig. 2A). PCA was
used to validate the dataset. PCA indicated that the six samples
from both groups were distinguishable, as shown by short distances
between samples in the SO group along PC1 and PC2 and similar
distances between samples in the NSL group (Fig. 2B). A total of 6,399 proteins were
identified across all groups by proteomics analysis (Table SI). Among these, 5,806 proteins
were quantified. Specifically, compared with the NSL group, 823
proteins were upregulated (fold-change >1.3) and 95 proteins
were downregulated (fold-change <0.5; Fig. 2C; Table SII). The majority of peptides in
the 7–20 amino acid range align with the expected pattern from
enzymatic hydrolysis and mass spectrometry fragmentation. The
identified peptide length distribution meets quality control
requirements (Fig. 2D). In
Fig. 2E, heatmap analysis showed
that the protein expression in SO group mice was altered compared
with the NSL group mice, which indicated broad proteome modulation
in mouse lungs. Each protein exhibited a distinct concentration
profile, where red and green represented upregulated and
downregulated proteins, respectively, while white indicated no
significant change in expression levels.
Lysosome and autophagy pathway are
enhanced in asthma associated with obesity
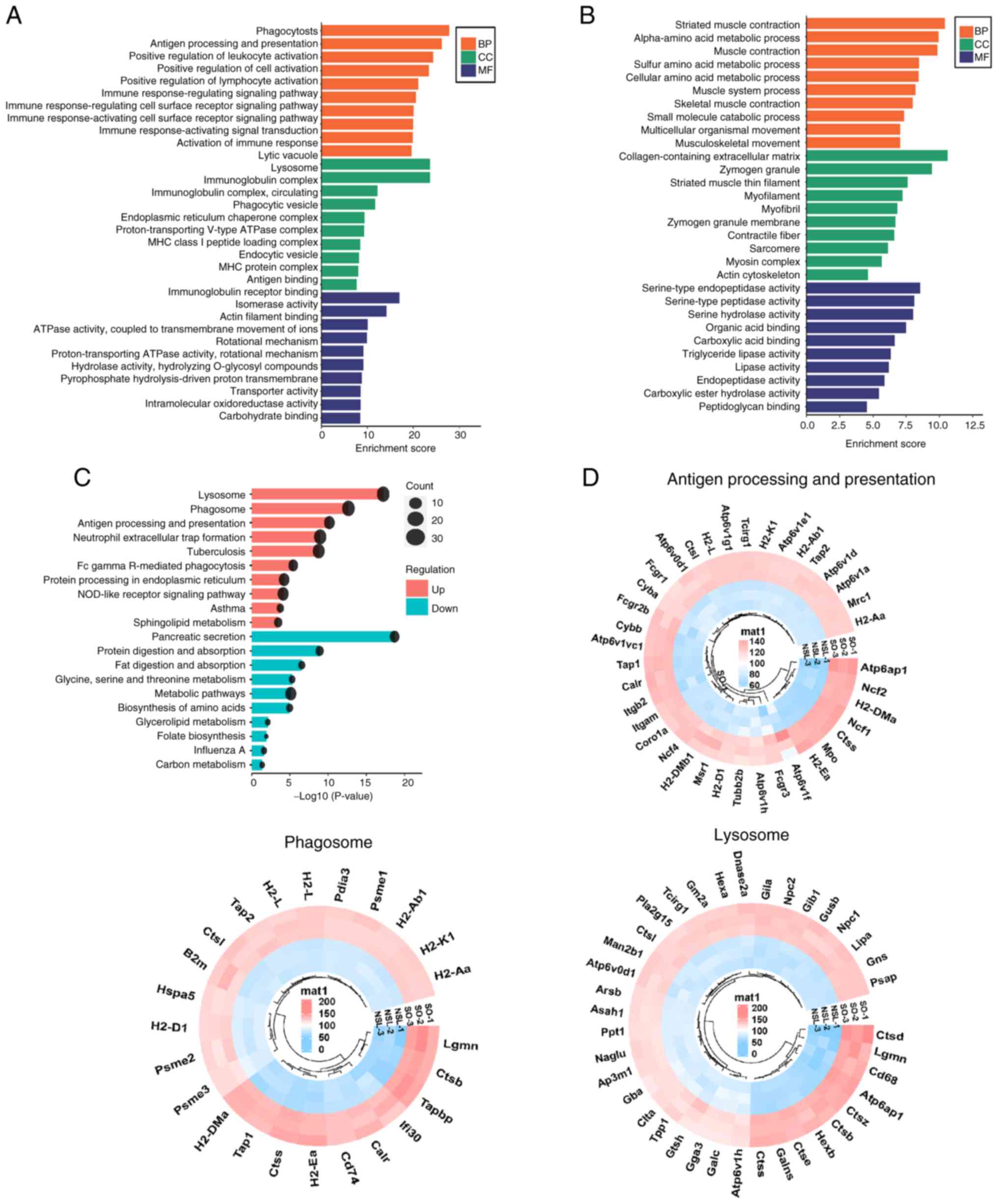
To determine the enriched biological processes
(BPs), cellular components (CCs) and molecular functions (MFs) of
significantly upregulated proteins, GO enrichment analysis was
performed (Fig. 3A). Based on
P-values obtained through Fisher's exact test, the top three BP
terms for upregulated proteins included ‘phagocytosis’, ‘antigen
processing and presentation’ and ‘positive regulation of cell
activation’. The top three CCs were ‘lytic vacuole’, ‘lysosome’ and
‘immunoglobulin complex’. The top three MFs associated with these
proteins were ‘antigen binding’, ‘immunoglobulin receptor binding’
and ‘isomerase activity’. The GO enrichment analysis revealed that
the upregulated DEPs were mainly associated with immunity.
(Fig. 3B). KEGG pathway enrichment
analysis of the identified proteins indicated the involvement of
pathways such as ‘Lysosome’, ‘Phagosome’, and ‘Sphingolipid
metabolism’ (Fig. 3C; Table SIII). The upregulated
pathway-associated genes were clustered in circular heatmaps to
display DEPs (Fig. 3D). In the
‘lysosome’ pathway and ‘antigen processing and presentation’
pathway, CTSD was significantly upregulated in the lung tissue of
obese mice with asthma.
Pyroptosis-associated genes are
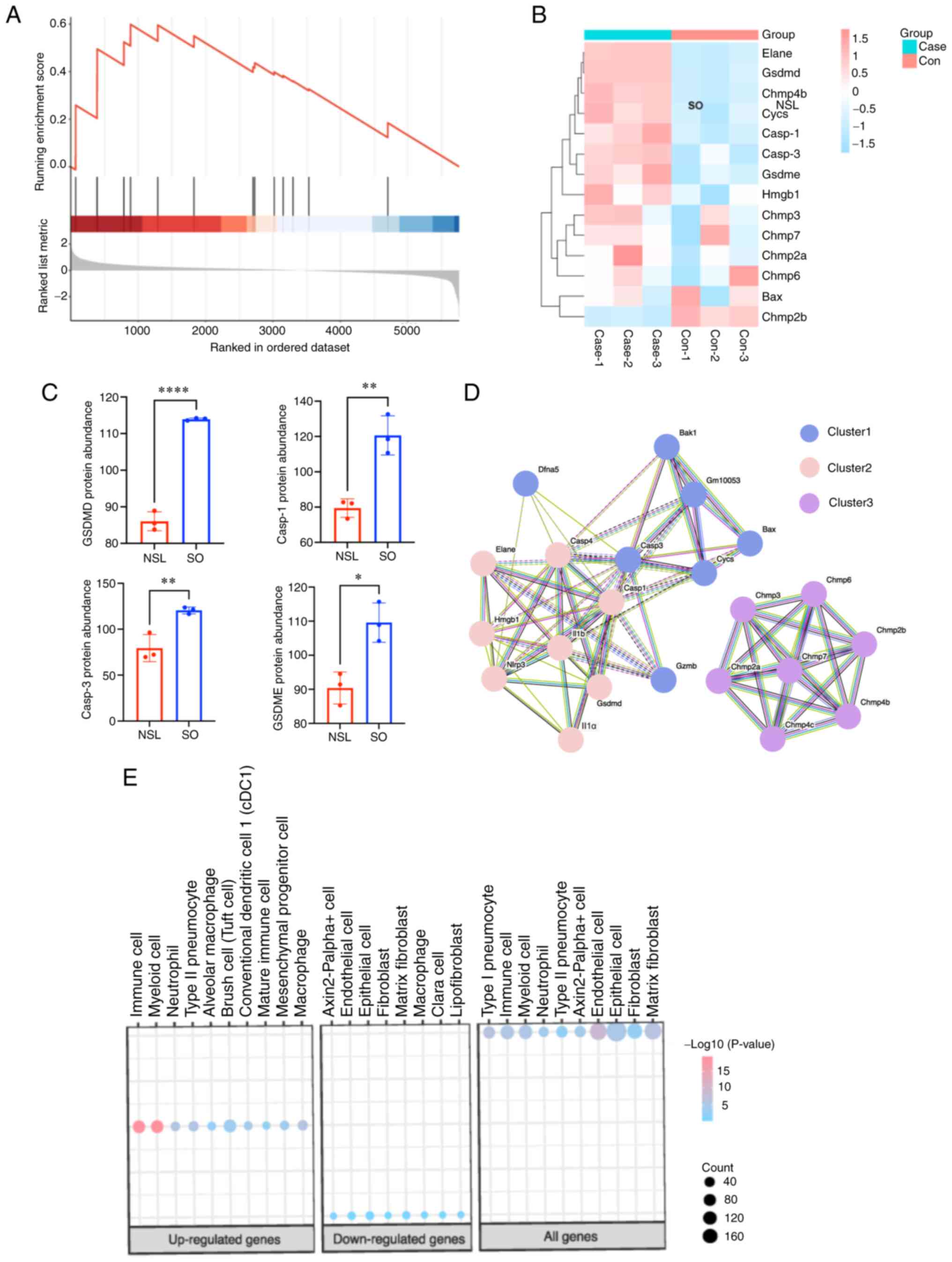
related to lysosomes and metabolism
Using the GSEA database, pyroptosis-associated
protein information was downloaded, proteins that were not
identified by proteomics of the present study were removed and
STRING database was used to analyze PPIs (Fig. 4A and B). GO enrichment analysis
showed that pyroptosis-associated proteins were associated with
lysosomes (Fig. 4C), including
CTSD and cathepsin B (CTSB). In addition, MCODE showed that protein
was associated with insulin receptors, such as ATPase H+
Transporting Accessory Protein 1 and T cell immune regulator 1
(Tcirg1). Furthermore, GSEA showed the pyrolytic pathway was
notably enhanced (Fig. 5A). The
heatmap of pyroptosis-associated genes showed that GSDMD, caspase-1
and −3 and GSDME protein were significantly upregulated (Fig. 5B and C). In addition, PPIs between
NLRP3 and pyroptosis-associated proteins were analyzed; NLRP3
interacted with multiple proteins, including caspase-1 and −3,
IL-1β and GSDMD (Fig. 5D). This
above interaction pattern suggests that NLRP3 could be involved in
the regulation or activation of these pyroptosis-related proteins,
indicating its potential role in cell pyroptosis. To identify the
specific cell types in the lungs that the upregulated and
downregulated differentially expressed proteins (DEPs) were
enriched in, we conducted a cell marker analysis. As in Fig. 5E, the upregulated DEPs were mainly
enriched in immune cells and myeloid cells. Conversely, the
downregulated DEPs were predominantly enriched in epithelial cells.
Interestingly, both the upregulated and downregulated DEPs showed
enrichment in macrophages cells (Fig.
5E). This suggests that macrophages play a crucial role in both
upregulated and downregulated differentially expressed proteins.
They are likely involved in regulating immune responses,
inflammation processes, and other related biological functions.
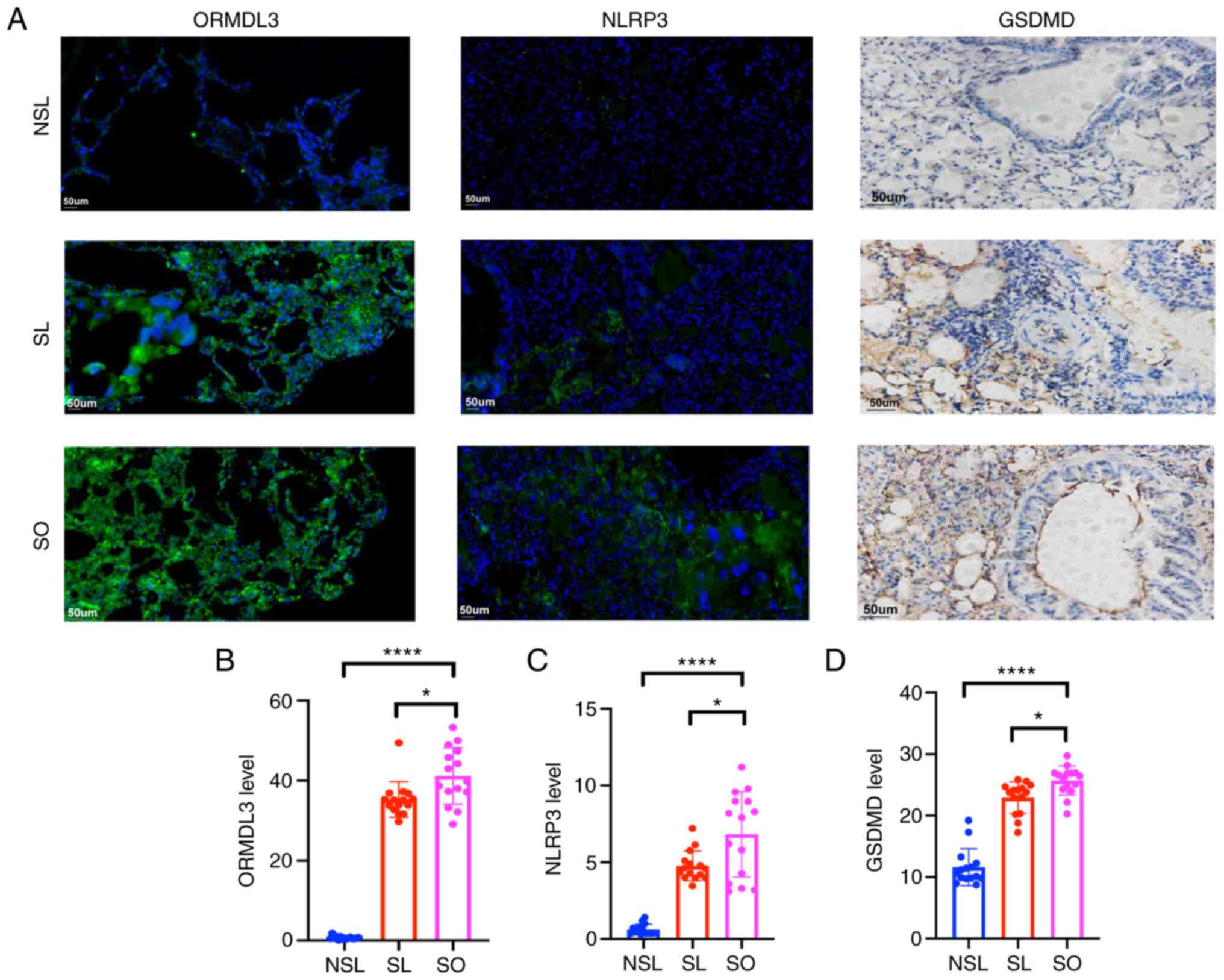
ORMDL3, NLRP3 and GSDMD expression
increases in asthma associated with obesity
Immunofluorescence staining showed that expression
of ORMDL3 and NLRP3 in the SL and SO groups was increased compared
with that in the NSL group (Fig.
6). Immunohistochemical staining showed that levels of GSDMD in
the SL and SO groups were increased compared with those in the NSL
group (Fig. 6).
CTSD, NLRP3 and GSDMD mRNA expression
is increased in HBE cells infected with ORMDL3-overexpressing
lentivirus
The fluorescence rate (indicating positive infection
rate) was 80% (Fig. 7A). Following
infection with ORMDL3-overexpressing lentivirus, mRNA expression of
ORMDL3, CTSD, NLRP3 and GSDMD was significantly increased in HBE
cells compared with the negative controls (Fig. 7B).
 | Figure 7.CTSD, NLRP3 and GSDMD mRNA expression
in HBE cells infected with ORMDL3-overexpressing lentivirus. (A)
HBE cells infected with ORMDL3-overexpressing lentivirus. Scale
bar, 100 or 300 µm. (B) CTSD, NLRP3 and GSDMD mRNA expression was
detected by reverse transcription-quantitative PCR. *P<0.05,
**P<0.01 and ***P<0.001. NC, negative control; CTSD,
cathepsin D; GSDMD, Gasdermin D; NLRP3, NOD-like receptor thermal
protein domain associated protein 3; HBE, human bronchial
epithelial; OE, overexpression; B, bright field; G, green
fluorescence field; ORMDL3, orosomucoid-like 3. |
Discussion
HBE cells serve as the primary barrier in the airway
and serve a key role in airway remodeling. Pyroptosis is a
proinflammatory cell death process and cell damage pathway that is
associated with airway remodeling (36). In the present study, higher
expression of ORMDL3 and pyroptosis-associated factors in the lung
tissue of obese mice with asthma was observed. ORMDL3 has been
shown to participate in airway remodeling in previous study
(37). Inflammation and high
expression of GSDMD are the hallmarks of pyroptosis (38). The present study indicated that
pyroptosis occurred in lung tissue of obese mice with asthma and
ORMDL3 and pyroptosis was associated with airway remodeling in
asthma associated with obesity.
There has been a growing focus on the role of the
NLRP3 inflammasome in asthma (39–41).
NLRP3 is the most widely studied member of the NLR family (42,43)
and is expressed in the mesenchyme and membrane of neutrophils,
macrophages, epithelial cells and other types of cell (43). When stimulated by microbial
infection or self-injury signals, the innate immune system inhibits
levels of NLRP3, IL-1β and IL-18, which can alleviate airway
inflammation (44), attenuate
airway hyperresponsiveness (45)
and effectively prevent progression of asthma (46). NLRP3, an important inflammation
factor, is activated in adipose tissue (47). When NLRP3 inflammation is
activated, the NLRP3 domain is exposed and NLRP3 oligomerizes to
form the NLRP3- apoptosis-associated speck-like protein containing
CARD (ASC) complex. Activation of caspase-1, IL-1β and IL-18, as
well as the formation of active N-terminal GSDMD, leads to cell
membrane perforation and subsequent release of inflammatory
factors. These processes promote cell rupture and induce cell
pyroptosis. Pyroptosis is a type of programmed cell death
associated with NLRP3 inflammation (48). In this study,
NLRP3/GSDMD-associated pyroptosis occurred in obese mice with
asthma, which is accompanied by morphological changes in lung
tissue. It is likely that the observed morphological alterations in
the lung tissue are linked to the inflammatory processes and
cellular damage caused by pyroptosis. These findings provide
valuable insights into the potential mechanisms underlying the
development and progression of obese asthma.
The proteomic results revealed a significant
increase in the expression of factors associated with cell
pyroptosis. GSEA indicated that the pyroptosis pathway was
significantly enhanced. Expression of GSDMD, GSDME and caspase-1
and −3 in the SO group was significantly higher than that in the
NSL group. Asthma associated with obesity primarily affected
‘lysosome’, ‘antigen processing and presentation’ and ‘phagosome’,
which increased pyroptosis-associated factors, such as GSDMD and
GSDME, eventually leading to cell pyroptosis. In addition,
immune-associated cells were activated, especially macrophages.
Macrophage pyroptosis serves an important role in lung injury and
inflammatory diseases, such as sepsis-related acute lung injury
(ALI), and Idiopathic pulmonary fibrosis (IPF) (49,50).
This suggests that macrophages may serve a vital role via
pyroptosis in airway remodeling in asthma associated with obesity.
Furthermore, ORMDL3 may influence the biological activity of
phagosomes by regulating the related proteins in ‘phagosomes’ KEGG
pathway (Fig. 3C). Caspases,
regulators of antioxidant defense and pathogen clearance, primarily
regulate phagosomal maturation and fusion with lysosomes (51). Emerging evidence suggests that
caspases also contribute to inflammatory cell death by inducing
rapid pyroptosis in infected cells (52,53).
Based on previous studies, Orm proteins have a direct impact on
asthma by regulating sphingomyelin (54,55).
In this study, it was observed that asthma with obesity also led to
the activation of the ‘sphingolipid metabolism’ pathway (Fig. 3C), as confirmed by the KEGG pathway
analysis. This finding suggests a potential link between the
dysregulation of sphingolipid metabolism and the development of
asthma with obesity. Furthermore, it is hypothesized that ORMDL3
may play a critical regulatory role in this potential link.
Sphingolipid are a key family of lipids involved in
membrane structure and intracellular signaling. Ceramide serves as
a key intermediate product of sphingomyelin metabolism and
functions as an inflammatory mediator (56). In obesity, high expression of
ORMDL3 in vivo and in vitro causes inflammation and
promotes ceramide production (21,22),
especially ceramide c24:0 > c24:1 > c16:0 in lung epithelial
cells (10). CTSD is a direct
downstream factor of ceramide (57). Notably, CTSD belongs to the
lysosomal cathepsin family. Entry of CTSD into the cytoplasm
increases mitochondrial permeability, releases cytochrome and
triggers caspases to induce apoptosis (58). In the present study, CTSD
expression was significantly increased in obese mice with asthma.
Furthermore, CTSD, NLRP3 and GSDMD expression was increased in HBE
cells transfected with ORMDL3-overexpressing lentivirus, which
indicated that CTSD may be a link between ORMDL3 and
NLRP3/GSDMD-associated pyroptosis. Thus, ORMDL3 may be the
initiating factor of pyroptosis. These finding suggested that
ORMDL3 may enhance cellular phagocytosis by activating caspases,
leading to pyroptosis.
In pyroptosis, CTSD specifically cleaves caspase-8
(59,60) and cleaves and activates GSDMD,
leading to the non-classical pyroptosis pathway (61). In addition, caspase-8 can interact
with ASC of caspase-1 during bacterial infection, thus activating
caspase-1 and the classical pyroptosis pathway. CTSD aggravates the
inflammatory reaction in pancreatitis by enhancing activation of
CTSB (62–64). CTSB was found to have
pyroptosis-promoting effects (65). Based on the aforementioned results,
it was hypothesized that CTSD is associated with pyroptosis of HBE
cells in asthma associated with obesity via direct or indirect
pathways. Previous study have reported that caspase-1 and −8 play
important roles in CTSD-activated pyroptosis (66). In our study, which involved
proteome analysis, we also observed an increase of caspase-1 and −8
in asthma associated with obesity. In addition, the present study
suggested that ORMDL3 may influence the biological activity of
lysosomes. CTSD participated in the lysosomal pathway and was
significantly upregulated in lung tissue of obese mice with asthma.
Furthermore, HBE cell experiments demonstrated that overexpression
of ORMDL3 led to an increase in CTSD expression. ORMDL3 positively
regulated pyroptosis-associated factors, including NLRP3 and GSDMD,
and pyroptosis-related genes were associated with lysosomes.
Although further research is required to understand the mechanisms
underlying these observations, the present study suggested that
ORMDL3 may play an important role in regulating biological activity
of lysosomes.
A limitation of the current study is that it did not
investigate protein and mRNA levels of NLRP3, GSDMD and CTSD
following transfection with small interfering (si)ORMDL3 to
determine the underlying mechanism. The focus of the present study
was on the effects of ORMDL3 overexpression and its role in asthma
associated with obesity. Further studies are needed to explore the
impact of siORMDL3 transfection on the aforementioned protein and
mRNA levels.
Overall, the present study indicated that ORMDL3
promoted upregulation of CTSD expression, which led to activation
of the NLRP3/GSDMD-related pyroptosis pathway and ultimately
contributed to airway remodeling. The present study aimed to
elucidate the regulatory mechanism of pyroptosis by ORMDL3 via the
CTSD/NLRP3/GSDMD pathway. The present findings provide insight into
the underlying mechanisms and may identify therapeutic targets to
combat airway remodeling in asthma associated with obesity.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Shandong Provincial
Natural Science Foundation (grant no. ZR2020MH003).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the ProteomeXchange Consortium
repository, proteomecentral.proteomexchange.org (accession no.
PXD043630).
Authors' contributions
YS designed the study and edited the manuscript. FL
and YZ performed experiments. FL wrote the manuscript. YG, CX, JY,
GL and QS analyzed data. FL, YS and QS confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by the Animal
Ethics Committee of Shandong Provincial Hospital affiliated with
Shandong First Medical University (approval no. 2020-1328) in
Shandong, China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peters U, Dixon AE and Forno E: Obesity
and asthma. J Allergy Clin Immunol. 141:1169–1179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akinbami LJ and Fryar CD: Current asthma
prevalence by weight status among adults: United States, 2001-2014.
NCHS Data Brief. 239:1–8. 2016.PubMed/NCBI
|
|
3
|
Bantulà M, Roca-Ferrer J, Arismendi E and
Picado C: Asthma and obesity: Two diseases on the rise and bridged
by inflammation. J Clin Med. 10:1692021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pathak MP, Patowary P, Goyary D, Das A and
Chattopadhyay P: β-caryophyllene ameliorated obesity-associated
airway hyperresponsiveness through some non-conventional targets.
Phytomedicine. 89:1536102021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lopes ACR, Zavan B, Corrêa YJC, Vieira TM,
Severs LJ, Oliveira LM and Soncini R: Impact of obesity and
ovariectomy on respiratory function in female mice. Respir Physiol
Neurobiol. 294:1037752021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barton JH, Ireland A, Fitzpatrick M,
Kessinger C, Camp D, Weinman R, McMahon D, Leader JK, Holguin F,
Wenzel SE, et al: Adiposity influences airway wall thickness and
the asthma phenotype of HIV-associated obstructive lung disease: A
cross-sectional study. BMC Pulm Med. 16:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta S, Lodha R and Kabra SK: Asthma,
GERD and obesity: Triangle of inflammation. Indian J Pediatr.
85:887–892. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lamkanfi M and Dixit VM: Mechanisms and
functions of inflammasomes. Cell. 157:1013–1022. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao Y: Angiogenesis and vascular functions
in modulation of obesity, adipose metabolism, and insulin
sensitivity. Cell Metab. 18:478–489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reddel HK, Bateman ED, Becker A, Boulet
LP, Cruz AA, Drazen JM, Haahtela T, Hurd SS, Inoue H, de Jongste
JC, et al: A summary of the new GINA strategy: A roadmap to asthma
control. Eur Respir J. 46:622–639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao W, Li L, Wang Y, Zhang S, Adcock IM,
Barnes PJ, Huang M and Yao X: Bronchial epithelial cells: The key
effector cells in the pathogenesis of chronic obstructive pulmonary
disease? Respirology. 20:722–729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cohen L, Xueping E, Tarsi J, Ramkumar T,
Horiuchi TK, Cochran R, DeMartino S, Schechtman KB, Hussain I,
Holtzman MJ, et al: Epithelial cell proliferation contributes to
airway remodeling in severe asthma. Am J Respir Crit Care Med.
176:138–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Fan G, Tao N and Sun T: Role of
pyroptosis in respiratory diseases and its therapeutic potential. J
Inflamm Res. 15:2033–2050. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai YM, Chiang KH, Hung JY, Chang WA, Lin
HP, Shieh JM, Chong IW and Hsu YL: Der f1 induces pyroptosis in
human bronchial epithelia via the NLRP3 inflammasome. Int J Mol
Med. 41:757–764. 2018.PubMed/NCBI
|
|
15
|
Zhuang J, Cui H, Zhuang L, Zhai Z, Yang F,
Luo G, He J, Zhao H, Zhao W, He Y and Sun E: Bronchial epithelial
pyroptosis promotes airway inflammation in a murine model of
toluene diisocyanate-induced asthma. Biomed Pharmacother.
125:1099252020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Xiao Z, Jiang Z, Jiang Y, Li W and
Wang M: Schisandrin B attenuates airway inflammation and airway
remodeling in asthma by inhibiting NLRP3 inflammasome activation
and reducing pyroptosis. Inflammation. 44:2217–2231. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Meng J, Wang C, Wang Y, Yang C and
Li Y: Hydrogen sulfide attenuates cigarette smoke-induced
pyroptosis through the TLR4/NF-κB signaling pathway. Int J Mol Med.
49:562022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng Y, Li M, Yangzhong X, Zhang X, Zu A,
Hou Y, Li L and Sun S: Pyroptosis in inflammation-related
respiratory disease. J Physiol Biochem. 78:721–737. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moffatt MF, Kabesch M, Liang L, Dixon AL,
Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et
al: Genetic variants regulating ORMDL3 expression contribute to the
risk of childhood asthma. Nature. 448:470–473. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding Z, Yu F, Sun Y, Jiao N, Shi L, Wan J
and Liu Q: ORMDL3 promotes angiogenesis in chronic asthma through
the ERK1/2/VEGF/MMP-9 pathway. Front Pediatr. 9:7085552021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Y, Zan W, Qin L, Han S, Ye L, Wang M,
Jiang B, Fang P, Liu Q, Shao C, et al: Ablation of ORMDL3 impairs
adipose tissue thermogenesis and insulin sensitivity by increasing
ceramide generation. Mol Metab. 56:1014232022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang YM: Orosomucoid-like protein 3,
rhinovirus and asthma. World J Crit Care Med. 10:170–182. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
James B, Milstien S and Spiegel S: ORMDL3
and allergic asthma: From physiology to pathology. J Allergy Clin
Immunol. 144:634–640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim TB, Kim SY, Moon KA, Park CS, Jang MK,
Yun ES, Cho YS, Moon HB and Lee KY: Five-aminoimidazole-
4-carboxamide-1-beta-4-ribofuranoside attenuates poly (I:C)-induced
airway inflammation in a murine model of asthma. Clin Exp Allergy.
37:1709–1719. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, He F, Chen L, Li Q, Jin S, Zheng
H, Lin J, Zhang H, Ma S, Mei J and Yu J: Resveratrol inhibits
pulmonary fibrosis by regulating miR-21 through MAPK/AP-1 pathways.
Biomed Pharmacother. 105:37–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
UniProt Consortium: UniProt: A worldwide
hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using principal
component analysis and heatmap. Nucleic Acids Res. 43:W566–W570.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cicaloni V, Pecorelli A, Tinti L, Rossi M,
Benedusi M, Cervellati C, Spiga O, Santucci A, Hayek J, Salvini L,
et al: Proteomic profiling reveals mitochondrial alterations in
Rett syndrome. Free Radic Biol Med. 155:37–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang S, Zeng Y, He X, Liu F, Pei P and
Zhang T: Folate-deficiency induced acyl-CoA synthetase short-chain
family member 2 increases lysine crotonylome involved in neural
tube defects. Front Mol Neurosci. 15:10645092022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen G, Cheng J, Yu H, Huang X, Bao H, Qin
L, Wang L, Song Y, Liu X and Peng A: Quantitative proteomics by
iTRAQ-PRM based reveals the new characterization for gout. Proteome
Sci. 19:122021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qian Z, Cai YD and Li Y: A novel
computational method to predict transcription factor DNA binding
preference. Biochem Biophys Res Commun. 348:1034–1037. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng
C, Luo T, Xu L, Liao G, Yan M, et al: CellMarker: A manually
curated resource of cell markers in human and mouse. Nucleic Acids
Res. 47:D721–D728. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu T, Zhou YT, Wang LQ, Li LY, Bao Q,
Tian S, Chen MX, Chen HX, Cui J and Li CW: NOD-like receptor
family, pyrin domain containing 3 (NLRP3) contributes to
inflammation, pyroptosis, and mucin production in human airway
epithelium on rhinovirus infection. J Allergy Clin Immunol.
144:777–787. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang R, Tan M, Xu J and Zhao X:
Investigating the regulatory role of ORMDL3 in airway barrier
dysfunction using in vivo and in vitro models. Int J
Mol Med. 44:535–548. 2019.PubMed/NCBI
|
|
38
|
de Vasconcelos NM, Van Opdenbosch N, Van
Gorp H, Martín-Pérez R, Zecchin A, Vandenabeele P and Lamkanfi M:
An apoptotic caspase network safeguards cell death induction in
pyroptotic macrophages. Cell Rep. 32:1079592020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim RY, Pinkerton JW, Essilfie AT,
Robertson AAB, Baines KJ, Brown AC, Mayall JR, Ali MK, Starkey MR,
Hansbro NG, et al: Role for NLRP3 inflammasome-mediated,
IL-1β-dependent responses in severe, steroid-resistant asthma. Am J
Respir Crit Care Med. 196:283–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Theofani E, Semitekolou M, Samitas K, Mais
A, Galani IE, Triantafyllia V, Lama J, Morianos I, Stavropoulos A,
Jeong SJ, et al: TFEB signaling attenuates NLRP3-driven
inflammatory responses in severe asthma. Allergy. 77:2131–2146.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leszczyńska K, Jakubczyk D and Górska S:
The NLRP3 inflammasome as a new target in respiratory disorders
treatment. Front Immunol. 13:10066542022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sehgal A, Behl T, Kaur I, Singh S, Sharma
N and Aleya L: Targeting NLRP3 inflammasome as a chief instigator
of obesity, contributing to local adipose tissue inflammation and
insulin resistance. Environ Sci Pollut Res Int. 28:43102–43113.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang WJ, Chen SJ, Zhou SC, Wu SZ and Wang
H: Inflammasomes and fibrosis. Front Immunol. 12:6431492021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guan M, Ma H, Fan X, Chen X, Miao M and Wu
H: Dexamethasone alleviate allergic airway inflammation in mice by
inhibiting the activation of NLRP3 inflammasome. Int
Immunopharmacol. 78:1060172020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Theofani E, Semitekolou M, Morianos I,
Samitas K and Xanthou G: Targeting NLRP3 inflammasome activation in
severe Asthma. J Clin Med. 8:16152019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen S, Yao L, Huang P, He Q, Guan H, Luo
Y, Zou Z, Wei S, Peng G, Yan J, et al: Blockade of the
NLRP3/caspase-1 axis ameliorates airway neutrophilic inflammation
in a toluene diisocyanate-induced murine asthma model. Toxicol Sci.
170:462–475. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Yuan Y, Huang ZX, Chen H, Lan R,
Wang Z, Lai K, Chen H, Chen Z, Zou Z, et al: GSDME-mediated
pyroptosis promotes inflammation and fibrosis in obstructive
nephropathy. Cell Death Differ. 28:2333–2350. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bergsbaken T, Fink SL and Cookson BT:
Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol.
7:99–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiao Y, Zhang T, Zhang C, Ji H, Tong X,
Xia R, Wang W, Ma Z and Shi X: Exosomal miR-30d-5p of neutrophils
induces M1 macrophage polarization and primes macrophage pyroptosis
in sepsis-related acute lung injury. Crit Care. 25:3562021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liang Q, Cai W, Zhao Y, Xu H, Tang H, Chen
D, Qian F and Sun L: Lycorine ameliorates bleomycin-induced
pulmonary fibrosis via inhibiting NLRP3 inflammasome activation and
pyroptosis. Pharmacol Res. 158:1048842020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Songane M, Khair M and Saleh M: An updated
view on the functions of caspases in inflammation and immunity.
Semin Cell Dev Biol. 82:137–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aachoui Y, Leaf IA, Hagar JA, Fontana MF,
Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A and Miao
EA: Caspase-11 protects against bacteria that escape the vacuole.
Science. 339:975–978. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Miao EA, Rajan JV and Aderem A:
Caspase-1-induced pyroptotic cell death. Immunol Rev. 243:206–214.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Debeuf N, Zhakupova A, Steiner R, Van
Gassen S, Deswarte K, Fayazpour F, Van Moorleghem J, Vergote K,
Pavie B, Lemeire K, et al: The ORMDL3 asthma susceptibility gene
regulates systemic ceramide levels without altering key asthma
features in mice. J Allergy Clin Immunol. 144:1648–1659. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Breslow DK, Collins SR, Bodenmiller B,
Aebersold R, Simons K, Shevchenko A, Ejsing CS and Weissman JS: Orm
family proteins mediate sphingolipid homeostasis. Nature.
463:1048–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Janneh AH and Ogretmen B: Targeting
sphingolipid metabolism as a therapeutic strategy in cancer
treatment. Cancers (Basel). 14:21832022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Heinrich M, Wickel M, Schneider-Brachert
W, Sandberg C, Gahr J, Schwandner R, Weber T, Saftig P, Peters C,
Brunner J, et al: Cathepsin D targeted by acid
sphingomyelinase-derived ceramide. EMBO J. 18:5252–5263. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bhadra K: A mini review on molecules
inducing caspase-independent cell death: A new route to cancer
therapy. Molecules. 27:64012022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Conus S, Pop C, Snipas SJ, Salvesen GS and
Simon HU: Cathepsin D primes caspase-8 activation by multiple
intra-chain proteolysis. J Biol Chem. 287:21142–21151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Di YQ, Han XL, Kang XL, Wang D, Chen CH,
Wang JX and Zhao XF: Autophagy triggers CTSD (cathepsin D)
maturation and localization inside cells to promote apoptosis.
Autophagy. 17:1170–1192. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Orning P, Weng D, Starheim K, Ratner D,
Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, et al: Pathogen
blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin
D and cell death. Science. 362:1064–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Aghdassi AA, John DS, Sendler M, Weiss FU,
Reinheckel T, Mayerle J and Lerch MM: Cathepsin D regulates
cathepsin B activation and disease severity predominantly in
inflammatory cells during experimental pancreatitis. J Biol Chem.
293:1018–1029. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xing Y, Wang JY, Li MY, Zhang ZH, Jin HL,
Zuo HX, Ma J and Jin X: Convallatoxin inhibits IL-1β production by
suppressing zinc finger protein 91 (ZFP91)-mediated pro-IL-1β
ubiquitination and caspase-8 inflammasome activity. Br J Pharmacol.
179:1887–1907. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Song Z, Zou J, Wang M, Chen Z and Wang Q:
A comparative review of pyroptosis in mammals and fish. J Inflamm
Res. 15:2323–2331. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu C, Yao Q, Hu T, Cai Z, Xie Q, Zhao J,
Yuan Y, Ni J and Wu QQ: Cathepsin B deteriorates diabetic
cardiomyopathy induced by streptozotocin via promoting
NLRP3-mediated pyroptosis. Mol Ther Nucleic Acids. 30:198–207.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen S, Zhou C, Yu H, Tao L, An Y, Zhang
X, Wang Y, Wang Y and Xiao R: 27-Hydroxycholesterol contributes to
lysosomal membrane permeabilization-mediated pyroptosis in
co-cultured SH-SY5Y cells and C6 cells. Front Mol Neurosci.
12:142019. View Article : Google Scholar : PubMed/NCBI
|