Introduction
Gastric cancer (GC) is one of the most common gastrointestinal (GI) tract cancers worldwide, ranked fifth for its incidence and fourth for mortality (1). Chemotherapy, immunotherapy and targeted therapy have contributed to increased survival in the last decade. However, even precise targeted therapy based on the cellular and molecular levels have faced problems such as acquired drug resistance (2) and side effects (3). An improved understanding of molecular mechanisms and discovery of innovative potential molecular biomarkers are essential to improve the therapeutic effects.
Keratin is the intermediate protein subunit involved in maintaining epithelial cell integrity and exerts various physiological functions including membrane trafficking, signaling, protein synthesis and cell motility. It has been reported that keratin proteins are associated with proliferation, apoptosis, migration and invasion of a number of cancers (4–6). Keratin 23 (KRT23) knockdown reduces cellular proliferation and decreases DNA damage response in colon cancer cells (7). MYC-mediated amplification of KRT23 promotes the cell proliferation of liver cancer (8). A recent study showed that KRT23 exerts an important role in ovarian cancer migration through epithelial-mesenchymal transition by adjusting TGF-β/Smad signaling pathway (9). While increasing evidence indicates that KRT23 exhibits pro-cancer effects in some tumors, its role in gastric and a number of other cancers has not been comprehensively studied.
Melatonin (MLT) is a neuroendocrine hormone synthesized by the pineal gland. As a hydrophobic small molecule, MLT mainly exerts biological effects though signal transduction systems by binding to specific receptors on target cells. MLT has a variety of physiological effects such as regulating the circadian rhythm, promoting sleep, enhancing immunity and scavenging free radicals, as well as anti-oxidation and anti-stress properties. MLT also displays antitumor activity including anti-proliferation, pro-apoptosis and anti-metastasis (10,11). Based on the fact that MLT effectively inhibits a variety of tumors, it has been used in combination with chemotherapy drugs to enhance the sensitivity of tumor cells to chemotherapy and inhibit tumor drug resistance. This is performed not only with traditional chemotherapy drugs but also with novel targeted therapies (12,13). MLT can also act as a potential sensitizer, sensitizing cancer cells to γ-ray ionizing radiation (14). In addition, MLT counteracts the adverse effects of cancer therapy by displaying neuroprotective effects, decreasing cisplatin-induced ototoxicity and attenuating chemotherapy-induced genotoxicity (15,16). Our previous work revealed that MLT promotes the apoptosis of GC cells through the mitochondrial pathway (17). MLT is also found to inhibit the proliferation of GC cells by reducing the accumulation of hypoxia-inducible factor-1 α and the expression of VEGF (18). However, it is currently unclear whether the anti-GC roles of MLT are related to keratin. These intriguing matters led us to further investigate whether KRT23 contributes to the anti-GC activity of MLT.
In the present study, expression profile chips were used to explore the mechanisms underlying MLT anti-GC effects. The expression of KRT23 in GC cells and normal gastric epithelial cell line GES-1 was analyzed. The function of KRT23 in GC cells was further investigated by inducing CRISPR/Cas9-mediated knockout of KRT23. Finally, the present study focused on the effects of MLT on KRT23 knockout (KRT23-KO) cells to investigate whether KRT23 knockout contributed to the anti-GC effects of MLT and its potential molecular mechanism.
Material and methods
Cell culture
The GC cell line AGS was purchased from the Cell Bank of Shanghai Institute for Biological Science, Chinese Academy of Sciences. The GC cell line SNU-216 was obtained from Nanjing Kangbai Biotechnology Co., Ltd. The normal gastric epithelial cell line GES-1 was kindly provided by Professor Changming Huang (Fujian Medical University Union Hospital). AGS cells were cultured in HyClone DME/F-12 (cat. no. SH30023; HyClone; Cytiva), 10% fetal bovine serum (cat. no. 04-001-1A; Biological Industries). The SNU-216 and GES-1 cells were cultured in HyClone RMPI Medium Modified (cat. no. SH30809.01; HyClone; Cytiva) medium supplemented with 10% fetal bovine serum.
Cell viability
Cell viability was evaluated using the MTS assay. GC cell lines AGS and SNU-216 were grown in 96-well plates at a density of 1,000 cells per well. After growing for 24 h, cells were changed with fresh medium containing 0, 1, 2, 3, 4 or 5 mM MLT (cat. no. M5250; MilliporeSigma) for 24, 48 or 72 h. After the intervention, culture medium was replaced with 20 µl of the CellTiter 96 AQueous One Solution Cell Proliferation Assay (cat. no. G3582; Promega Corporation). and 80 µl of serum-free medium for 2 h. The absorbance of each sample was measured at a wavelength of 490 nm.
RNA isolation and gene expression profiling
AGS cells were grown in 100 mm dishes at a density of 2.5×105 cells per dish. After a 24 h incubation, cells were treated with 2.5 mM MLT or 1% ethanol as control for another 24 h. Then, cells were washed with PBS three times. Total cellular RNA was extracted using the RNeasy Mini kit (cat. no. 74104; Qiagen GmbH). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). Fragmented cRNA was synthesized based on instructions provided by the manufacturer and then hybridized with the customized Affymetrix GeneChip PrimeView Human Gene Expression Array (Affymetrix; Thermo Fisher Scientific, Inc.) to reveal the expression pattern of 19,042 genes. GeneChips were scanned using Affymetrix Genechip Scanner 3000 7G (Affymetrix; Thermo Fisher Scientific, Inc.). The acquired images were processed using GenePix Pro 6.0 software (Axon Instruments. Inc) for grid alignment and data extraction. Expression profiling data measured in our study would be deposited into the Gene Expression Omnibus database and the accession numbers provided upon request.
Identification of differently expressed genes (DEGs) and functional enrichment analysis
The reproducibility-based pairwise difference and pairwise fold change methods were combined to identify reproducible DEGs between the treatment group and the control group based on small-scale cell line datasets (19). The two DEGs lists analyzed by two algorithms were combined based on the reproducibility of dysregulation directions. The initial step (the initial number of the top-ranked genes) of the algorithms was set as 300 and the consistency threshold was 90%, according to the suggestions of previous study (19). Functional enrichment analysis of DEGs was conducted using the Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.jp/ or http://www.genome.jp/kegg/). The Benjamini-Hochberg procedure (20) was applied to control the false discovery rate. The statistical significance was set to FDR <10%.
Reverse transcription-quantitative (RT-q) PCR
A RT-qPCR quantification assay was performed to confirm results of the expression microarrays. Total RNA was extracted from GC cells (1×106 cells) using TRIzol® reagent (cat. no. 15596-026; Thermo Fisher Scientific, Inc.) based on instructions provided by the manufacturer. cDNA synthesis was performed using the PrimeScript™ RT reagent kit (cat. no. RR037A; Takara Bio, Inc.) and qPCR was performed using the TB Green Fast qPCR Mix (cat. no. RR430B; Takara Bio, Inc.), according to the manufacturer's protocols. The PCR cycling conditions were as follows: One cycle at 95°C for 1 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 30 sec. A total of 18 differentially expressed genes were analyzed by RT-qPCR in triplicate and normalized to the mean of GAPDH (Table SI). Normalized gene expression values were calculated based on the 2−ΔΔCq method (21). These experiments were replicated three times.
Knockout of KRT23 by CRISPR/Cas9
CRISPR/Cas9 targeted knockout was performed to delete KRT23 (GenBank accession number: NM_015515.5). The CRISPR/Cas9 vector PX260 was kindly provided by Professor Pingsheng Liu (Institute of Biophysics, Chinese Academy of Sciences). The single guide (sg)RNA was designed to target exon 2 of the KRT23 gene using the CRISPR design web tool (http://crispr.mit.edu/). The KRT23 sgRNA targeted sequence used was GCGGCTTTCCAGCTTCATGTTGG, which was synthesized, annealed and inserted into the BbsI digestion site of PX260 vector. A total of 3×105 AGS or SNU-216 cells were added to the wells of a 6-well plate. When cell density reached 70–80% confluency, cells were transfected with KRT23 sgRNA expressing PX260 vector or PX260 control vector by Lipofectamine® 3000 (cat. no. L3000001; Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h at 37°C and cultured in complete medium for 42 h at 37°C. After 48 h, cells were screened with 0.5 µg/ml puromycin to select positive cells. After 4 days, cells were separated as single cells into 96-well plates for single cell cloning. After 2 weeks, clones were plated onto 24-well plates for expansion. Genomic DNA was extracted using the Genloci TNA extraction kit (cat. no. GP0156; Genloci Biotechnologies Inc.) and amplified by PCR using the following cycling parameters: Initial denaturation at 98°C for 2 min; 25 cycles of 98°C for 10 sec and 72°C for 15 sec, and a final extension step at 72°C for 5 min. PCR products were submitted for Sanger sequencing by Biosune Biotechnology Co., Ltd. KRT23 expression levels were detected in monoclonal cells using immunoblotting.
Immunofluorescence staining
Cells were grown on coverslips in 24-well plates at a density of 8×104 cells/well for 24 h. Cells were fixed in 4% paraformaldehyde for 30 min at room temperature (RT) and permeabilized in 0.5% Triton X-100 for 20 min at RT. Then, cells were blocked with 5% bovine serum albumin for 2 h at RT and incubated overnight at 4°C with anti-KRT23 (1:100; cat. no. sc-365892; Santa Cruz Biotechnology, Inc.). Afterwards, cells were incubated with Alexa Fluor 488 anti-mouse secondary antibody (1:200; cat. no. A32723; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at RT and counterstained with Hoechst (cat. no. C1026; Beyotime Institute of Biotechnology) for 10 min at room temperature. Cell images were captured through a fluorescence microscope (Zeiss Axio Observer A1; Zeiss AG).
Colony formation
For cell colony formation analysis, cells were grown at a density of 1,000 cells/well in 6-well plates in triplicate. After 7–10 days, colonies were fixed using 100% methanol for 15 min at room temperature, stained with crystal violet for 10 min at RT and scanned using ImageQuant LAS 4000 mini (Cytiva). The number of colonies (50 cells/colony) were counted using ImageJ 1.44p Launcher software (National Institutes of Health).
Cell cycle analysis
To assess the cell cycle, a total of 5×105 cells were seeded in 6-well plates for 24 h at 37°C. Cells were harvested and washed twice with PBS. Cells were resuspended in 75% ethanol overnight at 4°C and then collected and stained using PI/RNase Staining Buffer (cat. no. C09-550825; BD Biosciences) for 30 min at 4°C. FACSCalibur Flow cytometry (BD Biosciences) was used to detect the cell cycle and the data were analyzed using ModFit LT 3.1 software (Verity Software House, Inc.).
Western blotting
Cells were lysed in RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) supplemented with phosphatase and protease inhibitors (cat. no. 5892791001; Roche Diagnostics). Protein concentrations were quantified using the Enhanced BCA Protein Assay kit (cat. no. P0010S; Beyotime Institute of Biotechnology) based on instructions provided by the manufacturer. Protein samples of the same concentrations (25 µg protein loaded per lane) were separated using a 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (MilliporeSigma) using a trans-blot apparatus (Bio-Rad Laboratories Inc.). Membranes were then blocked in 0.5% bovine serum albumin for 1 h at RT and incubated with primary antibodies including anti-p38 (1:1,000; cat. no. 8690S; Cell Signaling Technology, Inc.), anti-phosphorylated (p)-p38 (1:1,000; cat. no. 4511S; Cell Signaling Technology, Inc.), anti-ERK1/2 (1:1,000; cat. no. 4696S; Cell Signaling Technology, Inc.), anti-p-ERK1/2 (1:1,000; cat. no. 4376S; Cell Signaling Technology, Inc.), anti-KRT23 (1:1,000; cat. no. ab156569; Abcam) and anti-GAPDH (1:1,000; cat. no. 5174S; Cell Signaling Technology, Inc.). Anti-GAPDH was used as a control. Membranes were incubated with alkaline phosphatase-conjugated secondary antibody, goat anti-rabbit IgG (1:5,000; cat. no. ab98505; Abcam) or goat anti-mouse IgG (1:1,000; cat. no. sc-2008; Santa Cruz Biotechnology, Inc.) at room temperature for 30 min. The signal of target proteins was visualized using CDP-Star (Roche Diagnostics, Germany) reagent. All protein bands were scanned using ImageQuant LAS 4000 mini (Cytiva). Band intensities were analyzed using ImageJ2× software (National Institutes of Health).
Electric cell substrate impedance sensing (ECIS)
The ECIS Ztheta (Applied BioPhysics, Inc.) system was used to simultaneously monitor the impedance of electrode 8W10E arrays (Applied BioPhysics, Inc.). To maintain electrode capacitance, 8W10E arrays were pre-treated with 10 mM cysteine for 15 min. Cells were seeded at a density of 1×104 cells/well in 400 µl of culture medium supplemented with 10% FBS. Electrical impedance analysis of cell growth was measured using the designated 11 frequencies from 62.5–64000 Hz (62.5, 125, 250, 500, 1,000, 2,000, 4,000, 8,000, 16,000, 32,000 and 64,000 Hz) at 10 min intervals. During these measurements, the electrode array was placed in an array station located in the incubator connected to the ECIS Ztheta station controller outside the incubator. ECIS software version 1.2.123 (Applied BioPhysics, Inc.) was already installed on the PC and connected to the ECIS instrument, ready for the time course measurement of impedance values. Data were normalized to baseline.
Statistical analysis
All data were reported as the mean of three biological replicates with standard deviation. The independent-samples t-test was applied for comparison between two groups and one-way ANOVA followed by Tukey's post hoc test was applied for comparison of multiple groups. Statistical analyses were performed with SPSS software (version 22.0; IBM Corp.) or GraphPad Prism 8 (Dotmatics). P<0.05 was considered to indicate a statistically significant difference.
Results
MLT inhibited the outgrowth of GC cells
GC cell lines AGS and SNU-216 were exposed to 0, 1, 2, 3, 4 or 5 mM MLT for 24, 48 or 72 h. Cell viability was assessed by MTS assay. MLT inhibited the proliferation of AGS and SNU-216 cells in a dose and time-dependent manner (Fig. 1). The half maximal inhibitory concentration (IC50) of MLT on AGS and SNU-216 cells at 24 h was 2.613 and 2.464 mM, respectively. In our previous studies, the commonly used concentration and exposure time of MLT to inhibit cell viability in GI cancer cells were 2.5–3 mM and 24 h (17,22). Based on these results, 2.5 mM and 24 h were selected for subsequent experiments.
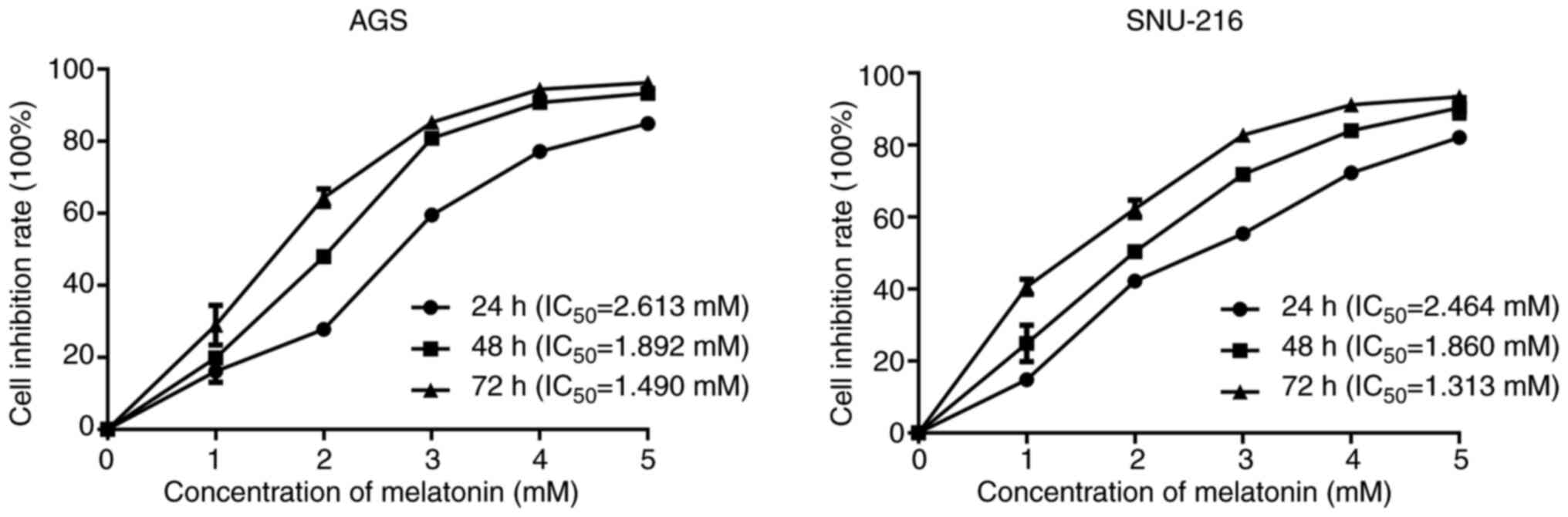 |
Figure 1.
Effects of melatonin on cell proliferation of gastric cancer cells AGS and SNU-216. Cell viability was detected by MTS assay.
|
Identification and functional analysis of DEGs in GC cells treated with MLT
To systematically explore the mechanisms behind the anti-GC effects of MLT, an expression profile chip was used to screen DEGs between MLT treated and untreated control AGS cells. Using the reproducibility-based pairwise difference method, 9,378 DEGs were identified whereas 6,926 DEGs were found based on the pairwise fold change method with a consistency threshold of 90% and an initial step of 300. Combining the two methods, a total of 6,388 DEGs with the same dysregulated directions were obtained. Among the 6,388 DEGs, 2,741 genes were upregulated and 3,647 genes were downregulated in AGS cells treated with MLT as compared with control cells (Table I). These genes were significantly enriched in the pathways of class O forkhead box transcription factor (FoxO), ErbB, lysosome, pyrimidine metabolism, RNA transport, spliceosome and cell cycle (Fig. 2A). To investigate the anticancer mechanism of MLT and confirm the accuracy of microarray data, 11 genes closely related to cancer development among the most significantly down- or upregulated DEGs were quantitated using RT-qPCR (Table SII). The expression patterns of these selected genes were consistent with the microarray results (Fig. 2B). A series of keratin molecules including KRT7, KRT20, KRT23, KRT75 and KRT81 mRNA levels in the human GC cell line AGS were downregulated after adding 2.5 mM of MLT for 24 h (P<0.05, Fig. 2C). mRNA expression levels of KRT23 in SNU-216 was also downregulated after adding 2.5 mM of MLT (Fig. 2D).
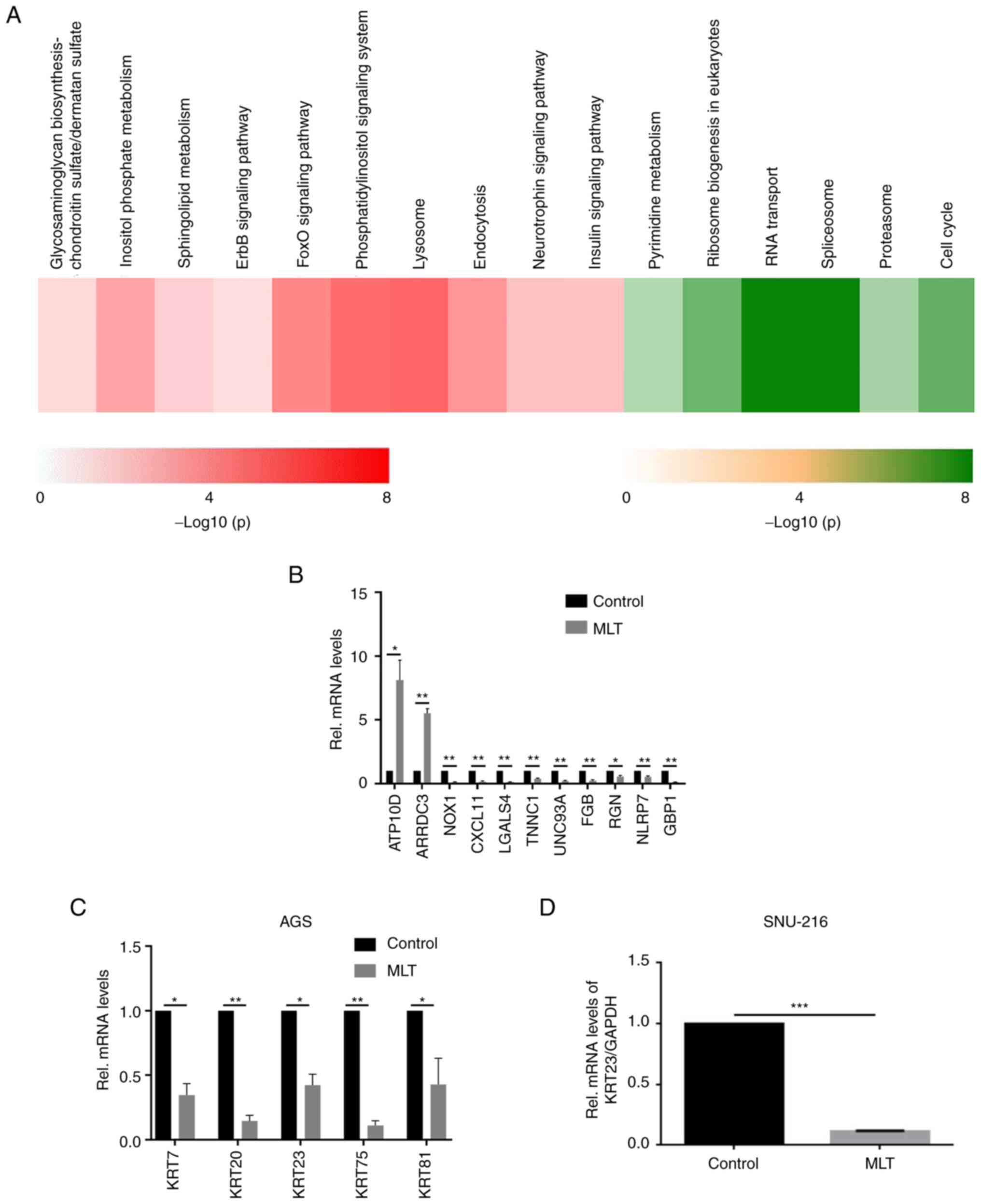 |
Figure 2.
Validation of microarray analysis. (A) Significantly enriched KEGG pathways associated with the upregulated (Red) and downregulated (Green) DEGs in AGS cells treated by MLT. (B) Expression of the highly DEGs were quantitated using reverse transcription-quantitative PCR in AGS GC cells with or without 2.5 mM of MLT treatment for 24 h. Expression of these selected genes showed a consistent pattern with microarray results. (C) Changes in the mRNA expression levels of KRT7, KRT20, KRT23, KRT75 and KRT81 in AGS GC cells with or without 2.5 mM of MLT treatment for 24 h. (D) Changes in KRT23 mRNA expression levels in SNU-216 GC cells with or without 2.5 mM of MLT treatment for 24 h. *P<0.05, **P<0.01 and ***P<0.001. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differently expressed genes; KRT, keratin; Rel, Relative; MLT, melatonin.
|
 |
Table I.
DEGs detected in AGS cells by the reproducibility-based PD and PFC.
|
Table I.
DEGs detected in AGS cells by the reproducibility-based PD and PFC.
| Series |
DEGs |
Upregulated |
Down |
Common |
Consistent |
Ratio |
P-value |
| PD |
9,738 |
2,741 |
3,647 |
6,388 |
6,388 |
1 |
0 |
| PFC |
6,926 |
|
|
|
|
|
|
Protein expression levels of KRT23 in both AGS and SNU-216 were further analyzed and confirmed that it was downregulated after adding 2.5 mM of MLT (Fig. 3A and B). Immunoblotting was performed to analyze the expression levels of KRT23 in the normal human gastric cell GES-1 and GC cells. Compared with GES-1, KRT23 protein was highly expressed in AGS and SNU-216 GC cells (Fig. 3C and D).
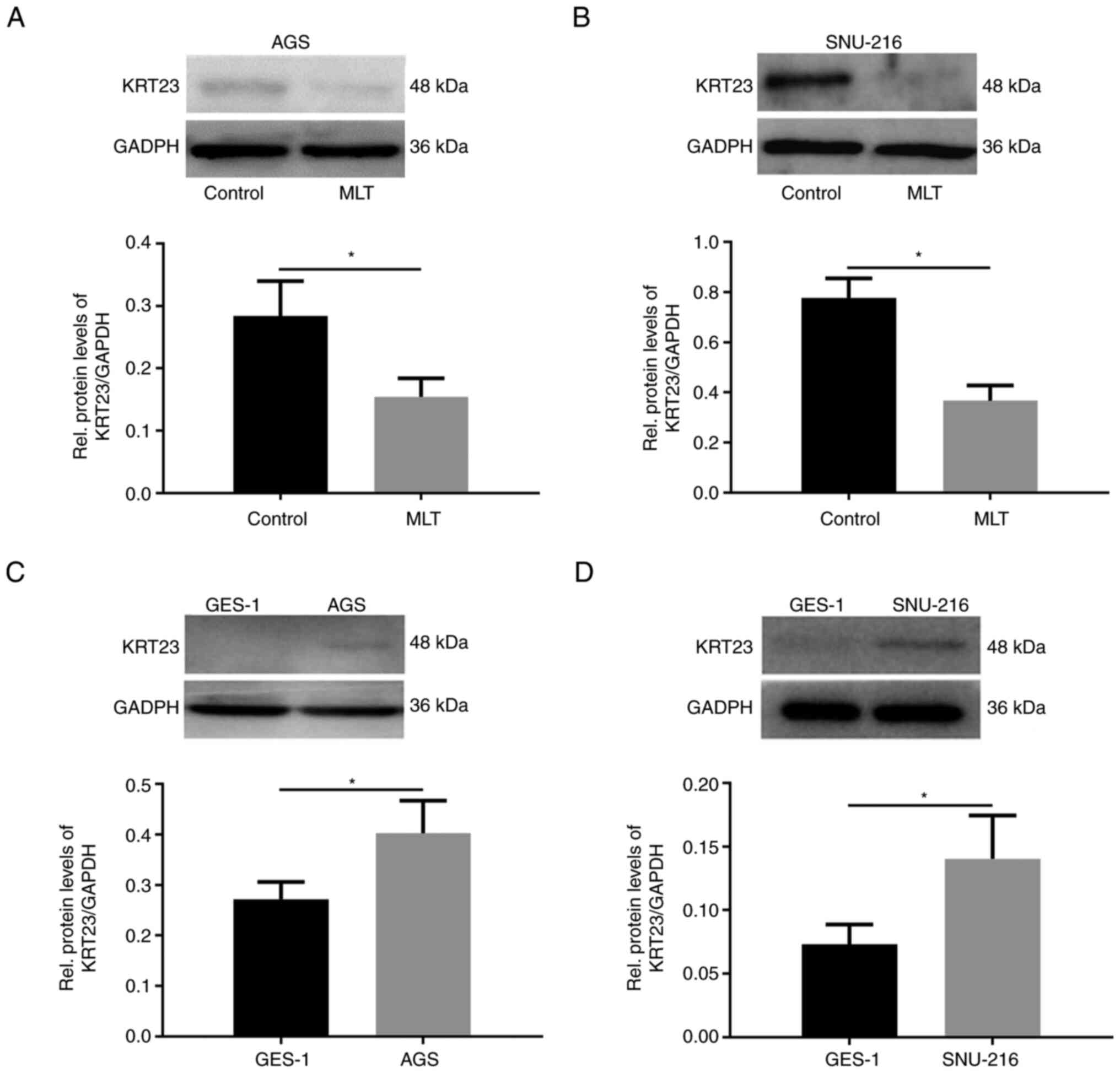 |
Figure 3.
Western blot analysis of KRT23 expression in GC cells. Western blot analysis of KRT23 expression levels in (A) AGS and (B) SNU-216 GC cells with or without 2.5 mM of MLT treatment for 24 h. Western blot analysis of KRT23 expression in (C) AGS and (D) SNU-216 GC cells compared with the normal human gastric cell GES-1. *P<0.05. KRT, keratin; GC, gastric cancer; MLT, melatonin.
|
Generation of KRT23 knockout GC cell lines
KRT23-KO GC cell lines were generated using CRISPR/Cas9 technology (Fig. 4A). Sequencing results were compared with the standard KRT23 sequence to identify the CRISPR-induced mutations (Fig. S1). The sgRNA selected led to 86 bases insertion in one allele of the second exon of KRT23-KO AGS cell line and one T base deletion in the KRT23-KO SNU-216 cell line (Fig. 4B). Immunoblotting was used to confirm KRT23 expression levels, which verified that KRT23 protein was not detectable in the KRT23-KO AGS and SNU-216 cell lines (Fig. 4C). To further identify the expression level of KRT23 in KRT23-KO and control cells, immunofluorescence staining was performed. Similar to immunoblotting, KRT23 deletion in KRT23-KO AGS and SNU-216 cells resulted in almost no staining of KRT23 (Fig. 4D).
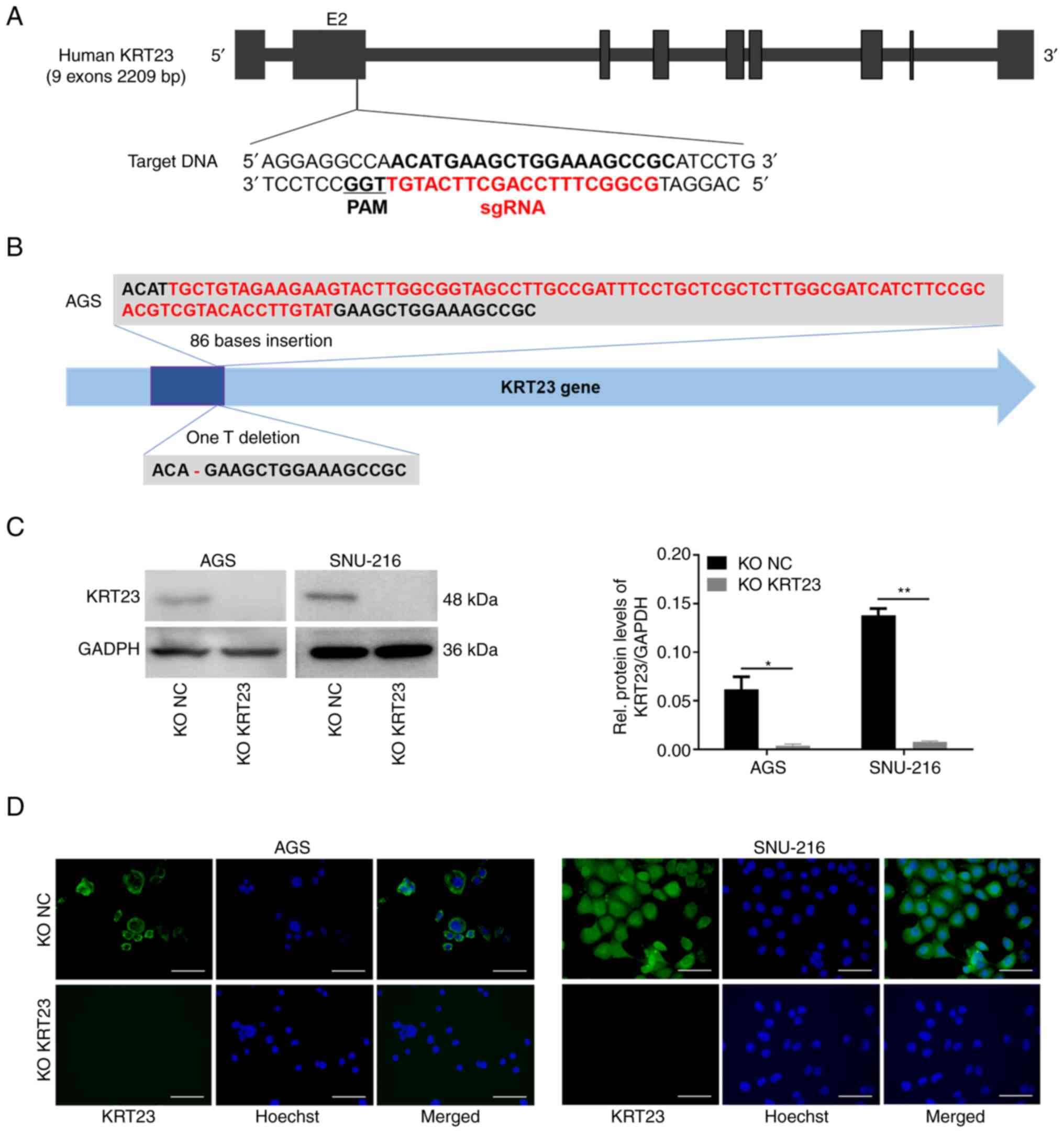 |
Figure 4.
KRT23 knockout using the CRISPR/Cas9 technique in GC cells. (A) Schematic depicting the KRT23 locus. Exon 2 of the KRT23 gene was the target site. The reference sequence in the targeted region and PAM motif was indicated. (B) Characterization of the mutations in KRT23-KO AGS or SNU-216 cells. (C) Verification of KRT23 knockout by western blotting. *P<0.05 and **P<0.01. (D) Immunofluorescence staining of KRT23-KO AGS or SNU-216 cells and corresponding control cells. Scale bars, 50 µm. KRT, keratin; GC, gastric cancer; KO, knockout; NC, negative control.
|
Knockout of KRT23 inhibits GC cell proliferation
The proliferation activity of cells was detected using the MTS assay and the corresponding growth curves were generated at different time points after 24, 48, 72, 96 or 120 h of inoculation. As demonstrated in Fig. 5A, the proliferation rate of AGS cells with KRT23-KO was considerably reduced when compared with the control group. Similarly, KRT23-KO SNU-216 cells proliferated at a lower rate than control cells (Fig. 5A). Next, the cells in each group were plated in 6-well plates with the same cell density. Results were collected and analyzed after 7–14 days and colonies with >50 cells in each group were calculated. The results showed that the clonal ability of GC cells was considerably decreased after KRT23 deletion (Fig. 5B). Then the number and proportion of cells in each stage of the cell cycle were detected using flow cytometry. Image fitting analysis was performed using ModFit software. KRT23-KO AGS cells had a higher number of G1 phase cells and a lower proportion of S phase cells as compared with the control group. In SNU-216 cells, KRT23 deletion enhanced the number of cells in G1 phase while decreasing the number of cells in S and G2/M phases (Fig. 5C). In addition, ECIS was used to monitor the growth of KRT23-KO AGS and SNU-216 cells for 5 consecutive days. In AGS cells, the electrical impedance of the NC group reached a peak at ~100 h, yet the KRT23-KO cells reached a peak at ~117 h and the peak value was lower compared with what was observed for the NC group. In SNU-216 cells, the electrical impedance of the NC group reached a peak of ~113 h, yet KRT23-KO cells reached highest impedance at ~125 h and the peak value was lower compared with what was measured in the NC group (Fig. 6A). The expression levels of p38, ERK1/2, p-p38 and p-ERK1/2 were further analyzed by western blotting. p-ERK1/2 and p-p38 expression levels were downregulated in the two KRT23-KO GC cell lines. Total protein levels for ERK1/2 and p38 were not substantially different from the control group (Fig. 6B).
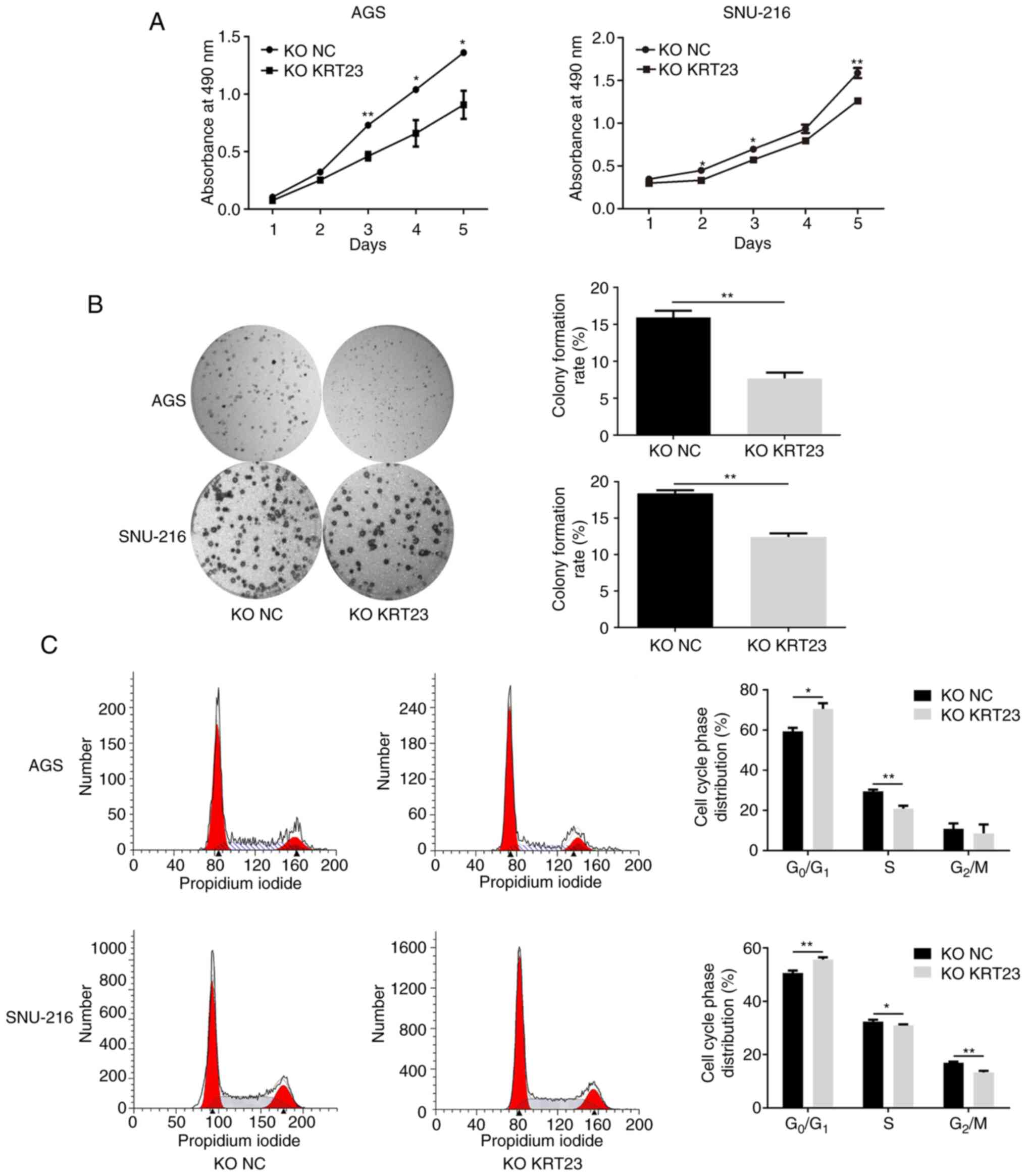 |
Figure 5.
The effect of KRT23 knockout on the proliferation of AGS/SNU-216 cells. (A) Growth curves of KRT23-KO AGS and SNU-216 cells with their control cells. (B) Representative photographs and colony formation assay of KRT23-KO AGS and SNU-216 cells and their control cells. (C) Cell cycle analysis for KRT23-KO AGS and SNU-216 cells and their control cells. KO NC vs. KO KRT23, *P<0.05 and **P<0.01. KRT, keratin; KO, knockout; NC, negative control.
|
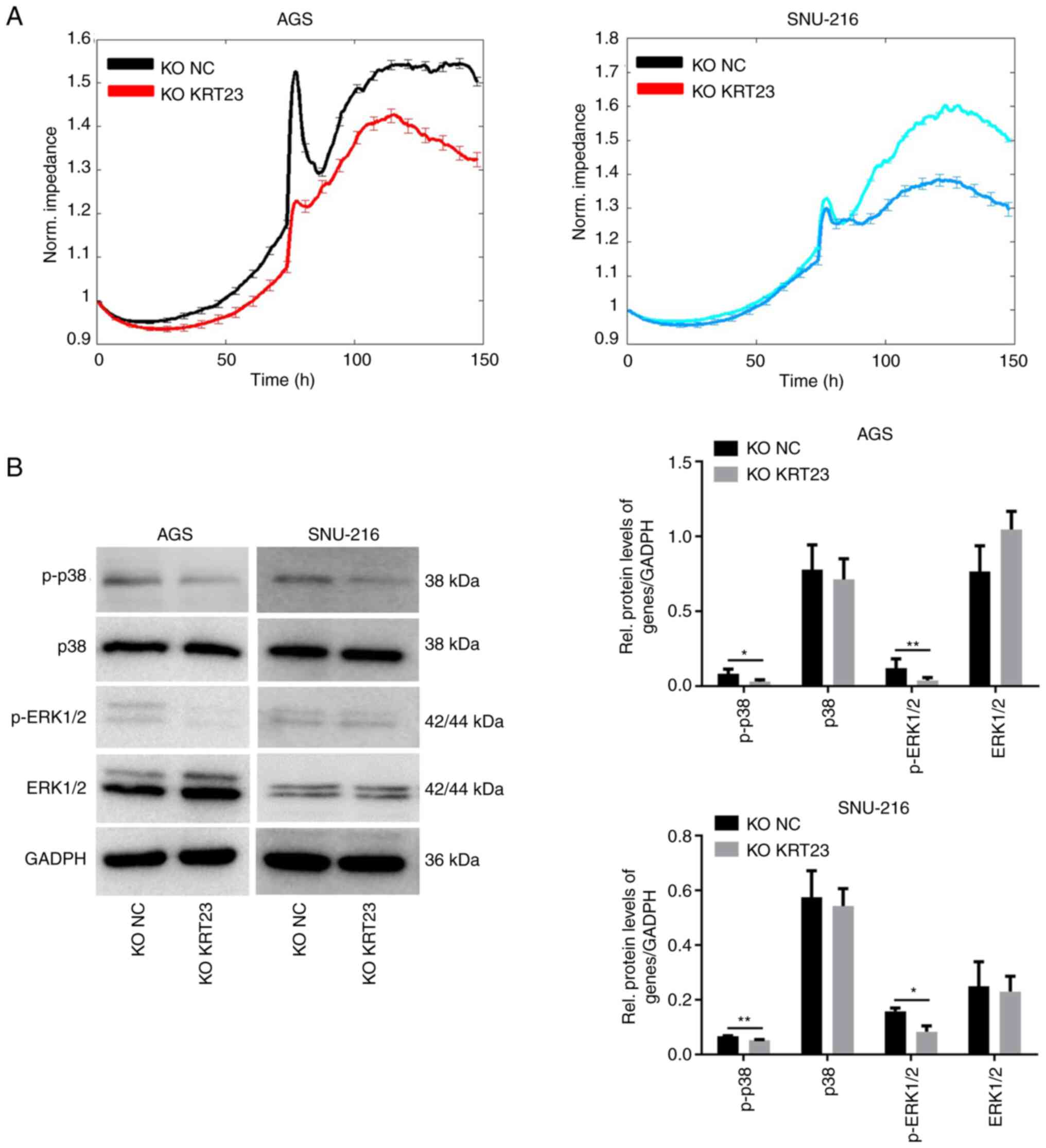 |
Figure 6.
Dynamic monitoring and western blot analysis of KRT23-KO GC cells. (A) Dynamic monitoring by ECIS revealed the growth status of KRT23-KO AGS and SNU-216 cells and their control cells for 5 consecutive days. (B) Western blot analysis of expression and activation of proteins in KRT23-KO AGS and SNU-216 cells and their control cells. *P<0.05 and **P<0.01. KRT, keratin; KO, knockout; GC, gastric cancer; NC, negative control; p-, phosphorylated.
|
Knockout of KRT23 enhances the inhibitory effects of MLT in GC cells
A total of 5×105 cells/well were added to 6-well plates for 24 h to perform colony formation experiments and cell cycle analysis. A total of 9×103 cells were plated in 96-well plates for 24 h to perform the MTS assay and then treated with 2.5 mM of MLT or vehicle control for 24 h. The effect of both MLT treatment and KRT23 knockout on GC cell proliferation was determined by colony formation, MTS and flow cytometry assays. Colony formation experiments revealed that both MLT intervention and KRT23 knockout decreased the number of colonies compared with the KO NC + Ctrl group (Fig. 7A). MTS assays revealed antiproliferative effects of MLT intervention on GC cells (Fig. 7B). MLT intervention raised the proportion of cells in G0/G1 phase, while decreasing the percentage of cells in S and G2 phases in AGS cell line, according to flow cytometry (Fig. 8A). Moreover, compared with the single MLT intervention group, a combination of MLT treatment with KRT23 depletion more profoundly suppressed colony formation, restrained cell multiplication and caused G0/G1 phase arrest (Figs. 7A and B and 8A). These data demonstrated that loss of KRT23 improved the anti-proliferative action of MLT on GC cells. ECIS monitoring demonstrated that KRT23 knockout further increased the inhibitory effects of MLT. A total of 8×104 cells/well were added to an electrode 8W10E array for 24 h. After adding MLT, ECIS was used to monitor the effect of MLT on KRT23-KO cells for 24 h. MLT intervention and KRT23 knockout both decreased normalized impedances. Moreover, compared with the single MLT intervention group, MLT treatment and KRT23 deletion acted synergistically to decrease normalized impedances (Fig. 8B).
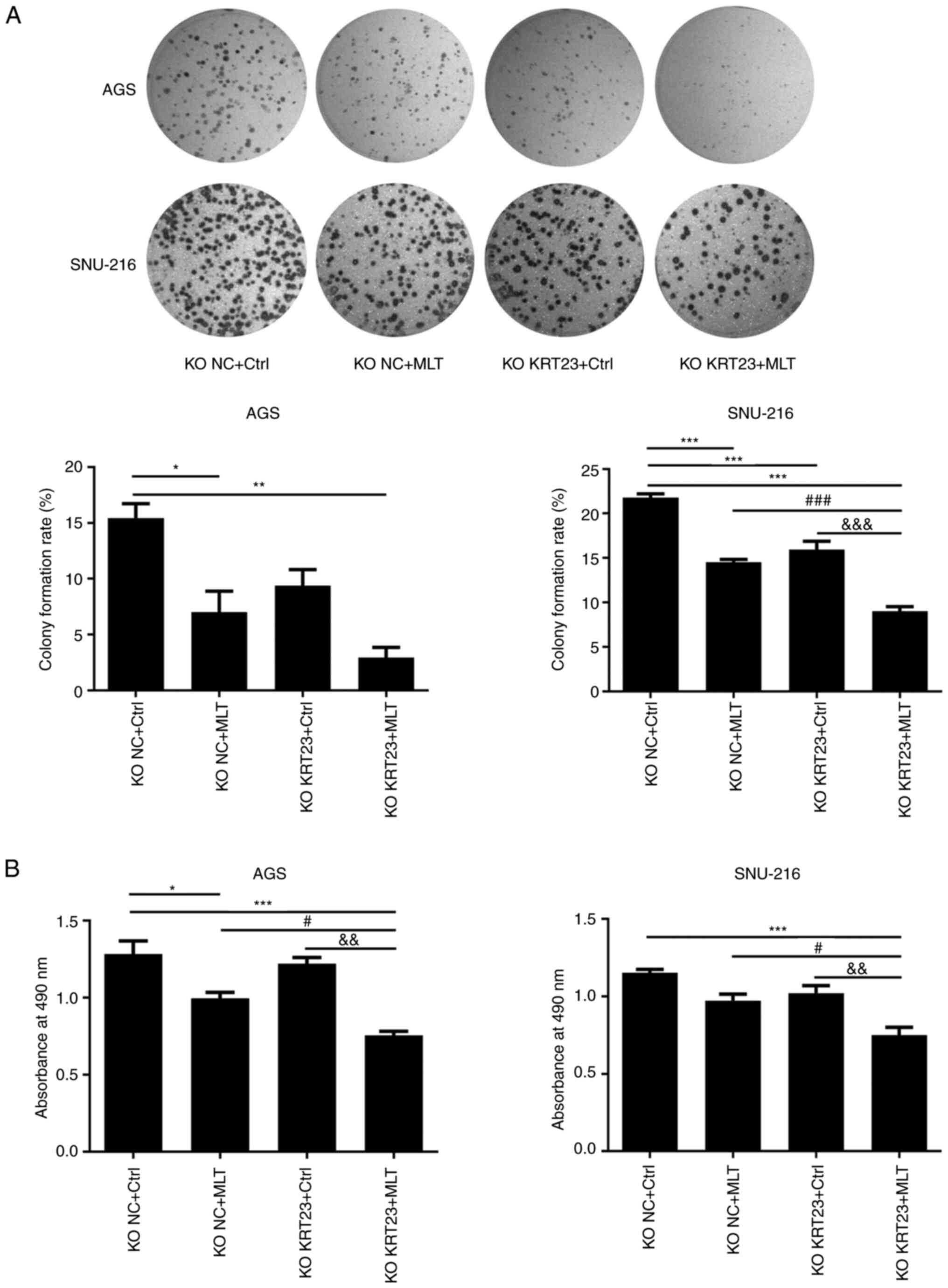 |
Figure 7.
KRT23 knockout enhanced anti-proliferative abilities of MLT on human GC cells. (A) Representative photographs and results of a colony formation assay with or without 2.5 mM of MLT treatment for 24 h. (B) Cell viability of AGS and SNU-216 cells with or without 2.5 mM of MLT treatment for 24 h. *P<0.05, **P<0.01 and ***P<0.001 vs. KO NC + Ctrl group; #P<0.05 and ###P<0.001 vs. the KO NC + MLT group; &&P<0.01 and &&&P<0.001 vs. the KO KRT23 + Ctrl group. KRT, keratin; MLT, melatonin; GC, gastric cancer; KO, knockout; NC, negative control; p-, phosphorylated.
|
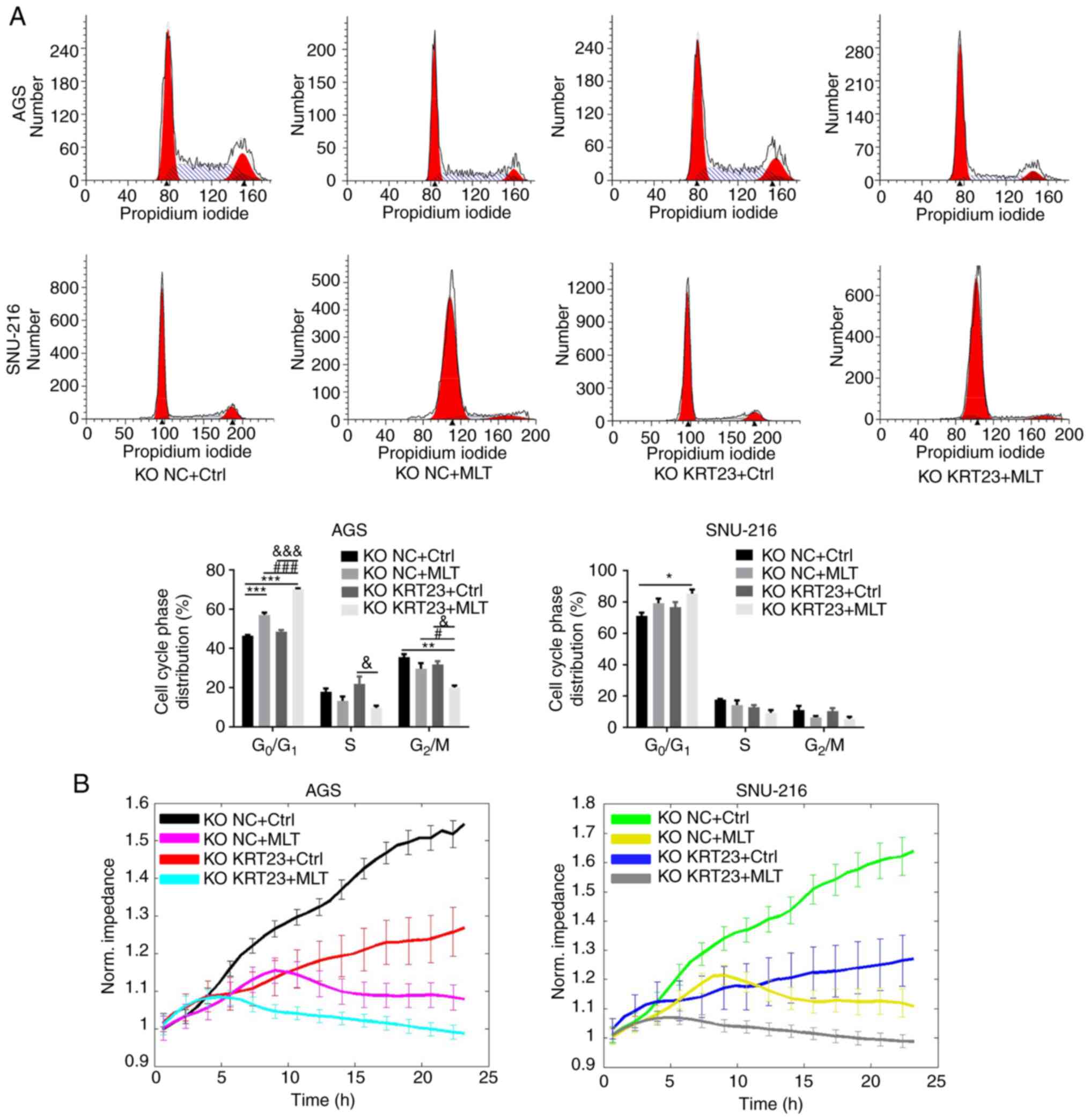 |
Figure 8.
Cell cycle analysis and dynamic monitoring of KRT23-KO cells treated with or without MLT. (A) Flow cytometric analysis of KRT23-KO GC cells with or without 2.5 mM of MLT treatment for 24 h. Percentages of cell populations at diverse stages of the cell cycle were listed within the panels. All histograms represented mean ± standard deviation values of three biological replicates. (B) Dynamic monitoring of KRT23 knockout cells treated with or without 2.5 mM of MLT for 24 h using the ECIS method. *P<0.05, **P<0.01 and ***P<0.001 vs. the KO NC + Ctrl group; #P<0.05 and ###P<0.001 vs. the KO NC + MLT group; &P<0.05 and &&&P<0.001 vs. the KO KRT23+Ctrl group. KRT, keratin; KO, knockout; NC, negative control; MLT, melatonin; GC, gastric cancer.
|
Knockout of KRT23 enhances the inhibitory effects of MLT via inhibiting p38/ERK phosphorylation in GC cells
The human KRT23 KO and control GC cell lines were treated with 2.5 mM MLT or ethanol control for 24 h. The expression levels of KRT23, p-p38, p38, p-ERK1/2 and ERK1/2 were further analyzed by immunoblotting. Immunoblotting revealed that both MLT intervention and KRT23 deletion decreased the expression of KRT23 in AGS and SNU-216 cell lines. Furthermore, p-p38 expression levels were downregulated in both KO NC + MLT group and KO KRT23 + Ctrl group compared with the KO NC + Ctrl group. MLT intervention or loss of KRT23 decreased p-ERK1/2 expression in AGS cell line. These results suggested that both MLT intervention and KRT23 knockout decreased the phosphorylation of p38 and ERK1/2 in GC cell lines. Compared with the single MLT intervention group, a combination of MLT treatment with KRT23 deletion further reduced the phosphorylation of p38 and ERK1/2. The total protein levels of ERK1/2 and p38 were not significantly different among groups (Fig. 9A and B).
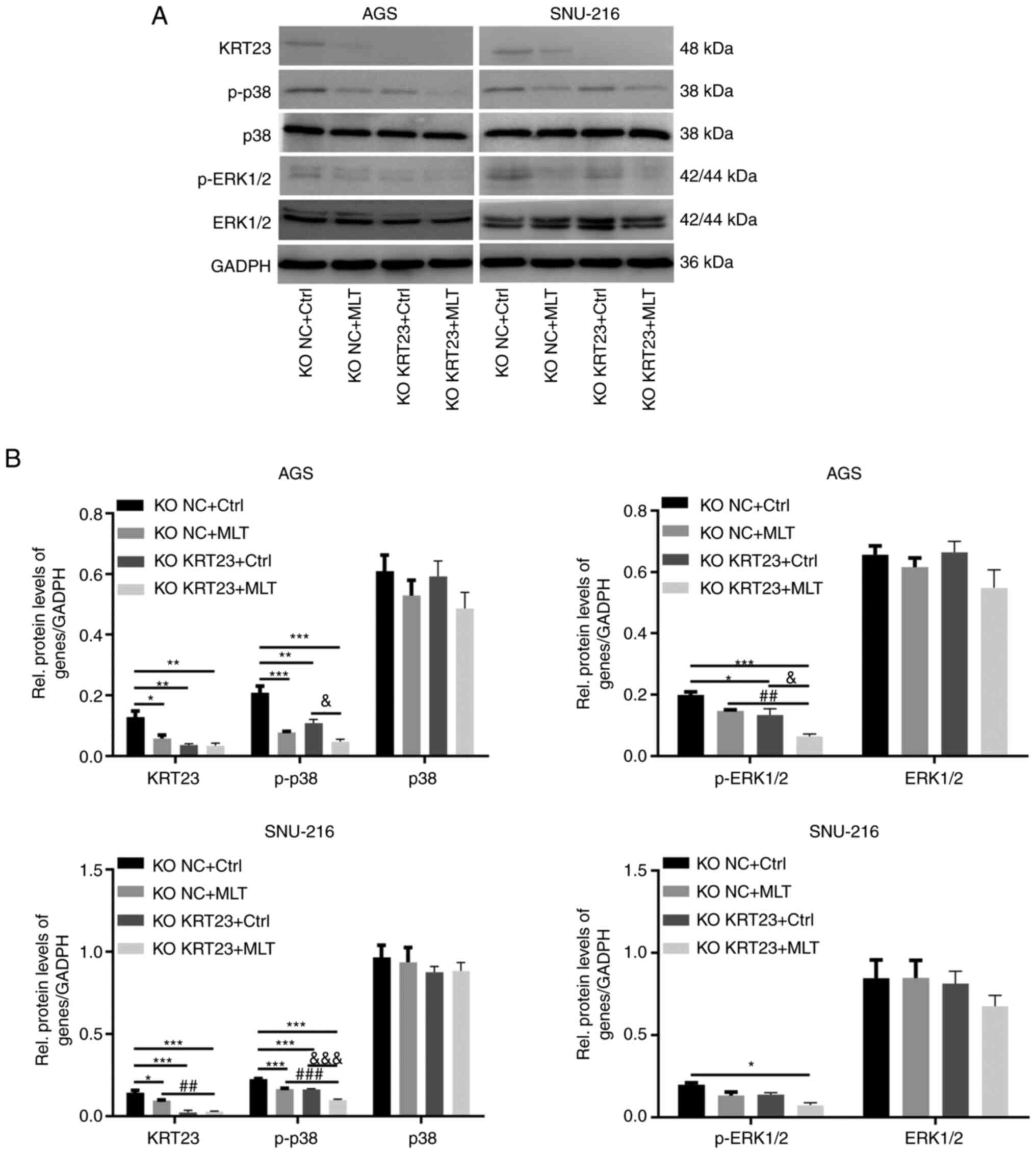 |
Figure 9.
KRT23 knockout enhanced the inhibitory effects of MLT on human GC cells via suppressing p38/ERK phosphorylation. (A) Western blot analysis of expression and activation of proteins in AGS and SNU-216 cells with or without 2.5 mM of MLT treatment for 24 h. (B) Statistical analysis of western blot results. *P<0.05, **P<0.01 and ***P<0.001 vs. the KO NC + Ctrl group; ##P<0.01 and ###P<0.001 vs. the KO NC + MLT group; &P<0.05 and &&&P<0.001 vs. the KO KRT23+Ctrl group. KRT, keratin; MLT, melatonin; GC, gastric cancer; KO, knockout; NC, negative control; p-, phosphorylated; Rel, Relative.
|
Discussion
MLT, the primary hormone produced by the pineal gland, is a versatile molecule with a variety of biological activities. The content of MLT in the GI tract is 400 times greater than the amount present in the pineal gland, 10 to 100 times higher than levels in serum and much higher than that which is observed in other tissues (23). This suggests a potential involvement of MLT in maintaining GI health and function (24). MLT shows direct beneficial effects on GI tissues through its antioxidant, immunomodulatory and mucosal microcirculation activities (25). It also exerts indirect effects through the brain-gut axis, including gastroprotection and gut microbiota-modulating (26,27). The protective and therapeutic potential of MLT has been confirmed in a number of GI diseases such as gastritis, gastric ulcer and irritable bowel syndrome (28–30). However, the nocturnal urinary excretion of MLT is depressed in patients with stomach and colon cancer (31). A significant reduction of circulating MLT is revealed in patients with GC (32). Decreased MLT synthesis/metabolism indexes are associated with reduced survival among patients with GC (33). A number of studies have explored the effects of MLT supplementation on GC, suggesting that MLT has multiple antitumor effects on GI tumors (34,35). It has also been shown to enhance the sensitivity of GC cells to chemotherapy (36,37) and reduce the side effects of chemotherapy (38). Its roles and underlying molecular mechanism in GC deserve further investigation.
The current study systematically dissects the global mechanism behind the anti-GC activity of MLT. Expression profile chips revealed that MLT influenced 3,647 downregulated and 2,741 upregulated DEGs in AGS cells. MLT was found to significantly downregulate DEGs that play a pro-cancer role in tumor occurrence and development, such as PSG11 (39), PSG2 (40), NOX1 (41) and CXCL11 (42). By contrast, microarray analyses also revealed upregulation of tumor suppressor genes as a result of MLT treatment, including ARRDC3 (43) and PTPN13 (44). The reverse regulation of these genes by MLT in cancer cells further demonstrated anti-tumor effects of MLT. In addition, the correlation between the anti-tumor effects of MLT and DEGs as well as the molecular mechanisms behind this have not been completely studied. This provides a large number of potential molecular targets and new ideas for subsequent research related to the anticancer effects of MLT.
Pathway analysis revealed that genes with reduced expression were significantly enriched in pyrimidine metabolism, RNA transport, spliceosome and cell cycle pathways. High expression genes were associated with ErbB, FoxO and lysosome signaling pathways. These pathways have been previously linked to cancer genesis and progression. FoxO signaling pathway has been shown to inhibit cell proliferation, invasion and death in prostate cancer (45). Pyrimidines reverse DNA damage induced by carbamoyl phosphate synthetase-1 silencing and rescue growth in non-small cell lung cancer (46). Tea polysaccharide induces apoptosis and suppresses HCT116 cell growth via lysosome-mediated cytotoxic autophagy (47). A study found that spliceosome-targeted therapies trigger apoptosis through extrinsic mechanisms in breast cancer cells (48). The findings of the present study also suggested that MLT might have anticancer properties via these mechanisms. This is consistent with our previous work on three types of GI cancers, which indicates that MLT exerted pro-apoptosis and anti-proliferation roles through FoxO, spliceosome, RNA transport and cell cycle signaling pathways (22).
A number of studies have discovered that a number of subtypes of keratin proteins are significantly expressed in a wide range of tumor tissues, associated with enhanced cancer stemness occurrence, development of tumors and cancer metastasis and aggressiveness (49–51). Notably, the present study found that multiple keratin subtypes including KRT7, KRT20, KRT23, KRT75 and KRT81 were downregulated by MLT in GC cells. As previously reported, KRT7 expression is related to a poor prognosis in colorectal carcinoma (52). KRT7 also increases proliferation and promotes migration of ovarian cancer (53). High KRT20 expression is significantly associated with aggressive urothelial carcinoma variants (54). KRT81 is abundantly expressed in melanoma tissues and is thought to have a role in melanoma proliferation, invasion and migration (55). Similarly, KRT23 is abundant in hepatocellular carcinoma and has been confirmed as an independent molecular marker and drug target for human liver tumors (8). KRT23 promotes colorectal cancer cell growth via activating telomerase reverse transcriptase expression (56). KRT23 knockdown significantly inhibits colon cancer cell proliferation and DNA damage repair pathways (7). Together, these observations indicate that KRT23 might play an important role in the development and progression of GI tumors. Moreover, a bioinformatic study revealed that KRT23 is also a specific prognostic factor for GC (57). The data of the present study further confirmed that the protein levels of KRT23 were substantially higher in GC cells than that in human normal gastric epithelial cell line GES-1. These results suggested that KRT23 had a close relationship with GC. Thus, KRT23 was considered as a molecular biomarker for GC and became the focus of current study.
CRISPR/Cas9-mediated KRT23 knockout model was constructed to explore the role of KRT23 in GC. KRT23 deletion inhibited proliferation and arrested cell cycle progression, decreasing the phosphorylation of ERK1/2, p38. A previous study revealed that stimulation of p38 MAPK signaling pathway promotes thyroid cancer proliferation (58). Increased phosphorylation of ERK promotes the growth of lung cancer cells (59). A number of treatments suppress cancer cell growth by inhibiting p38 or ERK signal pathways (60,61). Similar to the observations of the present study, KRT23 knockdown in colon cancer inhibits the occurrence and progression of colon cancer by inhibiting the p38/MAPK and ERK/MAPK signaling pathways (56).
A recent study reports that MLT-induced cytoskeleton reorganization inhibited the proliferation of melanoma cells (62). Consistently, the present study found that MLT inhibited the expression of the cytoskeletal filament KRT23. Moreover, KRT23 knockout plus MLT treatment significantly inhibited the proliferation of GC cells than MLT treatment alone. KRT23 deletion strengthened the anti-proliferation effect of MLT on GC cells. These data demonstrated that KRT23 deletion enhanced MLT's anti-GC action. It has been reported that MLT inhibits the migration and invasion of prostate cancer by inhibiting the p38, c-Jun, PLC signaling cascades (63). Similarly, MLT reduces lung cancer metastasis by suppressing p38/ERK, PLC and β-catenin signaling cascades (64). The nicotine-induced increase in motility and invasiveness of MCF7 cancer cells is abrogated by MLT through inhibition of ERK phosphorylation (65). Consistent with previous studies, the present study revealed that MLT treatment inhibited GC cell proliferation by suppressing p38/ERK phosphorylation. Moreover, a combination of MLT intervention with KRT23 deletion further strengthened the anti-proliferation effect of MLT on GC cells and decreased the phosphorylation of p38 and ERK1/2. Taken together, the present study indicated that MLT played a role as a potential KRT23 inhibitor and further loss of KRT23 enhanced MLT's anti-GC action via inhibiting p38/ERK phosphorylation.
However, there remained some limitations in the present study. The specific mechanism by which MLT affects the expression and function of KRT23 remains unclear. This will become the next focus of future research. In addition, the in vitro results need to be further verified in vivo.
In conclusion, KRT23 expression is significantly upregulated in GC cells. KRT23 is a novel GC oncogene contributing to GC progression. MLT acts as a potential KRT23 inhibitor and suppresses GC cell proliferation via reducing p38/ERK phosphorylation (Fig. 10). Targeting KRT23 and combining it with MLT may provide a new strategy for GC therapy.
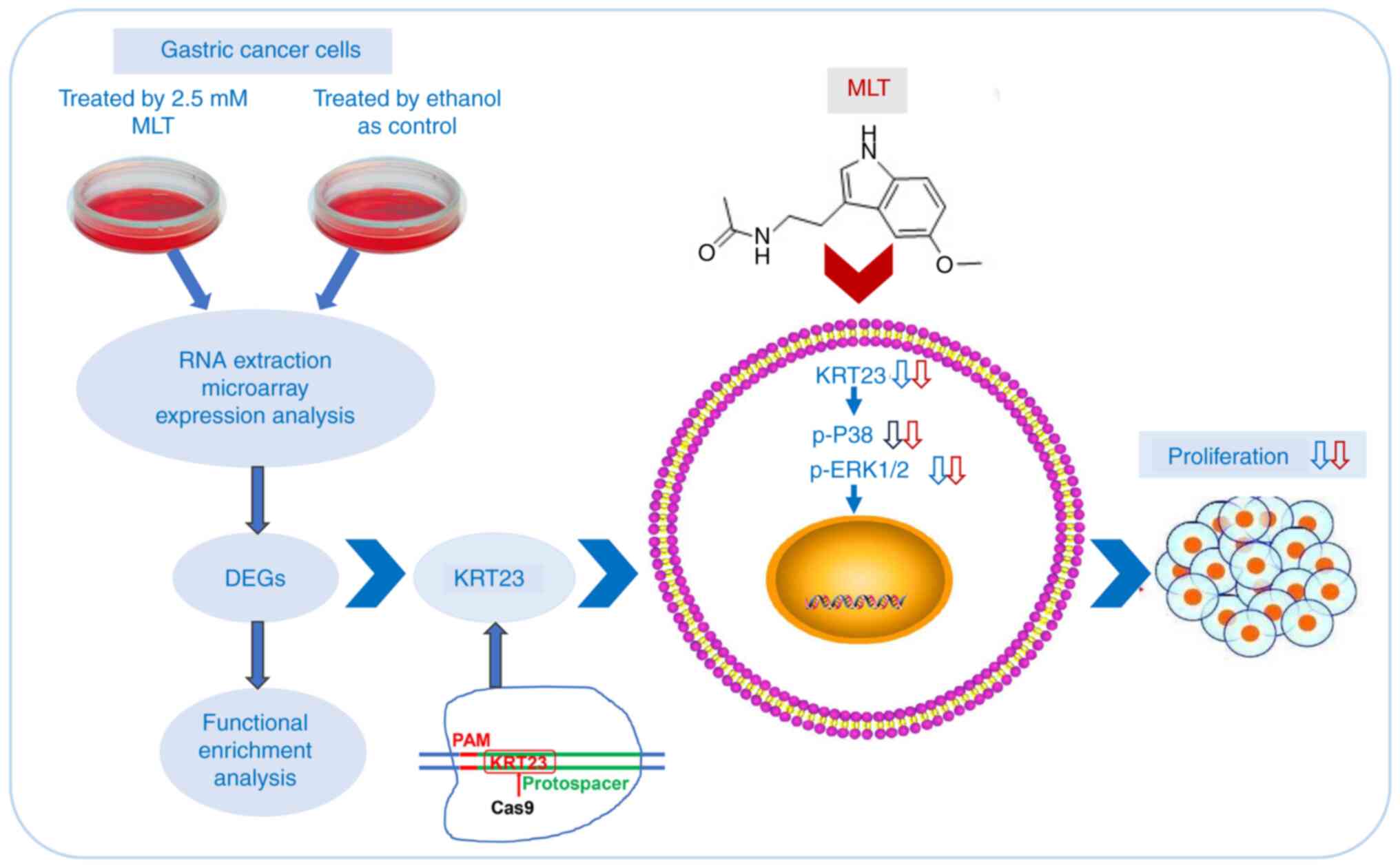 |
Figure 10.
Anti-GC mechanism of MLT was explored by expression profile chip, and KRT23 was revealed as a potential molecular target. KRT23 is a novel GC oncogene contributing to GC progression. KRT23 knockout contributes to the antitumor effects of MLT in GC via suppressing p38/ERK phosphorylation. GC, gastric cancer; MLT, melatonin; KRT, keratin; p-, phosphorylated; DEG, differently expressed genes.
|
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Pingsheng Liu (Institute of Biophysics, Chinese Academy of Sciences) for providing the CRISPR/Cas9 vector PX260 and Professor Changming Huang (Fujian Medical University Union Hospital) for kindly providing the GES-1 cells.
Funding
The present study was supported by grants from the Natural Science Foundation of Fujian Province (grant no. 2021J01679), the Special Fund of Fujian Provincial Department of Finance (grant no. 2020B011) and the Joint Funds for Innovation of Science and Technology of Fujian province (grant no. 2019Y9002).
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LL, RXZ and JS designed the present study and wrote and revised the manuscript. LL, MFL and JHL conducted the experiments and collected the data. HQS performed the high-throughput sequencing experiments and the bioinformatics analysis. ZGZ and DCL performed the literature search and analyzed the data. LSC, SSF and XPL performed the statistical analysis and produced the figures. LL, RXZ and JS confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang C, Yuan W, Lai C, Zhong S, Yang C, Wang R, Mao L and Chen Z and Chen Z: EphA2-to-YAP pathway drives gastric cancer growth and therapy resistance. Int J Cancer. 146:1937–1949. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Z, Ran H, Wang Z, Zhou S and Wang Y: Targeted and pH-facilitated theranostic of orthotopic gastric cancer via phase-transformation doxorubicin-encapsulated nanoparticles enhanced by low-intensity focused ultrasound (LIFU) with reduced side effect. Int J Nanomedicine. 14:7627–7642. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu Z, Yu S, Ye S, Shen Z, Gao L, Han Z, Zhang P, Luo F, Chen S and Kang M: Keratin 17 activates AKT signalling and induces epithelial-mesenchymal transition in oesophageal squamous cell carcinoma. J Proteomics. 211:1035572020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang PB, Chen Y, Ding GR, Du HW and Fan HY: Keratin 18 induces proliferation, migration, and invasion in gastric cancer via the MAPK signalling pathway. Clin Exp Pharmacol Physiol. 48:147–156. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen B, Xu X, Lin DD, Chen X, Xu YT, Liu X and Dong WG: KRT18 modulates alternative splicing of genes involved in proliferation and apoptosis processes in both gastric cancer cells and clinical samples. Front Genet. 12:6354292021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Birkenkamp-Demtroder K, Hahn SA, Mansilla F, Thorsen K, Maghnouj A, Christensen R, Øster B and Ørntoft TF: Keratin23 (KRT23) knockdown decreases proliferation and affects the DNA damage response of colon cancer cells. PLoS One. 8:e735932013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim D, Brocker CN, Takahashi S, Yagai T, Kim T, Xie G, Wang H, Qu A and Gonzalez FJ: Keratin 23 is a peroxisome proliferator-activated receptor alpha-dependent, MYC-amplified oncogene that promotes hepatocyte proliferation. Hepatology. 70:154–167. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren M, Gao Y, Chen Q, Zhao H, Zhao X and Yue W: The overexpression of keratin 23 promotes migration of ovarian cancer via epithelial-mesenchymal transition. Biomed Res Int. 2020:82187352020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Florido J, Martinez-Ruiz L, Rodriguez-Santana C, López-Rodríguez A, Hidalgo-Gutiérrez A, Cottet-Rousselle C, Lamarche F, Schlattner U, Guerra-Librero A, Aranda-Martínez P, et al: Melatonin drives apoptosis in head and neck cancer by increasing mitochondrial ROS generated via reverse electron transport. J Pineal Res. 73:e128242022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsiao SY, Tang CH, Chen PC, Lin TH and Chao CC: Melatonin inhibits EMT in bladder cancer by targeting autophagy. Molecules. 27:86492022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hasan M, Marzouk MA, Adhikari S, Wright TD, Miller BP, Matossian MD, Elliott S, Wright M, Alzoubi M, Collins-Burow BM, et al: Pharmacological, mechanistic, and pharmacokinetic assessment of novel melatonin-tamoxifen drug conjugates as breast cancer drugs. Mol Pharmacol. 96:272–296. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hao J, Fan W, Li Y, Tang R, Tian C, Yang Q, Zhu T, Diao C, Hu S, Chen M, et al: Melatonin synergizes BRAF-targeting agent vemurafenib in melanoma treatment by inhibiting iNOS/hTERT signaling and cancer-stem cell traits. J Exp Clin Cancer Res. 38:482019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Q, Sun Z, Du L, Xu C, Wang Y, Yang B, He N, Wang J, Ji K, Liu Y and Liu Q: Melatonin Sensitizes human colorectal cancer cells to gamma-ray ionizing radiation in vitro and in vivo. Int J Mol Sci. 19:39742018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
JG DEA, Serra LSM, Lauand L, Kuckelhaus SAS and Sampaio ALL: Protective effect of melatonin on cisplatin-induced ototoxicity in rats. Anticancer Res. 39:2453–2458. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Palmer ACS, Zortea M, Souza A, Santos V, Biazús JV, Torres ILS, Fregni F and Caumo W: Clinical impact of melatonin on breast cancer patients undergoing chemotherapy; effects on cognition, sleep and depressive symptoms: A randomized, double-blind, placebo-controlled trial. PLoS One. 15:e02313792020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song J, Ma SJ, Luo JH, Zhang H, Wang RX, Liu H, Li L, Zhang ZG and Zhou RX: Melatonin induces the apoptosis and inhibits the proliferation of human gastric cancer cells via blockade of the AKT/MDM2 pathway. Oncol Rep. 39:1975–1983. 2018.PubMed/NCBI
|
|
18
|
Wang RX, Liu H, Xu L, Zhang H and Zhou RX: Involvement of nuclear receptor RZR/RORgamma in melatonin-induced HIF-1alpha inactivation in SGC-7901 human gastric cancer cells. Oncol Rep. 34:2541–2546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ao L, Yan H, Zheng T, Wang H, Tong M, Guan Q, Li X, Cai H, Li M and Guo Z: Identification of reproducible drug-resistance-related dysregulated genes in small-scale cancer cell line experiments. Sci Rep. 5:118952015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benjamini Y and Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ao L, Li L, Sun H, Chen H, Li Y, Huang H, Wang X, Guo Z and Zhou R: Transcriptomic analysis on the effects of melatonin in gastrointestinal carcinomas. BMC Gastroenterol. 20:2332020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bubenik GA: Thirty four years since the discovery of gastrointestinal melatonin. J Physiol Pharmacol. 59 (Suppl 2):S33–S51. 2008.
|
|
24
|
Asghari MH, Moloudizargari M, Ghobadi E, Fallah M and Abdollahi M: Melatonin as a multifunctional anti-cancer molecule: Implications in gastric cancer. Life Sci. 185:38–45. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bubenik GA: Gastrointestinal melatonin: Localization, function, and clinical relevance. Dig Dis Sci. 47:2336–2348. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jaworek J, Brzozowski T and Konturek SJ: Melatonin as an organoprotector in the stomach and the pancreas. J Pineal Res. 38:73–83. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iesanu MI, Zahiu CDM, Dogaru IA, Chitimus DM, Pircalabioru GG, Voiculescu SE, Isac S, Galos F, Pavel B, O'Mahony SM and Zagrean AM: Melatonin-microbiome two-sided interaction in dysbiosis-associated conditions. Antioxidants (Basel). 11:22442022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo J, Song J, Zhang H, Zhang F, Liu H, Li L, Zhang Z, Chen L, Zhang M, Lin D, et al: Melatonin mediated Foxp3-downregulation decreases cytokines production via the TLR2 and TLR4 pathways in H. pylori infected mice. Int Immunopharmacol. 64:116–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choudhary P, Roy T, Chatterjee A, Mishra VK, Pant S and Swarnakar S: Melatonin rescues swim stress induced gastric ulceration by inhibiting matrix metalloproteinase-3 via down-regulation of inflammatory signaling cascade. Life Sci. 297:1204262022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen KH, Zeng BY, Zeng BS, Sun CK, Cheng YS, Su KP, Wu YC, Chen TY, Lin PY, Liang CS, et al: The efficacy of exogenous melatonin supplement in ameliorating irritable bowel syndrome severity: A meta-analysis of randomized controlled trials. J Formos Med Assoc. 122:276–285. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kvetnoi IM and Levin IM: Daily excretion of melatonin in patients with cancer of the stomach and large intestine. Vopr Onkol. 33:29–32. 1987.PubMed/NCBI
|
|
32
|
Colombo F, Iannotta F, Fachinetti A, Giuliani F, Cornaggia M, Finzi G, Mantero G, Fraschini F, Malesci A, Bersani M, et al: Changes in hormonal and biochemical parameters in gastric adenocarcinoma. Minerva Endocrinol. 16:127–139. 1991.(In Italian). PubMed/NCBI
|
|
33
|
Lv JW, Zheng ZQ, Wang ZX, Zhou GQ, Chen L, Mao YP, Lin AH, Reiter RJ, Ma J, Chen YP and Sun Y: Pan-cancer genomic analyses reveal prognostic and immunogenic features of the tumor melatonergic microenvironment across 14 solid cancer types. J Pineal Res. 66:e125572019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Song J, Li Y, Lin C, Chen Y, Zhang X and Yu H: Melatonin inhibited the progression of gastric cancer induced by Bisphenol S via regulating the estrogen receptor 1. Ecotoxicol Environ Saf. 259:1150542023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang K, Cai R, Fei S, Chen X, Feng S, Zhang L, Liu H, Zhang Z, Song J and Zhou R: Melatonin enhances anti-tumor immunity by targeting macrophages PD-L1 via exosomes derived from gastric cancer cells. Mol Cell Endocrinol. 568–569. 1119172023.
|
|
36
|
Shi X, Li H, Dan Z, Shu C, Zhu R, Yang Q, Wang Y and Zhu H: Melatonin potentiates sensitivity to 5-fluorouracil in gastric cancer cells by upregulating autophagy and downregulating myosin light-chain kinase. J Cancer. 14:2608–2618. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng L, Li S, He K, Kang Y, Li T, Li C, Zhang Y, Zhang W and Huang Y: Melatonin regulates cancer migration and stemness and enhances the anti-tumour effect of cisplatin. J Cell Mol Med. 27:2215–2227. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lissoni P, Barni S, Mandala M, Ardizzoia A, Paolorossi F, Vaghi M, Longarini R, Malugani F and Tancini G: Decreased toxicity and increased efficacy of cancer chemotherapy using the pineal hormone melatonin in metastatic solid tumour patients with poor clinical status. Eur J Cancer. 35:1688–1692. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao H, Jian D, Cheng X, Tang H and Ma J: Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol. 21:3012021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Marrero-Rodríguez D, Taniguchi-Ponciano K, Subramaniam M, Hawse JR, Pitel KS, Arreola-De la Cruz H, Huerta-Padilla V, Ponce-Navarrete G, Figueroa-Corona MDP, Gomez-Virgilio L, et al: Krüppel-Like Factor 10 participates in cervical cancer immunoediting through transcriptional regulation of Pregnancy-Specific Beta-1 Glycoproteins. Sci Rep. 8:94452018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gu L, Feng C, Li M, Hong Z, Di W and Qiu L: Exosomal NOX1 promotes tumor-associated macrophage M2 polarization-mediated cancer progression by stimulating ROS production in cervical cancer: A preliminary study. Eur J Med Res. 28:3232023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Han S, Wu B, Zhong C, Shi Y, Lv C, Fu L, Zhang Y, Lang Q, Liang Z, et al: CXCL11 correlates with immune infiltration and impacts patient immunotherapy efficacy: A pan-cancer analysis. Front Immunol. 13:9512472022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wedegaertner H, Pan WA, Gonzalez CC, Gonzalez DJ and Trejo J: The alpha-arrestin ARRDC3 is an emerging multifunctional adaptor protein in cancer. Antioxid Redox Signal. 36:1066–1079. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li JP, Zhang XM, Liu BC, Ren SG, Zhao XH and Liu YJ: Comprehensive analysis of the PTPN13 expression and its clinical implication in breast cancer. Neoplasma. 70:188–198. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shan Z, Li Y, Yu S, Wu J, Zhang C, Ma Y, Zhuang G, Wang J, Gao Z and Liu D: CTCF regulates the FoxO signaling pathway to affect the progression of prostate cancer. J Cell Mol Med. 23:3130–3139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, Bezwada D, Rodriguez-Canales J, Villalobos P, Lin YF, et al: CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature. 546:168–172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou Y, Zhou X, Huang X, Hong T, Zhang K, Qi W, Guo M and Nie S: Lysosome-mediated cytotoxic autophagy contributes to tea polysaccharide-induced colon cancer cell death via mTOR-TFEB signaling. J Agric Food Chem. 69:686–697. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bowling EA, Wang JH, Gong F, Wu W, Neill NJ, Kim IS, Tyagi S, Orellana M, Kurley SJ, Dominguez-Vidaña R, et al: Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 184:384–403.e21. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tsai FJ, Lai MT, Cheng J, Chao SC, Korla PK, Chen HJ, Lin CM, Tsai MH, Hua CH, Jan CI, et al: Novel K6-K14 keratin fusion enhances cancer stemness and aggressiveness in oral squamous cell carcinoma. Oncogene. 38:5113–5126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Perone Y, Farrugia AJ, Rodriguez-Meira A, Győrffy B, Ion C, Uggetti A, Chronopoulos A, Marrazzo P, Faronato M, Shousha S, et al: SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ERalpha breast cancer. Nat Commun. 10:21152019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao S, Huang HY, Han X, Ye Y, Qin Z, Zhao G, Li F, Hu G, Hu L and Ji H: Keratin 14-high subpopulation mediates lung cancer metastasis potentially through Gkn1 upregulation. Oncogene. 38:6354–6369. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hrudka J, Fiserova H, Jelinkova K, Matej R and Waldauf P: Cytokeratin 7 expression as a predictor of an unfavorable prognosis in colorectal carcinoma. Sci Rep. 11:178632021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
An Q, Liu T, Wang MY, Yang YJ, Zhang ZD, Liu ZJ and Yang B: KRT7 promotes epithelialmesenchymal transition in ovarian cancer via the TGFβ/Smad2/3 signaling pathway. Oncol Rep. 45:481–492. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Eckstein M, Wirtz RM, Gross-Weege M, Breyer J, Otto W, Stoehr R, Sikic D, Keck B, Eidt S, Burger M, et al: mRNA-expression of KRT5 and KRT20 defines distinct prognostic subgroups of muscle-invasive urothelial bladder cancer correlating with histological variants. Int J Mol Sci. 19:33962018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang K, Liang Y, Zhang W, Zeng N, Tang S and Tian R: KRT81 knockdown inhibits malignant progression of melanoma through regulating interleukin-8. DNA Cell Biol. 40:1290–1297. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang N, Zhang R, Zou K, Yu W, Guo W, Gao Y, Li J, Li M, Tai Y, Huang W, et al: Keratin 23 promotes telomerase reverse transcriptase expression and human colorectal cancer growth. Cell Death Dis. 8:e29612017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Min L, Zhao Y, Zhu S, Qiu X, Cheng R, Xing J, Shao L, Guo S and Zhang S: Integrated analysis identifies molecular signatures and specific prognostic factors for different gastric cancer subtypes. Transl Oncol. 10:99–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang QG, Xiong CF and Lv YX: Kin17 facilitates thyroid cancer cell proliferation, migration, and invasion by activating p38 MAPK signaling pathway. Mol Cell Biochem. 476:727–739. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhong C, Chen C, Yao F and Fang W: ZNF251 promotes the progression of lung cancer by activating ERK signaling. Cancer Sci. 111:3236–3244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xiong L, Guo W, Yang Y, Gao D, Wang J, Qu Y and Zhang Y: Tectoridin inhibits the progression of colon cancer through downregulating PKC/p38 MAPK pathway. Mol Cell Biochem. 476:2729–2738. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Niu M, Zhang B, Li L, Su Z, Pu W, Zhao C, Wei L, Lian P, Lu R, Wang R, et al: Targeting HSP90 inhibits proliferation and induces apoptosis through AKT1/ERK pathway in lung cancer. Front Pharmacol. 12:7241922021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Alvarez-Artime A, Cernuda-Cernuda R, Francisco Artime N, Cepas V, Gonzalez-Menendez P, Fernadez-Vega S, Quiros-Gonzalez I, Sainz RM and Mayo JC: Melatonin-induced cytoskeleton reorganization leads to inhibition of melanoma cancer cell proliferation. Int J Mol Sci. 21:5482020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang SW, Tai HC, Tang CH, Lin LW, Lin TH, Chang AC, Chen PC, Chen YH, Wang PC, Lai YW and Chen SS: Melatonin impedes prostate cancer metastasis by suppressing MMP-13 expression. J Cell Physiol. 236:3979–3990. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chao CC, Chen PC, Chiou PC, Hsu CJ, Liu PI, Yang YC, Reiter RJ, Yang SF and Tang CH: Melatonin suppresses lung cancer metastasis by inhibition of epithelial-mesenchymal transition through targeting to Twist. Clin Sci (Lond). 133:709–722. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Proietti S, Catizone A, Masiello MG, Dinicola S, Fabrizi G, Minini M, Ricci G, Verna R, Reiter RJ, Cucina A and Bizzarri M: Increase in motility and invasiveness of MCF7 cancer cells induced by nicotine is abolished by melatonin through inhibition of ERK phosphorylation. J Pineal Res. 64:e124672018. View Article : Google Scholar : PubMed/NCBI
|
























