Introduction
Recurrent miscarriage is used to refer to more than
three pregnancy failures before 20 weeks of gestation. Recurrent
miscarriage affects 1–2% of couples globally and ~50% of cases have
no identifiable cause worldwide (1). The incidence of recurrent miscarriage
is ~1/300 pregnancies (2).
Recurrent miscarriage is a complex pathological condition that can
be caused by gestational infection, defective hormone homeostasis,
genetic defects, poor endometrial differentiation or other
unexplained factors (3). Placenta
formation is an essential step for a successful pregnancy; it
involves a well-orchestrated interplay between the blastocyst and
the receptive endometrium (4,5).
Thus, defective placentation can lead to complications, such as
preeclampsia or recurrent implantation failure (6,7).
Uterine endometrial differentiation and trophoblast
invasion are crucial for early placenta formation. During
implantation, the trophectoderm of the blastocyst invades the
maternal uterine wall followed by the generation of
cytotrophoblasts (8). These
trophoblast cells undergo epithelial-mesenchymal transition (EMT)
and invade the maternal uterine stroma layer (9). The migration and invasion of
trophoblasts mediate the formation of the placenta and the
remodeling of the maternal spiral arterial walls. Thus, defective
trophoblast proliferation and invasion cause several complications,
including preeclampsia and recurrent miscarriage (9).
MicroRNAs (miRNAs/miRs) are non-coding RNAs
comprised of 20–24 nucleotides that post-transcriptionally mediate
gene translation (10). Through
binding to the 3′-untranslated region, miRNAs mediate mRNA
translation or stability. Hairpin pre-miRNAs are exported from the
nucleus to the cytoplasm where they are processed by the
ribonuclease Dicer (11). Mature
miRNAs then bind to the RNA-induced silencing complex to mediate
mRNA stability (12).
Dysregulation of miRNAs has been widely studied in several
diseases, and in neuronal development and cancer progression. For
example, miR-450a is a tumor suppressor that inhibits ovarian
cancer cell growth, EMT and metastasis (13). In addition, miR-195 facilitates
de novo lipogenesis to inhibit breast cancer cell migration
and invasion (14). Moreover,
miRNAs mediate female reproduction (15,16).
Overexpression of miR-30-5p has been reported to induce ferroptosis
of trophoblasts, thus causing preeclampsia (17). Furthermore, miR-141 and miR-200a
inhibit the expression of endocrine gland-derived vascular
endothelial growth factor, thus preventing preeclampsia (18). Furthermore, aberrant expression of
miRNA signatures, including miR-20b-5p, miR-155-5p and miR-718, is
associated with recurrent miscarriage (19). Despite miRNAs being involved in
early pregnancy, the underlying molecular mechanism remains
unclear.
The centrosome is composed of a pair of centrioles,
mother and daughter centrioles, and the outer protein matrix
pericentriolar materials. The centrosome, the primary
microtubule-organizing center of cells, maintains cell shape, and
directional migration or segregation of duplicated chromosomes into
daughter cells equally during the M phase (20,21).
In addition to regulating microtubule dynamics, the centrosome
functions as a center to coordinate cell cycle progression
(22). Cyclin E localizes to the
centrosome to promote G1/S transition (23). Disruption of centrosome integrity
by depleting centrosomal proteins arrests the cells in the G0/G1
phase (24). Thus, precise control
of centrosome function and homeostasis maintains cell cycle
progression.
During the quiescent stage or under stress, the
primary cilia grow from the mother centriole of the centrosome. The
primary cilia are composed of the basal body (mother centriole),
microtubule-based axoneme and the outer ciliary membranes (24,25).
Unlike motile cilia, primary cilia are immotile antenna-like
structures that conduct extracellular signaling during development
(26). Several signaling
receptors, such as receptor tyrosine kinase, G protein-coupled
receptors or hedgehog signaling, reside on the ciliary membrane to
coordinate environmental cues (27,28).
Dysfunction of primary cilia causes many disorders during
development and differentiation, which are collectively termed
ciliopathies (26). Primary cilia
also play important roles in successful pregnancy. Primary cilia
are observed in the placenta during the first trimester of
pregnancy. These primary cilia activate extracellular
signal-regulated kinase (ERK) signaling to promote placenta
formation (29,30). Thus, defective primary cilia
formation leads to preeclampsia and other complications such as
gestational diabetes mellitus (30).
Previous studies have shown that miR-155-5p is
dysregulated in recurrent miscarriage; however, the underlying
molecular mechanism remains unclear (19,31).
The present study, we will investigate whether miR-155-5p inhibits
trophoblast cell growth and invasion. MiR-155-5p mimic or inhibitor
will be transfected into trophoblast cells followed by examining
cell migration and invasion. In addition, to uncover the underlying
molecular mechanism, the centrosome homeostasis and primary
ciliogenesis were also examined. Thus, the data will uncover the
role of miR-155-5p in mediating centrosomal functions and
trophoblast cell growth and invasion.
Materials and methods
Cell culture and drug treatment
The human immortalized endometrial stroma
telomerase-immortalized human endometrial stromal cells (T-HESCs)
and HTR-8/SVneo (HTR8) trophoblast cells were maintained in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 1% glutamine, 1% sodium pyruvate and 1%
penicillin/streptomycin under a humidified atmosphere containing 5%
CO2 at 37°C. The HTR8 cell line was established using
extravillous cytotrophoblasts freshly isolated from a
first-trimester placenta and transfected with a plasmid containing
the simian virus 40 large T antigen (32). This cell line contains two
populations, one of epithelial and one of mesenchymal origin
(33). For triggering
decidualization in vitro, T-HESCs were treated with 0.3 mM
8-Bromo-cAMP (cat. no. BML-CN115; Enzo Life Sciences) and 10 µM
medroxyprogesterone 17-acetate (MPA) at 37°C for 72 h. To block
centrosome amplification, cells were treated with 250 nM Centrinone
(LCR-263, cat. no. HY-18682; MedChemExpress) at 37°C for 72 h. To
activate p38, cells were treated with 1 µM sodium salicylate (cat.
no. S3007; MilliporeSigma) at 37°C for 72 h.
miRNA and small interfering RNA
(siRNA) transfection
A commercially available siRNA against
autophagy-related 16-like 1 (ATG16L1; SMARTpool; GE Healthcare
Dharmacon, Inc.), and miRNA mimics or inhibitors (miRIDIAN
microRNA; GE Healthcare Dharmacon, Inc.) were purchased from
Horizon Discovery. The siRNA and miRNA sequences were as follows:
SMARTpool-siATG16L1 (5′-UGUGGAUGAUUAUCGAUUA-3′;
5′-GGCACACACUCACGGGACA-3′; 5′-GCAUUGGAUUAACGGAAAC-3′;
5′-GUUAUUGAUCUCCGAACAA-3′); miR-20b-5p mimic
(5′-CAAAGUGCUCAUAGUGCAGGUAG-3′); miR-155-5p mimic
(5′-UUAAUGCUAAUCGUGAUAGGGGU-3′); miR-718 inhibitor
(5′-CUUCCGCCCCGCCGGGCGUCG-3′); and miR-155-5p inhibitor
(5′-UUAAUGCUAAUCGUGAUAGGGGU-3′). The negative controls of miRNA
mimics and inhibitors were commercially available. The miRNA mimic
negative control (5′-UCACAACCUCCUAGAAAGAGUAGA-3′; cat. no.
MIMAT0000039) was purchased from Horizon Discovery (GE Healthcare
Dharmacon, Inc.). miRNA inhibitor negative control
(5′-UUGUACUACACAAAAGUACUG-3′; cat. no. MIMAT0000295) was purchased
from Horizon Discovery (GE Healthcare Dharmacon, Inc.). The
scrambled siRNA with the target sequence 5′-GAUCAUACGUGCGAUCAGA-3′
was purchased from Sigma (MilliporeSigma).
Cells were transfected with 50 nM miRNA or siRNA
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C for 72 h. Samples were collected and analyzed
immediately. To investigate the role of p38 in modulating primary
cilia, SB203580 (working concentration, 10 µM; Abcam) was used to
inhibit the function of p38 at 37°C for 72 h.
Western blotting
Cells were trypsinized and lysed with CelLytic™ MT
cell lysis reagent (MilliporeSigma) containing protease inhibitor
for 10 min on ice. Lysates were centrifuged at 13,000 × g for 10
min at 4°C and the supernatant was collected. The protein
concentration was determined using the Bio-Rad protein quantity
assay (Bio-Rad Laboratories, Inc.) by BioSpectrometer® (Eppendorf
AG, Germany). The samples (50 µg/lane) were mixed with 2X sample
buffer and heated to 100°C for 10 min. Equal amounts of proteins
were separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis at 150 V for 90 min. Following gel separation, the
samples were transferred to PVDF membranes at 20 V overnight. Next,
the membrane was blocked with 3% BSA (MilliporeSigma) dissolved in
TBST (0.005% Tween) at room temperature for 1 h and incubated with
the following antibodies: Anti-Akt (cat. no. #4691; Cell Signaling
Technology, Inc.), anti-phospho-Akt (Thr308; cat. no. #9275; Cell
Signaling Technology, Inc.), anti-ATG16L1 (cat. no. #8089S; Cell
Signaling Technology, Inc.), anti-cleaved-caspase 3 (cat. no.
#9664; Cell Signaling Technology, Inc.), anti-cleaved-poly
(ADP-ribose) polymerase (PARP; cat. no. #9542; Cell Signaling
Technology, Inc.), anti-cyclin A (cat. no. ab38; Abcam),
anti-cyclin D (cat. no. 2G3G5; Thermo Fisher Scientific, Inc.),
anti-cyclin E (cat. no. #4132; Cell Signaling Technology, Inc.),
anti-1A/1B light chain 3B (LC3A/B; cat. no. #12741; Cell Signaling
Technology, Inc.), anti-matrix metalloproteinase 2 (MMP2; cat. no.
#40994; Cell Signaling Technology, Inc.), anti-p38 (cat. no. #9212;
Cell Signaling Technology, Inc.), anti-phospho-p38 (cat. no.
09-272; Merck KgaA), anti-p44/42 mitogen-activated protein kinase
(ERK1/2; cat. no. #4695; Cell Signaling Technology, Inc.),
anti-phospho-ERK1/2 (cat. no. #4370; Cell Signaling Technology,
Inc.) and anti-actin (cat. no. ab8227; Abcam) in 1:10,000 dilutions
at 4°C overnight. After washing with TBST for 30 min, the membrane
was incubated with an HRP-conjugated secondary antibody (ab205718,
Abcam) in 1:7,000 at room temperature for 1 h. Subsequently, the
signals were detected by the ECL kit (MilliporeSigma) with actin as
the loading control and analyzed by ImageJ (version 1.53k, National
Institutes of Health, USA).
Immunofluorescence microscopy
Cells were fixed and permeabilized in ice-cold pure
methanol (MilliporeSigma) for 5 min at −20 °C. After
permeabilization, cells were incubated in blocking buffer
[containing 0.2% Triton X-100, 0.05% Tween-20 and normal goat serum
(MilliporeSigma)] for 1 h at room temperature and incubated with
the following antibodies: Anti-acetylated tubulin (cat. no. T6793;
MilliporeSigma), anti-ADP-ribosylation factor-like 13B (ARL13B;
cat. no. 17711-1-AP; Proteintech Group, Inc.), anti-centrosomal
protein 164 (CEP164; cat. no. NBP1-81445; Novus Biologicals, LLC),
anti-intraflagellar transport 88 (IFT88; cat. no.13967-1-AP;
Proteintech Group, Inc.) and anti-γ-tubulin (cat. no. T6557;
MilliporeSigma) in 1:700 dilutions at 4°C for overnight. After
which, the cells were washed for 30 min with PBS three times and
were then incubated with FITC- or Cy3-conjugated goat anti-mouse or
anti-rabbit IgG secondary antibodies (cat. nos. ab175473 and
ab150077; Abcam) in 1:700 dilutions in the presence of
6-diamino-2-phenylindole at room temperature for 1 h at room
temperature. The cells were then further washed for 30 min with PBS
three times. The coverslips were overlaid on 50% glycerol in PBS
and the signals were observed through an Axio imager D2
fluorescence microscope and analyzed by ZEN 2 blue edition software
(Zeiss AG).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted using TRI reagent
(MilliporeSigma) and Direct-zol™ RNA miniprep (Zymo Research
Corp.). RNA (1 µg) was then reverse transcribed into cDNA using an
RT kit (sensiFAST; Bioline) according to manufacturer's protocol.
Fast SYBR Green Master Mix (MilliporeSigma) was used to perform
qPCR with a final concentration of 0.25 nM gene-specific primers.
The sequences of the primers were as follows: E-cadherin forward:
5′-CCGAGAGCTACACGTTC-3′, reverse: 5′-TCTTCAAAATTCACTCTGCC-3′;
fibronectin-1 forward: 5′-CCATAGCTGAGAAGTGTTTTG-3′, reverse
5′-CAAGTACAATCTACCATCATCC-3′; MMP2 forward:
5′-GTGATCTTGACCAGAATACC-3′, reverse: 5′-GCCAATGATCCTGTATGTG-3′;
thrombospondin 1 (THBS1) forward: 5′-GTGACTGAAGAGAACAAAGAC-3′,
reverse 5′-CAGCTATCAACAGTCCATTC-3′; and zinc-finger E-box-binding
homeobox 1 (ZEB-1) forward: 5′-AAAGATGATGAATGCGAGTC-3′, reverse:
5′-TCCATTTTCATCATGACCAC-3′. PCR signals were compared among groups
after normalization, with GAPDH forward: 5′-TCGGAGTCAACGGATTTG-3′,
reverse: 5′-CAACAATATCCACTTTACCAGAG-3′) used as the internal
reference, calculated using the 2−ΔΔCq method (18,34,35).
The thermocycling conditions were as follows: 95 °C for 10 min.
Next, they were denatured at 95°C for 15 sec and annealing the
primer with DNA template at 60°C for 1 min. These cycles were
repeated for 40 times.
Wound healing assay
HTR8 cells were plated on 6-well plates and
transfected with miR-155-5p mimic and/or inhibitor prior to the
migration assay. A wound was created by scratching the cells with a
sterile 200-µl pipette tip, and fresh medium (serum-free) was added
to the cells after washing with PBS. The microscopy images of the
cells migrating into the wound area were captured at ×10
magnification under an optical microscope at 0 and 12 h after
scraping, in order to record the wound closure area. Wound closure
was calculated using the following formula: Wound closure
(%)=100-(initial area of wound-nth h area of the wound)/(initial
area of the wound).
Cell invasion assay
The cell invasion assay was performed using Falcon
cell culture inserts (pore size, 8 µm) according to the
manufacturer's protocol. Matrigel (1 mg/ml at 37°C; Corning, Inc.)
was loaded onto the upper chambers of the inserts for 30 min,
whereas the lower chamber contained a serum-loaded medium as an
attractant. Cells were seeded and overlaid on the Matrigel at a
density of 3×104 cells/well in serum-free medium. Upon
termination of the incubation at 37°C for 24 h, invasive cells were
fixed in 100% methanol for 4 min and stained with Giemsa's azure
eosin methylene blue solution (Merck KGaA) at room temperature for
2 h at room temperature and cell numbers were counted in six
high-power fields per insert under an optical microscope. The
experiments were performed in triplicate and repeated three
times.
Cell growth assay
HTR8 cells were plated at a density of
1×105 cells on a 6-well plate. After incubating cells
for 24, 48 and 72 h, cells were trypsinized and resuspended with 1
ml PBS for cell counting with hemocytometer. All treatments were
carried out in triplicate, with each experiment performed three
times.
EdU assay
HTR8 cells were seeded on 25×25-mm coverslips in
6-well tissue culture plates and transfected with the miR-155-5p
mimic and/or inhibitor. The cell proliferation ability was measured
using Click-iT® EdU Imaging Kits (Thermo Fisher Scientific, Inc.).
Briefly, 1 µl 10 µM EdU working solution (Component A) was added
and incubated at 37°C for 1 h. After which, the cells were
permeabilized in ice-cold methanol for 5 min. Subsequently, 1X
Click-iT® EdU buffer was added, as per the manufacturer's
instructions, and incubated at 37°C for 1 h. The cells were then
washed for 30 min with PBS three times and the signals were
observed under an Axio imager D2 fluorescence microscopy (Zeiss
AG).
ELISA
Supernatants from different experimental groups were
analyzed using insulin-like growth factor binding protein-1
(IGFBP1; cat. no. ab233618, Abcam) and prolactin (cat. no.
ab226901; Abcam) ELISA kits according to the manufacturer's
instructions. Briefly, supernatants were added to each well
followed by addition of 50 µl antibody cocktails and shaking at 30
× g at 37°C for 1 h. Then, each well was washed with 350 µl PBS
three times. After washing, 50 µl TMB development solution was
added to each well, followed by the termination of the reaction via
the addition of a stop solution. The solution was measured at 450
nm.
Bioinformatics
The predicted target gene of miR-20b-5p was
conducted by 2.0 of the miRNA Pathway Dictionary Database
(miRPathDB), which was freely accessible at https://mpd.bioinf.uni-sb.de/. MiRPathDB 2.0 performed
analysis of the underlying metabolic pathway and potential miRNA
targets, and with customized application, generated the relation
chart of predicted centrosome-associated target genes of
miR-20b-5p.
Statistical analysis
Values from the experimental assays are expressed as
the mean ± SEM of ≥3 independent experimental repeats. Differences
between two groups or >2 groups were compared using unpaired
Student's t-test or one-way ANOVA with Dunnett's post hoc test,
respectively. Data were analyzed using GraphPad Prism version no.9
(Dotmatics). P<0.05 was considered to indicate a statistically
significant difference. All experiments are repeated at least for
three times.
Results
miR-20b-5p, miR-155-5p or miR-718
inhibitor do not affect decidualization
Aberrant expression levels of miR-20b-5p, miR-155-5p
and miR-718 have been observed in women who suffer from recurrent
miscarriage (19). To investigate
the underlying molecular mechanisms, uterine endometrial
differentiation, and trophoblast growth and invasion, which are
essential events for successful pregnancy, were examined. It was
first checked whether aberrant expression levels of these miRNAs
affected endometrial stromal cell growth. Overexpression of
miR-20b-5p and miR-155-5p were used to mimic the upregulation of
these two miRNAs, and transfection with a miR-718 inhibitor was
used to induce downregulation of miR-718 in T-HESCs. T-HESCs were
transfected with miR-20b-5p, miR-155-5p and miR-718 inhibitor
(Fig. 1A-C), and cell numbers were
counted. Transfection with all three miRNAs had no effects on
T-HESC growth (Fig. 1D),
suggesting that miR-20b-5p, miR-155-5p and miR-718 inhibitor did
not affect endometrial stromal cell growth. After which, the
ability of uterine endometrial stromal differentiation, known as
decidualization, was examined. Decidualization was induced by
treating T-HESCs with cAMP and MPA for 3 days, followed by
examining the decidual markers IGFBP1 and prolactin. Prolactin and
IGFBP1 levels were significantly increased upon cAMP and MPA
co-treatment (Fig. 1E and F).
After which, the effects of miR-20b-5p, miR-155-5p and miR-718
inhibitor on decidualization were examined. Transfection of cells
with miR-20b-5p, miR-155-5p or miR-718 inhibitor did not affect the
levels of prolactin and IGFBP1 (Fig.
1G and H), suggesting that these miRNAs did not disturb
endometrial decidualization.
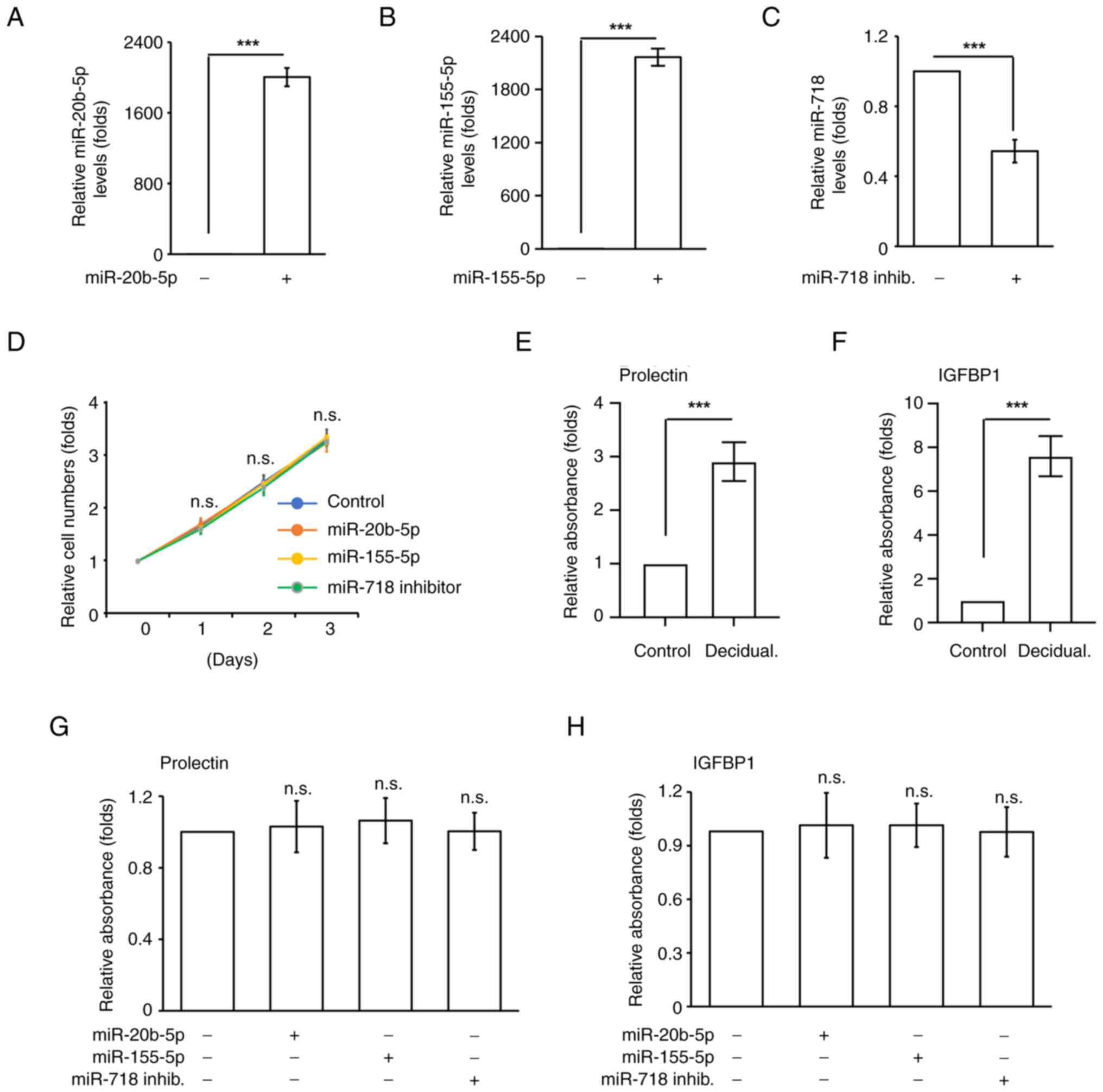 | Figure 1.miR-155-5p, miR-20b-5p and miR-718
inhibitor do not affect uterine endometrial stroma cell growth and
function. Expression levels of (A) miR-155-5p, (B) miR-20b-5p and
(C) miR-718. Quantitative results of cells transfected with
miR-155-5p, miR-20b-5p, or miR-718 inhib. (D) Overexpression of
miR-155-5p, miR-20b-5p or miR-718 inhibitor did not affect uterine
endometrial stroma cell growth. Quantitative results of relative
cell numbers of human uterine endometrial stroma cells transfected
with miR-155-5p, miR-20b-5p or miR-718 inhib. for 1, 2 and 3 days.
(E-H) Overexpression of miR-155-5p, miR-20b-5p or miR-718 inhib. do
not affect uterine stroma decidualization. (E and F) Treatment of
cAMP and MPA induced uterine stroma decidualization. Decidual
markers (E) prolactin and (F) IGFBP1 were upregulated when treating
cells with cAMP + MPA. (G and H) Overexpression of miR-155-5p,
miR-20b-5p or miR-718 inhib. does not affect (G) prolactin and (H)
IGFBP1 levels during decidualization. Data are presented as mean ±
SD from three independent experiments. (***P<0.001). All ‘-’
symbols in the figure represent the negative control transfection.
n.s., no significance; inhib., inhibitor; cAMP, cyclic adenosine
monophosphate; MPA, medroxyprogesterone 17-acetate; IGFBP1,
insulin-like growth factor binding protein-1. |
Overexpression of miR-155-5p inhibits
trophoblast cell growth
Trophoblast proliferation contributes to
establishing the maternal-fetal interface. It was assessed whether
miR-20b-5p, miR-155-5p and miR-718 inhibitor affected trophoblast
cell growth. The immortalized human trophoblast HTR8 cell line was
used in the present study. Overexpression of miR-20b-5p and
inhibition of miR-718 did not affect HTR8 cell growth; however,
when cells overexpressed miR-155-5p, the number of cells was
significantly reduced and apoptotic bodies were observed,
suggesting that miR-155-5p inhibited trophoblast cell growth
(Fig. 2A and B). The levels of
cyclins affect cell cycle progression; therefore, the expression
levels of different cyclins were assessed. Overexpression of
miR-155-5p did not affect the expression levels of cyclin A and
cyclin D; however, the levels of cyclin E were reduced, and this
phenotype was rescued by co-transfecting cells with the miR-155-5p
inhibitor (Fig. 2D). The
expression levels of miR-155-5p were also confirmed by performing
RT-qPCR (Fig. 2C). These findings
indicated that miR-155-5p reduced the expression of cyclin E.
Cyclin E regulates S phase entry (23); therefore, whether miR-155-5p
decreases the ability of HTR8 cells to enter the S phase was
assessed by EdU incorporation assay. Overexpression of miR-155-5p
reduced the proportion of EdU+ cells, and this phenotype
was rescued by co-transfecting cells with the miR-155-5p inhibitor
(Fig. 2E and F). Thus, miR-155-5p
may reduce the S phase entry of HTR8 cells. The ability of cells to
enter the M phase was next examined by assessing the mitotic index.
Overexpression of miR-155-5p reduced the mitotic index of HTR8
cells, and this was rescued by co-transfecting cells with
miR-155-5p inhibitor (Fig. 2G).
Taken together, miR-155-5p inhibited trophoblast HTR8 cell growth
by reducing S phase and M phase entry.
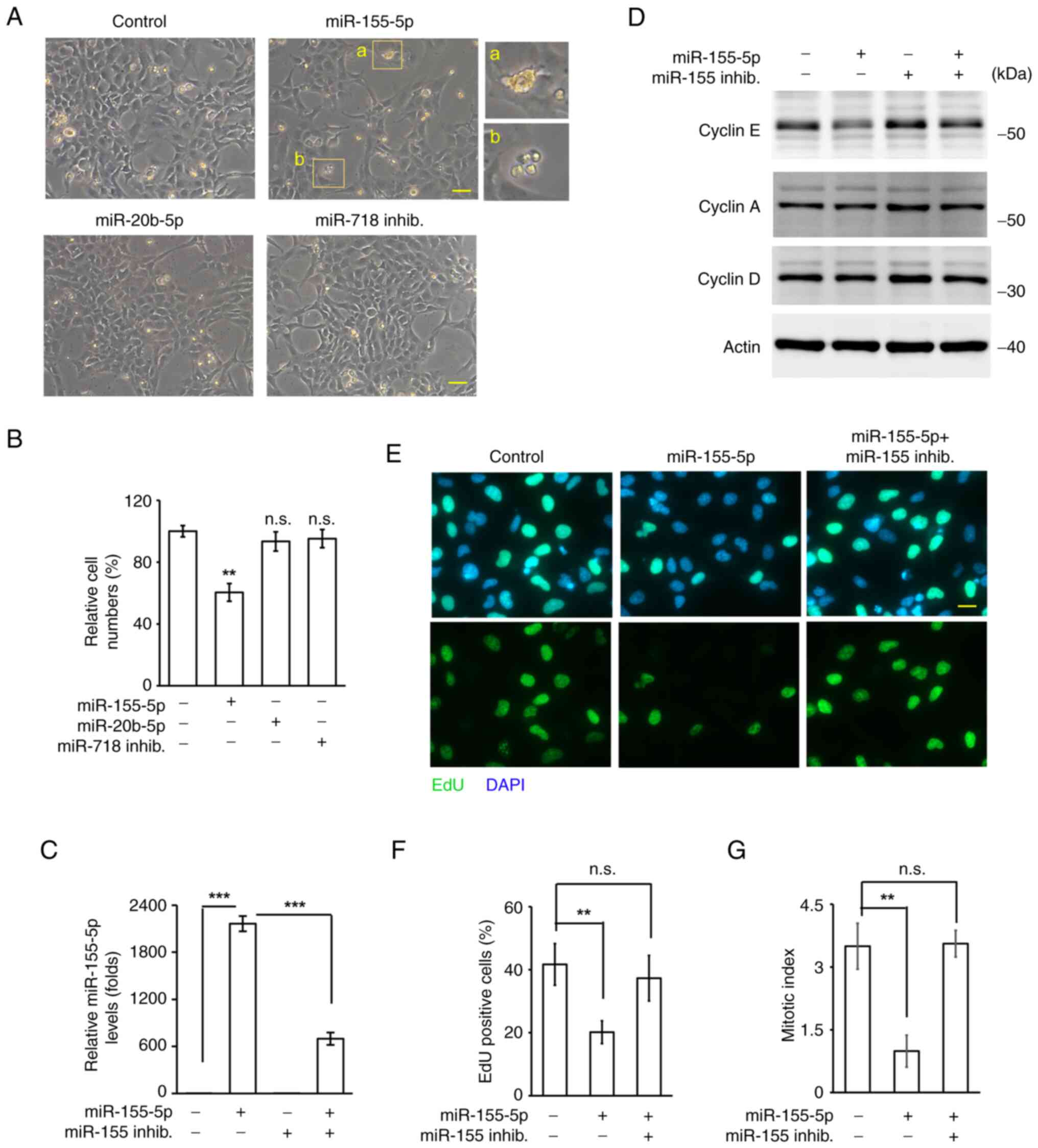 | Figure 2.miR-20b-5p inhibits trophoblast HTR8
cell migration and invasion. (A and B) Overexpression of miR-155-5p
inhibits HTR8 cell growth. (A) Overexpression of miR-155-5p, not
miR-20b-5p or miR-718 inhib., reduced trophoblast HTR8 cell growth.
Enlarged views of a and b are apoptotic bodies. Scale bar, 20 µm
(B) Quantitative results of (A). (C) Relative levels of miR-155-5p
in miR-155-5p with or without miR-155-5p overexpressing cells. (D)
Cyclin E levels reduced in miR-155-5p-overexpressing cells.
Extracts of miR-155-5p or miR-155-5p inhib. transfected cells were
analyzed by western blotting with antibodies against cyclin E,
cyclin A, cyclin D and actin (loading control). Overexpression of
miR-155-5p reduced S phase entry. (E) EdU-positive cells were
reduced in miR-155-5p-overexpressing cells. Scale bar, 10 µm. (F)
Quantitative results of (E). (G) Overexpression of miR-155-5p
reduced M phase entry. Mitotic index was reduced in
miR-155-5p-overexpressing cells. Data are presented as mean ± SD
from three independent experiments (**P<0.01, ***P<0.001).
All ‘-’ symbols in the figure represent the negative control
transfection. n.s., no significance; inhib., inhibitor; HTR8,
HTR-8/SVneo. |
In addition to cell cycle progression, some
apoptotic bodies were observed in miR-155-5p-overexpressing cells;
therefore, cell apoptosis was assessed. Cleaved-caspase 3 and
cleaved-PARP, which are markers of apoptosis, were examined in
miR-155-5p-overexpressing cells. Overexpression of miR-155-5p
increased the levels of cleaved-caspase 3 and cleaved-PARP
(Fig. 3A and B), and this
phenotype was rescued by co-transfection with the miR-155-5p
inhibitor. These data suggested that miR-155-5p induced cell
apoptosis.
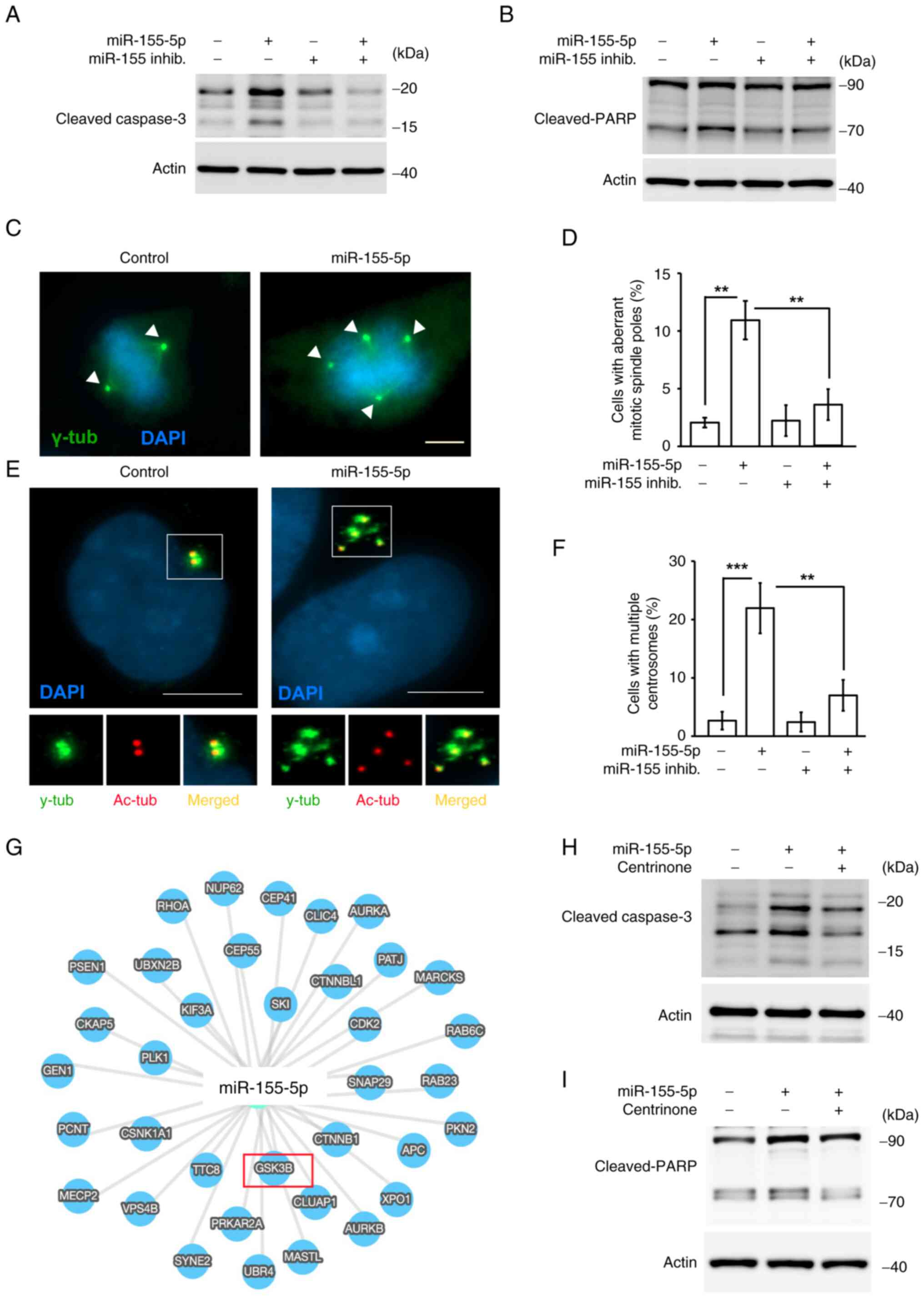 | Figure 3.Overexpression of miR-155-5p
facilitates centrosome amplification and cellular apoptosis. (A and
B) Overexpression of miR-155-5p induced apoptosis. Markers of
cellular apoptosis, (A) cleaved-caspase 3 and (B) cleaved-PARP,
were examined by western blotting in the presence of miR-155-5p or
miR-155-5p inhib. (C and D) Overexpression of miR-155-5p leads to
aberrant mitotic spindle poles. (C) Immunofluorescence staining of
control or miR-155-5p-overexpressing cells with antibody against
γ-tub (green). The arrowhead indicates mitotic spindle poles. DNA
was stained with DAPI (scale bar, 5 µm). (D) Quantitative results
of (C). (E) Immunofluorescence staining of control, miR-155-5p-, or
miR-155-5p inhib.-overexpressing cells with antibodies against
acetylated tubulin (centrioles, red) and γ-tub (pericentriolar
material, green). DNA was stained with DAPI (scale bar, 10 µm). (F)
Quantitative results of (E). (G) GSK3β was a putative candidate of
miR-155-5p targeted gene shown by in silicon prediction. (H-I)
Inhibition of centrosome amplification by treating cells with
centrinone reduced cellular apoptosis. Extracts of
miR-155-5p-overexpressing cells in the presence or absence of
centrinone were analyzed by western blotting assay with antibodies
against (H) cleaved-caspase 3, (I) cleaved-PARP or actin. Data are
presented as mean ± SD from three independent experiments
(**P<0.01, ***P<0.001). inhib., inhibitor; PARP, poly
(ADP-ribose) polymerase; γ-tub, γ-tubulin; Ac-tub, acetylated
tubulin. |
Overexpression of miR-155-5p induces
centrosome amplification
Aberrant mitosis triggers cell apoptosis (36); therefore, whether
miR-155-5p-induced apoptosis by aberrant mitosis was assessed. The
mitotic spindle poles were examined by staining cells with
γ-tubulin. Under normal conditions, cells contained two mitotic
spindle poles and the chromosomes were aligned in the middle of the
cells (Fig. 3C, left panel);
however, when cells overexpressed miR-155-5p, multiple mitotic
spindle poles (>2 γ-tubulin spots) and misaligned chromosomes
were observed (Fig. 3C, right
panel; and Fig. 3D). This
phenotype was rescued when cells were co-transfected with the
miR-155-5p inhibitor (Fig. 3D).
Thus, these data suggested that miR-155-5p triggered aberrant
mitosis. Aberrant mitosis is caused by centrosome amplification
(cells with >3 centrosomes) during interphase (37); therefore, the centrosome copy
numbers during interphase were next assessed. Under normal
conditions, cells contained two centrosomes (Fig. 3E, left panel); however,
overexpression of miR-155-5p induced centrosome amplification
(Fig. 3E, right panel).
Co-transfection with the miR-155-5p inhibitor alleviated
miR-155-5p-induced centrosome amplification (Fig. 3F), supporting the hypothesis that
miR-155-5p induced centrosome amplification. Whether centrosome
amplification contributed to apoptosis upon miR-155-5p
overexpression was next examined. Overexpression of miR-155-5p
increased the levels of cleaved-caspase 3 and cleaved-PARP;
however, inhibition of centrosome amplification by treating cells
with the centrosome inhibitor centrinone rescued these phenotypes
(Fig. 3H and I), suggesting that
centrosome amplification promoted cell death. Using bioinformatics
analysis, several genes, such as GSK3β, PLK3, or KIF3A, were
predicted as the targets of miR-155-5p. GSK3β was identified as a
putative candidate of miR-155-5p (Fig.
3G). A previous study showed that the downregulation of GSK3β
led to the accumulation of β-catenin followed by induction of
centrosome amplification (38,39).
Thus, miR-155-5p may target GSK3β to induce centrosome
amplification. Taken together, miR-155-5p induced centrosome
amplification, thus triggering trophoblast cell apoptosis.
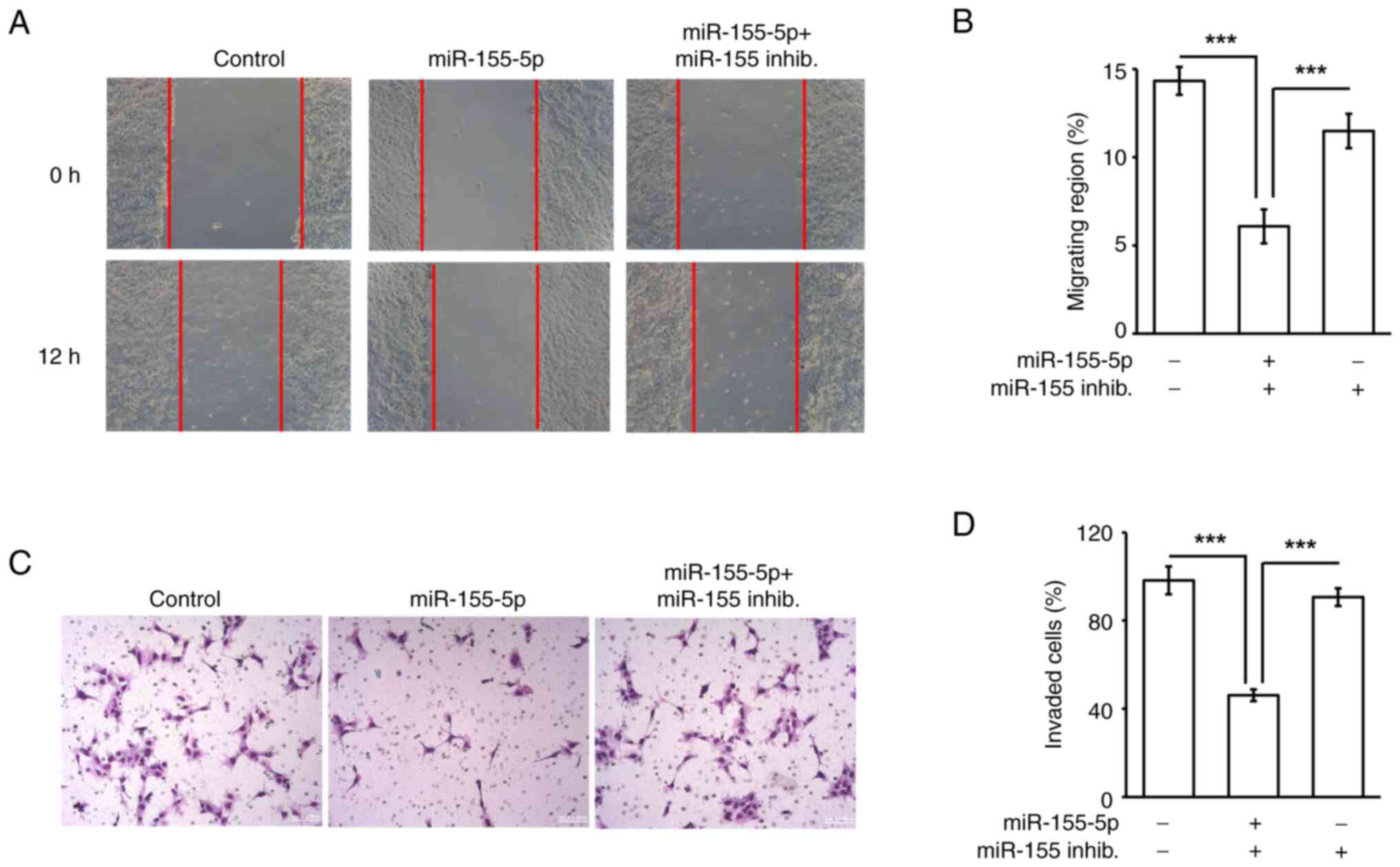
Overexpression of miR-155-5p inhibits
trophoblast migration and invasion
Trophoblast cell migration and invasion are critical
for placenta formation during early pregnancy (30); thus whether overexpression of
miR-155-5p impeded trophoblast migration and invasion was examined.
Since miR-155-5p inhibited HTR8 cell growth, seeding cell numbers
were adjusted according to the growth assay. A total of
5×105 control and 7×105
miR-155-5p-transsfected cells were seeded to perform the migration
and invasion assays. First, the effects of miR-155-5p on
trophoblast cell migration was assessed by performing a wound
healing assay. Overexpression of miR-155-5p inhibited trophoblast
cell migration, and this phenotype was rescued by co-transfecting
cells with the miR-155-5p inhibitor (Fig. 4A and B), suggesting that
upregulation of miR-155-5p reduced trophoblast cell migration.
Next, trophoblast cell invasion was examined. Consistent with the
results of the cell migration assay, the invasive ability of
trophoblast cells was reduced by miR-155-5p overexpression
(Fig. 4C and D), suggesting that
miR-155-5p inhibited cell invasion. Collectively, miR-155-5p
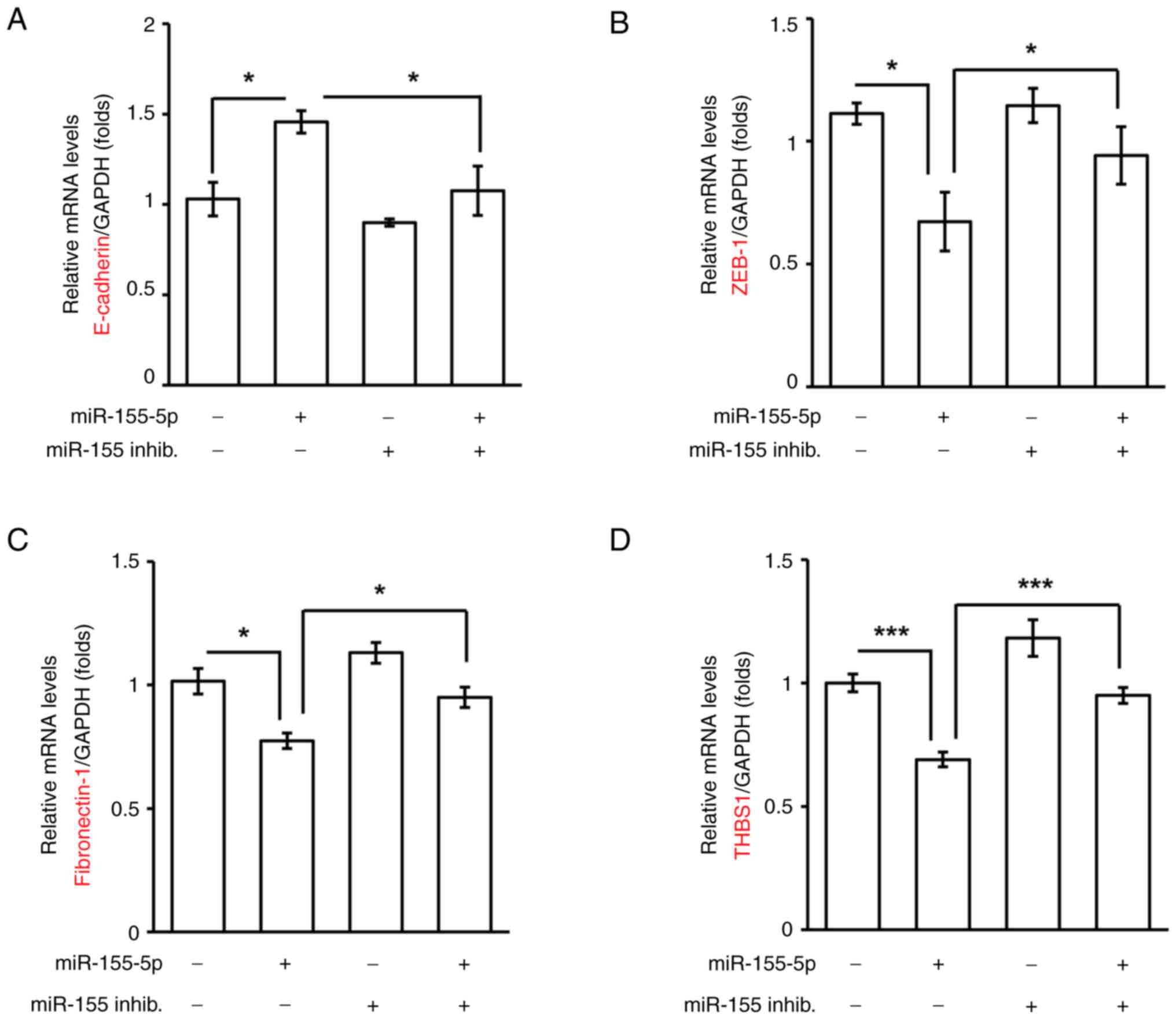
inhibited HTR8 trophoblast cell migration and invasion. EMT is key
for trophoblast cell invasion during early pregnancy (18). Since miR-155-5p inhibited
trophoblast cell invasion, the EMT was examined in the present
study. In miR-155-5p-overexpressing cells, the epithelial marker
E-cadherin was increased, whereas the mesenchymal markers ZEB-1,
fibronectin-1 and THBS1 were significantly decreased, suggesting
that miR-155-5p inhibited EMT (Fig.
5A-D). By contrast, co-transfection with the miR-155-5p
inhibitor rescued the expression levels of the EMT markers. Taken
together, miR-155-5p inhibited trophoblast cell invasion by
reducing EMT.
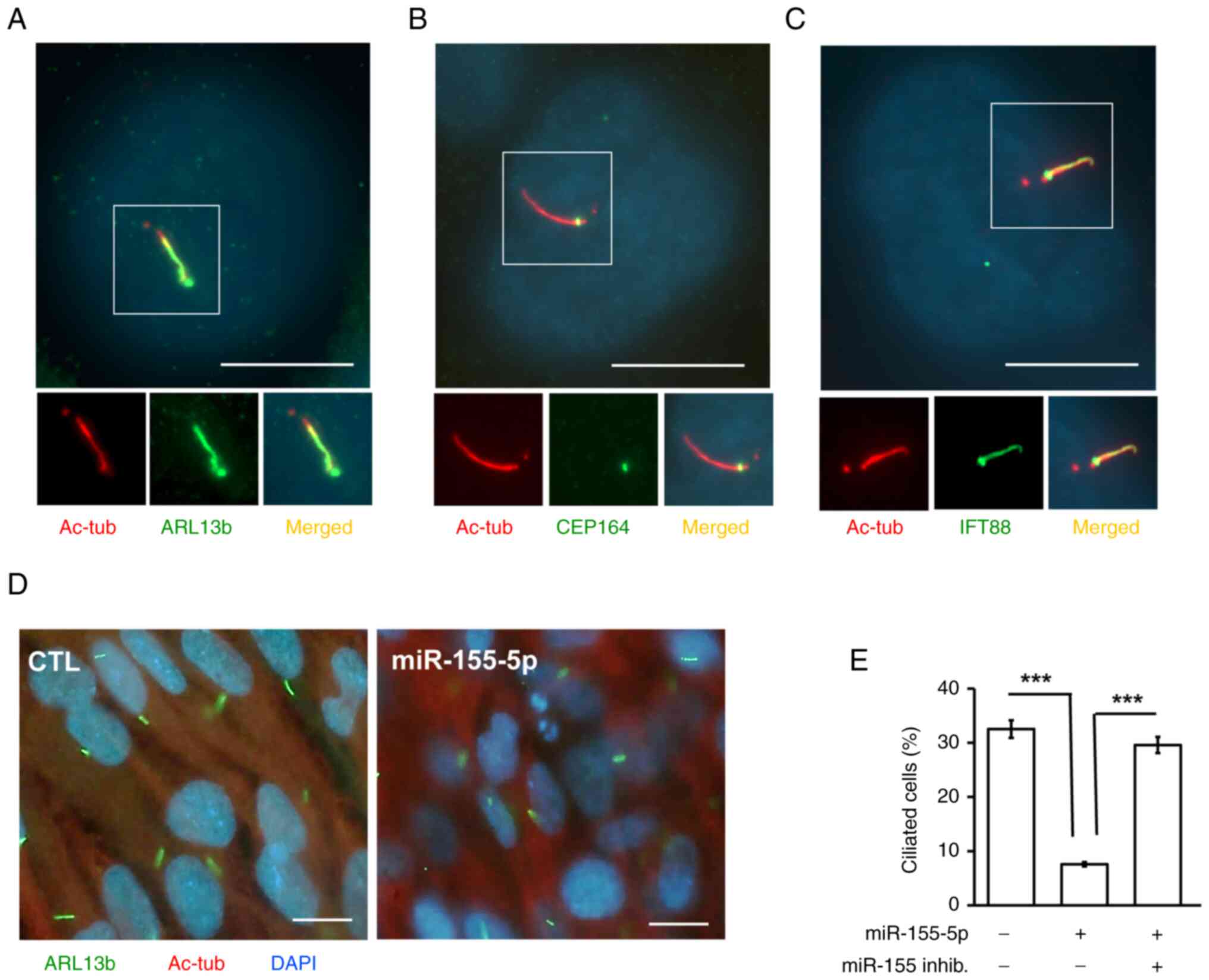
Overexpression of miR-155-5p inhibits
p38 activation and primary ciliogenesis
Primary cilia promote trophoblast cell invasion;
therefore, whether miR-155-5p affected primary cilia formation was
assessed. HTR8 cells grew primary cilia, as evidenced by several
markers, including markers of the axoneme (acetylated tubulin),
ciliary membrane (ARL13B), intraflagellar transporter (IFT88) and
basal body (CEP164; Fig. 6A-C).
Overexpression of miR-155-5p reduced the proportions of ciliated
cells, and this phenotype was rescued by co-transfection of cells
with the miR-155-5p inhibitor (Fig. 6D
and E). These data suggested that miR-155-5p reduced primary
cilia formation. Next, which signaling contributed to primary cilia
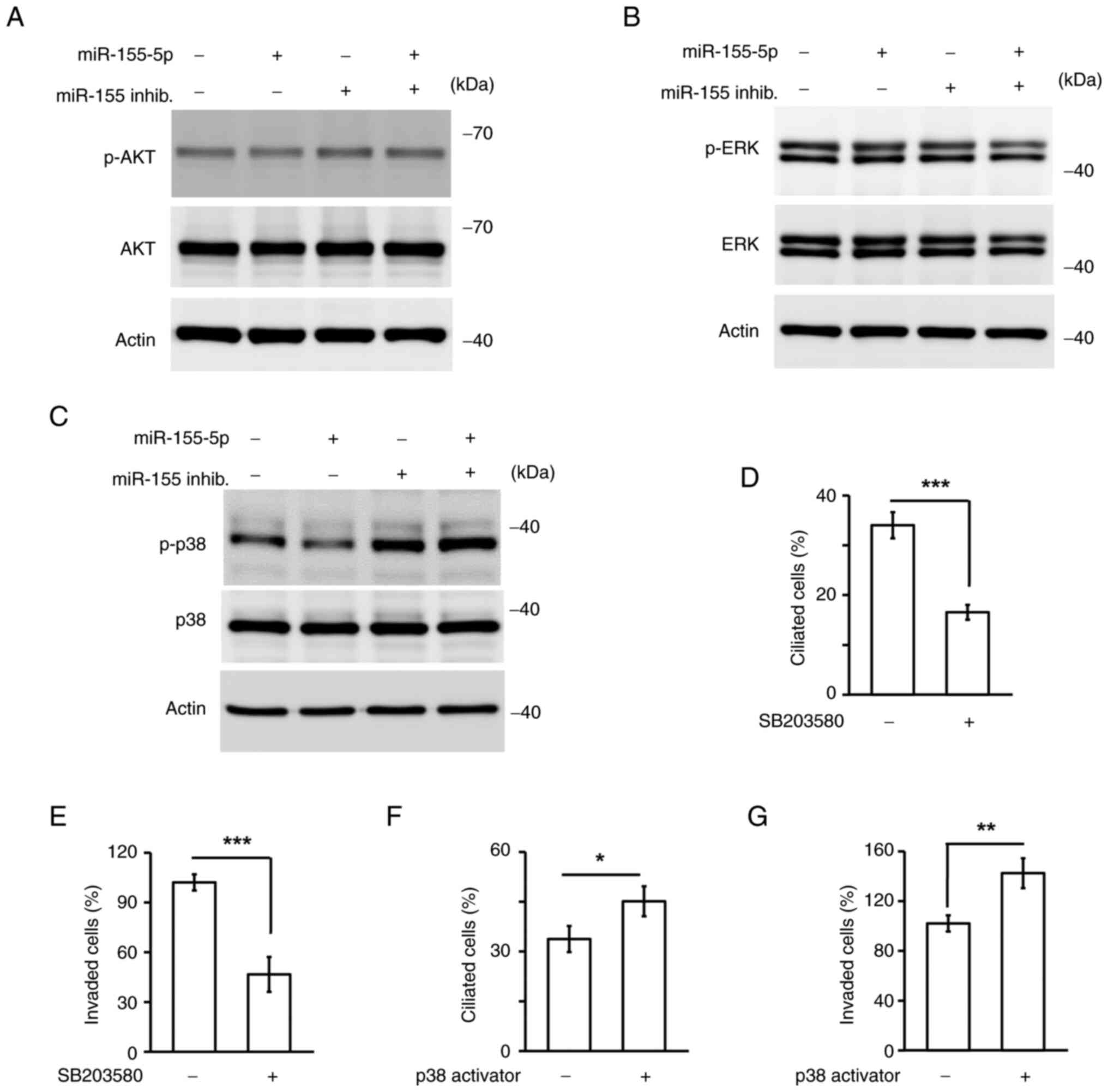
formation was assessed. AKT, ERK and p38 are known to regulate
primary cilia formation; therefore it was next assessed whether
these signaling pathways were activated and participated in
miR-155-5p-mediated primary cilia formation. Overexpression of
miR-155-5p did not affect AKT and ERK activation; however,
phospho-p38 (active form) was reduced, and this phenotype was
rescued by co-transfecting cells with the miR-155-5p inhibitor
(Fig. 7A-C). Thus, miR-155-5p
inhibited p38 activation. It was then examined whether p38 reduced
primary cilia formation. HTR8 cells grew primary cilia; however, in
response to treatment of HTR8 cells with the p38 inhibitor
SB203580, the proportions of ciliated cells were decreased
(Fig. 7D). It was also observed
that the p38 inhibitor reduced invasive ability (Fig. 7E), suggesting that p38 activity was
required for trophoblast invasion. To confirm this finding further,
miR-155-5p-overexpressing cells were treated with the p38 activator
and found activation of p38 restored the primary cilia formation
and invasive ability (Fig. 7F and
G). Taken together, miR-155-5p inactivated p38, thus reducing
primary cilia formation and trophoblast invasion.
Overexpression of miR-155-5p regulates
autophagy in trophoblast cells
Autophagic homeostasis maintains trophoblast cell
growth and invasion. It was then checked whether autophagy is
mediated by miR-155-5p. The levels of LC3 II were increased in
miR-155-5p-overexpressing cells, whereas this phenotype was rescued
by co-transfection with the miR-155-5p inhibitor, suggesting that
miR-155-5p induced autophagy (Fig.
8A). Next, whether autophagy contributed to cell death was
assessed. ATG16L1, an important regulator of autophagy, was
depleted by transfecting HTR8 cells with siRNA against ATG16L1
(Fig. 8B). In addition, the cell
death and cell cycle pathways were examined. Depletion of ATG16L1
reduced cleaved-caspase 3 and cleaved-PARP, whereas the cyclin E
expression was increased in ATG16L1-deficient cells (Fig. 8C and D). These data indicated that
autophagy triggered cell death and inhibited cell cycle
progression. Degradation of the extracellular matrix by MMP2
promotes cell invasion; therefore, the effect of miR-155-5p on MMP2
expression was examined. Overexpression of miR-155-5p reduced the
mRNA and protein expression levels of MMP2, whereas co-transfection
with the miR-155-5p inhibitor rescued the expression of MMP2
(Fig. 8E and F). Thus, miR-155-5p
may inhibit MMP2 expression. Next, whether MMP2 was regulated by
autophagy was assessed. The levels of MMP2 were increased in
ATG16L1-deficient cells (Fig. 8D),
suggesting that autophagy inhibited cell invasion. To confirm
whether autophagy regulated trophoblast cell growth and invasion,
autophagy was inhibited by treating cells with the selective
inhibitor of autophagy, 3-methyladenine (3-MA). Treatment with 3-MA
induced HTR8 cell invasion (Fig.
8G). These data suggested that autophagy is required for
trophoblast cell growth and invasion. Taken together, miR-155-5p
induced autophagy, reducing cell growth and invasion.
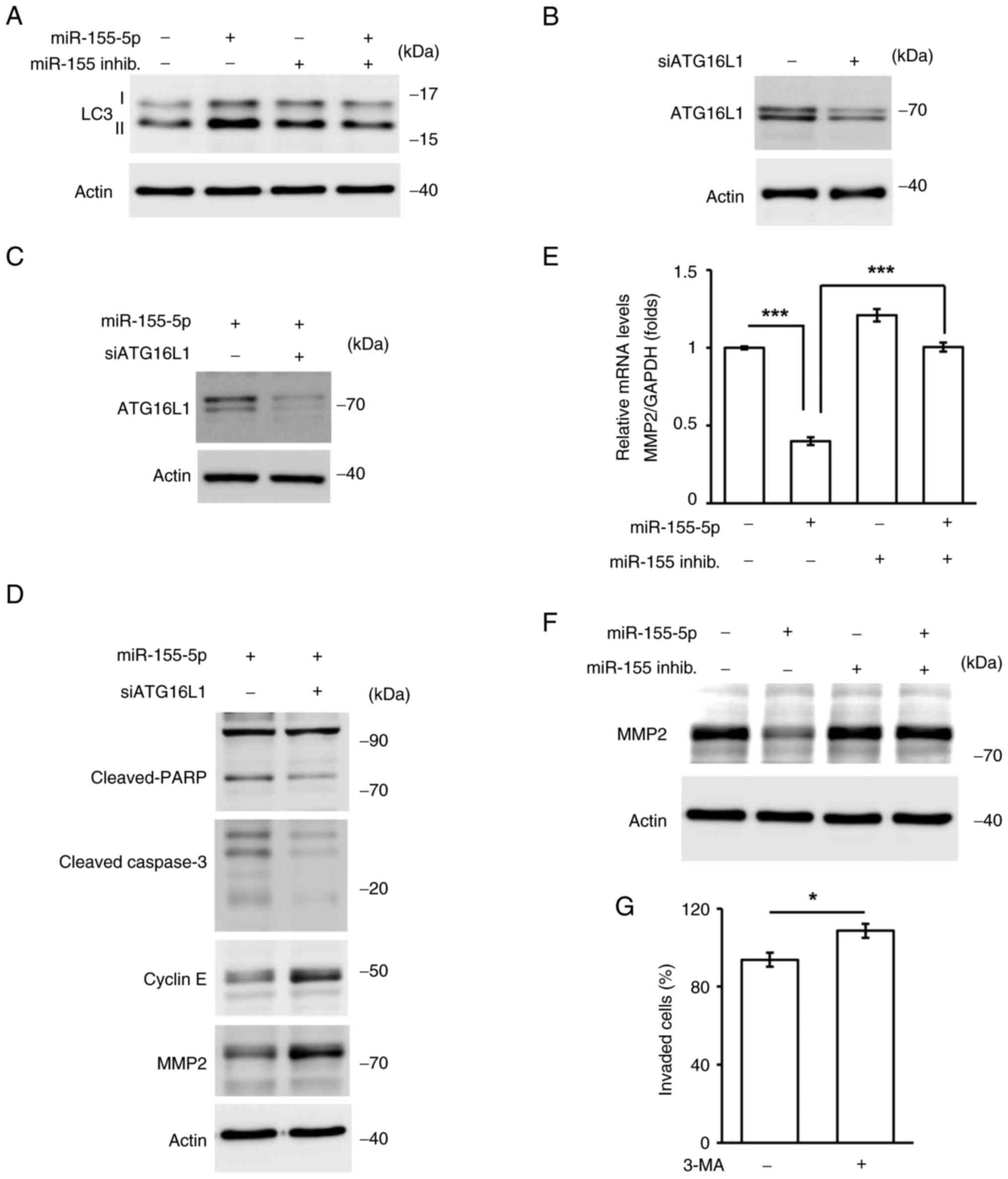 | Figure 8.miR-155-5p regulates autophagy for
trophoblast cell growth and invasion. (A) Overexpression of
miR-155-5p induced autophagy. Extracts of miR-155-5p- or miR-155-5p
inhibitor-transfecting cells were analyzed by western blotting
assay with antibodies against LC3I/II and actin. (B-D) Autophagy
inhibited trophoblast cell growth and invasion. (B-C) ATG16L1 was
depleted efficiently. Extracts of cells transfected with siRNA
against ATG16L1 in the (B) absence or (C) presence of miR-155-5p
were analyzed with antibodies against ATG16L1 and actin. (D)
Depletion of ATG16L1 inhibited cell apoptosis and promoted MMP2
expressions. Extraction of ATG16L1-depleting HTR8 cells were
analyzed by western blotting with antibodies against
cleaved-caspase 3, cleaved-PARP, cyclin E, MMP2 and actin. (E and
F) Overexpression of miR-155-5p inhibited MMP2 expression. (E) mRNA
levels of MMP2 were reduced in miR-155-5p-transfected cells. (F)
Protein levels of MMP2 were reduced in miR-155-5p-overexpressing
HTR8 cells. Extracts of miR-155-5p or miR-155-5p inhib. transfected
cells were analyzed by western blotting with antibodies against
MMP2 and actin (loading control). (G) Treatment of cells with 1 mM
3-MA induced cell invasion. All experiments were repeated at least
for three times. *P<0.05, ***P<0.001. All ‘-’ symbols in the
figure represent the negative control transfection. inhib.,
inhibitor; LC3I/II, light chain 3 I/II; ATG16L1, autophagy-related
16-like 1; HTR8, HTR-8/SVneo; PARP, poly (ADP-ribose) polymerase;
3-MA, 3-methyladenine. |
Discussion
Recurrent miscarriage leads to infertility in women,
and defective trophoblast cell growth and invasion are major causes
of recurrent miscarriage (1,40).
Dysregulation of miR-155-5p has been identified in women with
recurrent miscarriage; however, the underlying molecular mechanisms
remain to be elucidated. In the present study, it was demonstrated
that miR-155-5p blocked trophoblast cell growth and invasion by
affecting centrosome homeostasis and primary cilia formation, and
overexpression of miR-155-5p induced centrosome amplification,
followed by the promotion of cell death. Furthermore, miR-155-5p
inhibited primary cilia formation by reducing p38 activation, thus
inhibiting trophoblast cell invasion. Notably, these events were
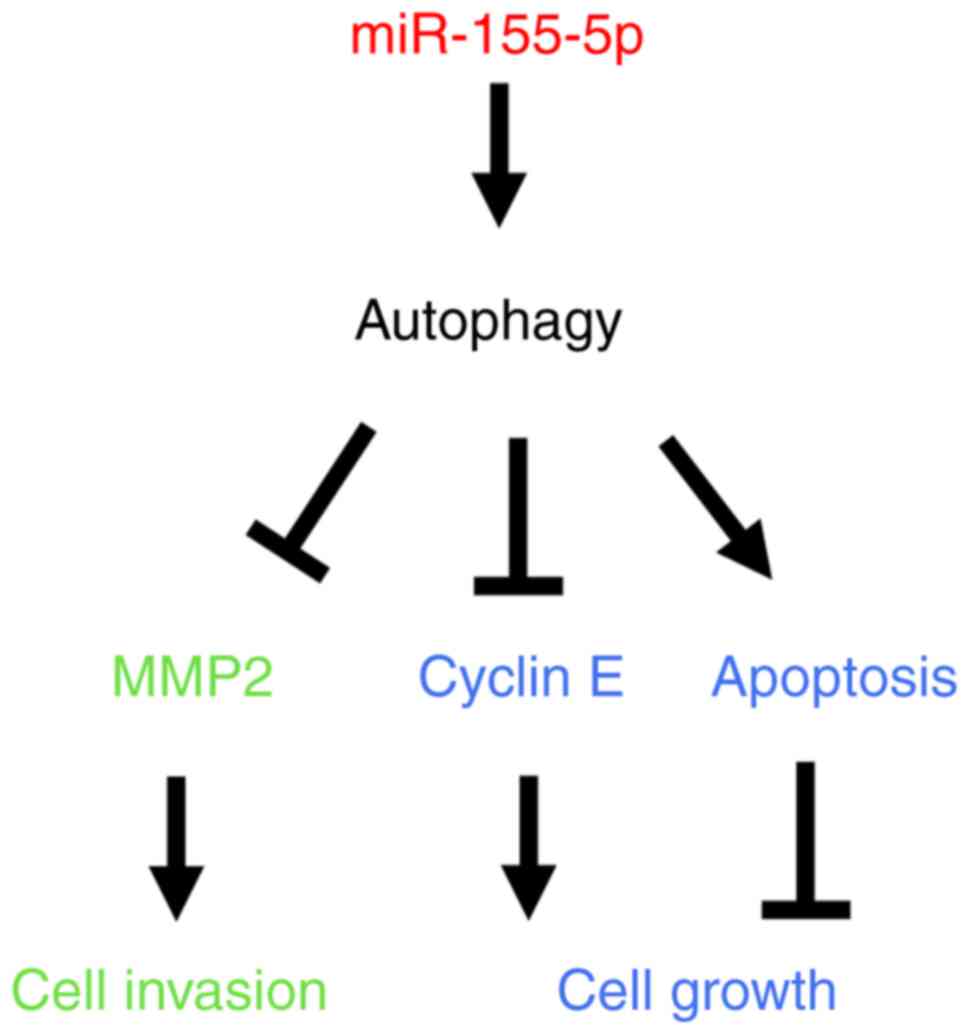
regulated by autophagy (Fig. 9).
Thus, the potential molecular mechanisms by which miR-155-5p
contributes to recurrent miscarriage may have been elucidated.
The present study found that miR-155-5p inactivated
p38, thus reducing primary cilia formation; however, the link
between p38 activation and primary cilia formation remains less
studied. Sex-determining region Y-box 9 (SOX9) belongs to the SOX
transcription factor family. SOX9 maintains organ development and
differentiation during mammalian embryo development (41). Recent studies have shown that SOX9
also participates in controlling primary cilia formation (42,43).
Primary cilia are markedly reduced in SOX9-knockout mice, implying
that SOX9 maintains primary cilia formation (44). Notably, p38 activation maintains
the mRNA stability of SOX9 (45).
Furthermore, depletion or inhibition of p38 via siRNA alleviates
the phosphorylation and protein stability of SOX9, supporting the
hypothesis that p38 activation is crucial for SOX9 expression and
activation (45). In the present
study, miR-155-5p inhibits p38 activation and inhibition of p38
reduced primary cilia formation in trophoblast cells. It was
therefore hypothesized that miR-155-5p may inactivate p38, thus
reducing SOX9 expression and inhibiting primary cilia formation.
This hypothesis requires further confirmation in the future.
During the progression of glioma, the exosomes
secreted by glioma stem-like cells contain a high abundance of
miR-155-5p. These miR-155-5p-containing exosomes are taken up by
the glioma and suppress the levels of acetyl-CoA thioesterase 12, a
tumor suppressor, thus promoting glioma cell proliferation
(46). By contrast, high
miR-155-5p levels reduce hepatocellular carcinoma cell growth
(47). By targeting collagen
triple helix repeat-containing 1, miR-155-5p can inhibit cell cycle
progression, invasion and migration, and promote cellular apoptosis
in vitro and in vivo (47). In addition, miR-155-5p indirectly
modulates the malignancy of liver cancer by regulating
Wnt/β-catenin signaling (47). The
present study showed that miR-155-5p inhibited cell cycle
progression and induced apoptosis in trophoblast cells; however,
miR-155-5p inhibited trophoblast cell invasion for placenta
formation by inhibiting the EMT and MMP2 expression. Thus,
miR-155-5p may serve contradictory roles in different tissues. In
glioma or triple-negative breast cancer, miR-155-5p promotes cell
growth; however, miR-155-5p inhibits liver cancer and trophoblast
cell proliferation and invasion (16,48).
These data imply that distinct targeted genes or regulatory
complexes may cause the tissue type-specific functions of
miR-155-5p. Future studies should focus on clarifying and
identifying the underlying distinct molecular mechanisms.
Preeclampsia is characterized by hypertension and
proteinuria during pregnancy (16). Patients with preeclampsia suffer
from organ dysfunction and the symptoms end after delivery
(16). During pregnancy,
trophoblast cells invade the maternal decidua and remodel the
uterine spiral arteries to increase uteroplacental blood perfusion
(40). Insufficient invasion of
trophoblast cells causes preeclampsia; notably, upregulation of
miR-155-5p is associated with preeclampsia (49). The expression of miR-155-5p has
been shown to be increased in preeclamptic placentas compared with
the control group (49); however,
the underlying molecular mechanism remains unclear. The present
study demonstrated that upregulation of miR-155-5p reduced
trophoblast invasion by reducing primary cilia formation.
Furthermore, previous studies have shown that loss of primary cilia
is observed in the preeclamptic placentas (18,30).
It was thus speculated that, during the progression of
preeclampsia, upregulation of miR-155-5p may inhibit trophoblast
cell EMT and invasion, therefore reducing the establishment of
maternal-fetal interfaces; therefore, targeting miR-155-5p may be a
good strategy to reduce the severity of preeclampsia or prevent
recurrent miscarriage.
The data are obtained from in vitro cell
culture models. To overcome this limitation and further confirm the
pathophysiological role of miR-155-5p, the miR-155-5p-overexpressed
transgenic mice can be generated and examine the implantation and
pregnancy in vivo.
In conclusion, the present study uncovered the role
of miR-155-5p in contributing to recurrent miscarriage.
Overexpression of miR-155-5p induced centrosome amplification, thus
reducing cell cycle progression and inducing cellular apoptosis. In
addition, miR-155-5p reduced primary cilia formation, thereby
inhibiting EMT and trophoblast invasion. Thus, miR-155-5p not only
functions as a biomarker of recurrent miscarriage but may be
considered a good candidate to prevent recurrent miscarriage.
Acknowledgements
The authors would like to thank Ms. Pey-Wen Liu
(Core Research Laboratory, College of Medicine, National Cheng Kung
University, Tainan, Taiwan) for technical support.
Funding
The present study was supported by the Ministry of Science and
Technology of Taiwan (MOST109-2320-B-006-042-MY3 and
NSTC112-2320-B-006-040) and the Chi Mei Medical Center
(CMNCKU11108).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
PJS and CYW conceived the study and wrote, reviewed
and edited the manuscript. YCT, TNK, RCL, HLT, YYC and PRL designed
experiments. YCT, TNK, HLT, YYC and PRL performed experiments. RCL,
HLT and YYC analyzed data. YCT and RCL wrote, reviewed and edited
the manuscript. All authors have read and approved the final
manuscript. CYW and RCL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
ATG16L1
|
autophagy-related 16-like 1
|
|
CEP164
|
centrosomal protein 164
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
IGFBP1
|
insulin-like growth factor binding
protein-1
|
|
IFT88
|
intraflagellar transport 88
|
|
MPA
|
medroxyprogesterone 17-acetate
|
|
SOX9
|
sex-determining region Y-box 9
|
|
T-HESC
|
telomerase-immortalized human
endometrial stromal cell
|
|
THBS1
|
thrombospondin 1
|
References
|
1
|
Bashiri A, Halper KI and Orvieto R:
Recurrent implantation failure-update overview on etiology,
diagnosis, treatment and future directions. Reprod Biol Endocrinol.
16:1212018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ford HB and Schust DJ: Recurrent pregnancy
loss: Etiology, diagnosis, and therapy. Rev Obstet Gynecol.
2:76–83. 2009.PubMed/NCBI
|
|
3
|
Rai R and Regan L: Recurrent miscarriage.
Lancet. 368:601–611. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
You Y, Stelzl P, Joseph DN, Aldo PB,
Maxwell AJ, Dekel N, Liao A, Whirledge S and Mor G: TNF-α regulated
endometrial stroma secretome promotes trophoblast invasion. Front
Immunol. 12:7374012021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang S, Lin H, Kong S, Wang S, Wang H,
Wang H and Armant DR: Physiological and molecular determinants of
embryo implantation. Mol Aspects Med. 34:939–980. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan-Castillo B, Gazze SA, Thomas S, Lucas
C, Margarit L, Gonzalez D, Francis LW and Conlan RS: Morphophysical
dynamics of human endometrial cells during decidualization.
Nanomedicine. 14:2235–2245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vento-Tormo R, Efremova M, Botting RA,
Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polański
K, Goncalves A, et al: Single-cell reconstruction of the early
maternal-fetal interface in humans. Nature. 563:347–353. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carter AM, Enders AC and Pijnenborg R: The
role of invasive trophoblast in implantation and placentation of
primates. Philos Trans R Soc Lond B Biol Sci. 370:201400702015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meakin C, Barrett ES and Aleksunes LM:
Extravillous trophoblast migration and invasion: Impact of
environmental chemicals and pharmaceuticals. Reprod Toxicol.
107:60–68. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jouravleva K, Golovenko D, Demo G, Dutcher
RC, Hall TMT, Zamore PD and Korostelev AA: Structural basis of
microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB. Mol
Cell. 82:4049–4063.e6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Basavarajappa D, Uebbing S, Kreiss M,
Lukic A, Suess B, Steinhilber D, Samuelsson B and Rådmark O: Dicer
up-regulation by inhibition of specific proteolysis in
differentiating monocytic cells. Proc Natl Acad Sci USA.
117:8573–8583. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Muys BR, Sousa JF, Plaça JR, de Araújo LF,
Sarshad AA, Anastasakis DG, Wang X, Li XL, de Molfetta GA, Ramão A,
et al: miR-450a acts as a tumor suppressor in ovarian cancer by
regulating energy metabolism. Cancer Res. 79:3294–3305. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1.
Sci Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Skalis G, Katsi V, Miliou A, Georgiopoulos
G, Papazachou O, Vamvakou G, Nihoyannopoulos P, Tousoulis D and
Makris T: MicroRNAs in preeclampsia. Microrna. 8:28–35. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, He Y, Wang JX, Chen MH, Xu JJ,
Jiang MH, Feng YL and Gu YF: miR-30-5p-mediated ferroptosis of
trophoblasts is implicated in the pathogenesis of preeclampsia.
Redox Biol. 29:1014022020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang CY, Tsai PY, Chen TY, Tsai HL, Kuo PL
and Su MT: Elevated miR-200a and miR-141 inhibit endocrine
gland-derived vascular endothelial growth factor expression and
ciliogenesis in preeclampsia. J Physiol. 597:3069–3083. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen CH, Lu F, Yang WJ, Yang PE, Chen WM,
Kang ST, Huang YS, Kao YC, Feng CT, Chang PC, et al: A novel
platform for discovery of differentially expressed microRNAs in
patients with repeated implantation failure. Fertil Steril.
116:181–188. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lai PY, Wang CY, Chen WY, Kao YH, Tsai HM,
Tachibana T, Chang WC and Chung BC: Steroidogenic factor 1 (NR5A1)
resides in centrosomes and maintains genomic stability by
controlling centrosome homeostasis. Cell Death Differ.
18:1836–1844. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen TY, Lien WC, Cheng HL, Kuan TS, Sheu
SY and Wang CY: Chloroquine inhibits human retina pigmented
epithelial cell growth and microtubule nucleation by downregulating
p150glued. J Cell Physiol. 234:10445–10457. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin RC, Chao YY, Lien WC, Chang HC, Tsai
SW and Wang CY: Polo-like kinase 1 selective inhibitor BI2536
(dihydropteridinone) disrupts centrosome homeostasis via ATM-ERK
cascade in adrenocortical carcinoma. Oncol Rep. 50:1672023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferguson RL and Maller JL: Centrosomal
localization of cyclin E-Cdk2 is required for initiation of DNA
synthesis. Curr Biol. 20:856–860. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen TY, Lin TC, Kuo PL, Chen ZR, Cheng
HL, Chao YY, Syu JS, Lu FI and Wang CY: Septin 7 is a centrosomal
protein that ensures S phase entry and microtubule nucleation by
maintaining the abundance of p150glued. J Cell Physiol.
236:2706–2724. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chao YY, Huang BM, Peng IC, Lee PR, Lai
YS, Chiu WT, Lin YS, Lin SC, Chang JH, Chen PS, et al: ATM- and
ATR-induced primary ciliogenesis promotes cisplatin resistance in
pancreatic ductal adenocarcinoma. J Cell Physiol. 237:4487–4503.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hildebrandt F, Benzing T and Katsanis N:
Ciliopathies. N Engl J Med. 364:1533–1543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamamoto Y and Mizushima N: Autophagy and
ciliogenesis. JMA J. 4:207–215. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Truong ME, Bilekova S, Choksi SP, Li W,
Bugaj LJ, Xu K and Reiter JF: Vertebrate cells differentially
interpret ciliary and extraciliary cAMP. Cell. 184:2911–2926.e18.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsai YC, Kuo TN, Chao YY, Lee PR, Lin RC,
Xiao XY, Huang BM and Wang CY: PDGF-AA activates AKT and ERK
signaling for testicular interstitial Leydig cell growth via
primary cilia. J Cell Biochem. 124:89–102. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang CY, Tsai HL, Syu JS, Chen TY and Su
MT: Primary cilium-regulated EG-VEGF signaling facilitates
trophoblast invasion. J Cell Physiol. 232:1467–1477. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu X, Liu H, Luo W, Wang X, Wang H and Li
L: Di-2-ethylhexyl phthalate-induced miR-155-5p promoted lipid
metabolism via inhibiting cAMP/PKA signaling pathway in human
trophoblastic HTR-8/Svneo cells. Reprod Toxicol. 114:22–31. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Graham CH, Hawley TS, Hawley RG,
MacDougall JR, Kerbel RS, Khoo N and Lala PK: Establishment and
characterization of first trimester human trophoblast cells with
extended lifespan. Exp Cell Res. 206:204–211. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abou-Kheir W, Barrak J, Hadadeh O and
Daoud G: HTR-8/SVneo cell line contains a mixed population of
cells. Placenta. 50:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tan HX, Yang SL, Li MQ and Wang HY:
Autophagy suppression of trophoblast cells induces pregnancy loss
by activating decidual NK cytotoxicity and inhibiting trophoblast
invasion. Cell Commun Signal. 18:732020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Orth JD, Loewer A, Lahav G and Mitchison
TJ: Prolonged mitotic arrest triggers partial activation of
apoptosis, resulting in DNA damage and p53 induction. Mol Biol
Cell. 23:567–576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang CY, Kao YH, Lai PY, Chen WY and Chung
BC: Steroidogenic factor 1 (NR5A1) maintains centrosome homeostasis
in steroidogenic cells by restricting centrosomal DNA-dependent
protein kinase activation. Mol Cell Biol. 33:476–484. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bahmanyar S, Guiney EL, Hatch EM, Nelson
WJ and Barth AIM: Formation of extra centrosomal structures is
dependent on beta-catenin. J Cell Sci. 123:3125–3135. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang CY, Lai PY, Chen TY and Chung BC:
NR5A1 prevents centriole splitting by inhibiting centrosomal DNA-PK
activation and β-catenin accumulation. Cell Commun Signal.
12:552014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Illsley NP, DaSilva-Arnold SC, Zamudio S,
Alvarez M and Al-Khan A: Trophoblast invasion: Lessons from
abnormally invasive placenta (placenta accreta). Placenta.
102:61–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ming Z, Vining B, Bagheri-Fam S and Harley
V: SOX9 in organogenesis: Shared and unique transcriptional
functions. Cell Mol Life Sci. 79:5222022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Edelman HE, McClymont SA, Tucker TR,
Pineda S, Beer RL, McCallion AS and Parsons MJ: SOX9 modulates
cancer biomarker and cilia genes in pancreatic cancer. Hum Mol
Genet. 30:485–499. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu WP, Cui YL, Chen LL, Ding K, Ding CH,
Chen F, Zhang X and Xie WF: Deletion of Sox9 in the liver leads to
hepatic cystogenesis in mice by transcriptionally downregulating
Sec63. J Pathol. 254:57–69. 2021.PubMed/NCBI
|
|
44
|
Schepers GE, Teasdale RD and Koopman P:
Twenty pairs of sox: Extent, homology, and nomenclature of the
mouse and human sox transcription factor gene families. Dev Cell.
3:167–170. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tew SR and Hardingham TE: Regulation of
SOX9 mRNA in human articular chondrocytes involving p38 MAPK
activation and mRNA stabilization. J Biol Chem. 281:39471–39479.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bao Z, Zhang N, Niu W, Mu M, Zhang X, Hu S
and Niu C: Exosomal miR-155-5p derived from glioma stem-like cells
promotes mesenchymal transition via targeting ACOT12. Cell Death
Dis. 13:7252022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen G, Wang D, Zhao X, Cao J, Zhao Y,
Wang F, Bai J, Luo D and Li L: miR-155-5p modulates malignant
behaviors of hepatocellular carcinoma by directly targeting CTHRC1
and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling.
Cancer Cell Int. 17:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang LW, Wu XJ, Liang Y, Ye GQ, Che YC, Wu
XZ, Zhu XJ, Fan HL, Fan XP and Xu JF: miR-155 increases stemness
and decitabine resistance in triple-negative breast cancer cells by
inhibiting TSPAN5. Mol Carcinog. 59:447–461. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu HY, Liu K and Zhang JL:
LINC00240/miR-155 axis regulates function of trophoblasts and M2
macrophage polarization via modulating oxidative stress-induced
pyroptosis in preeclampsia. Mol Med. 28:1192022. View Article : Google Scholar : PubMed/NCBI
|