Introduction
Knee osteoarthritis (KOA) is a common disease
affecting the quality of life of middle-aged and elderly
individuals (1). With an
ever-aging society, the incidence of KOA is increasing annually
(2). Clinical manifestations
include joint pain, swelling, limited movement and joint friction,
and the end stage of the disease can result in disability (3). The cause of KOA is primarily related
to sex, age, obesity, heredity factors, mechanical force and
congenital joint anomalies amongst other factors. KOA is the result
of a normal coupling imbalance between the degradation and
synthesis of chondrocytes, extracellular matrix and cartilage. It
is caused by a combination of mechanical and biological factors
leading to an imbalance in the normal coupling of degradation and
synthesis of the subchondral bone, and the degeneration of
articular cartilage is the characteristic and basic pathological
change of KOA (4–6).
Chondrocytes, the core components of cartilage
tissue, have the function of maintaining the normal structure and
the physiology of cartilage. When the number of chondrocytes
decreases, for example due to autophagy, senescence and apoptosis,
the structure and the function of the cartilage are affected
(7–10). Poly [ADP-ribose] polymerase-1
(PARP-1)-dependent cell death, known as parthanatos, (11–13)
is caused by the overactivation of PARP1, which catalyzes the
catabolism of intracellular nicotinamide adenine dinucleotide (NAD)
to produce poly [ADP-ribose] (PAR), which is translocated to the
cytoplasm and binds to the outer surface of the mitochondria,
causing the release of apoptosis-inducing factor (AIF) and the
recruitment of macrophage migration inhibitory factor (MIF) to the
nucleus, where it acts as a DNA endonuclease in synergy with
nucleic acid exonuclease G to induce chromatin condensation and DNA
breaks, generating fragments ~50 kb in length and inducing
apoptosis in parthanatos (12,14,15).
No research has revealed the association of PARP1/AIF with
chondrocyte apoptosis.
Rongjin Niantong Fang (RJNTF) originates from ‘Qing
Gong Pei Fang Ji Cheng’ which was compiled by the academic Chen
Keji, and consists of six component herbs, Achyranthes
bidentata Blume (Niu Xi), Angelica sinensis (Oliv) Diels
(Dang Gui), Heracleum hemsleyanum Diels (Du Huo),
Hansenia weberbaueriana (Fedde ex H. Wolff) Pime (Qiang
Huo), Saposhnikovia divaricata (Turcz.) Schischk (Fang Feng)
and Glycyrrhiza uralensis Fisch (Gan Cao). As a well-known
Traditional Chinese folk medicine, it is used to invigorate the
circulation of swelling, relieve pain caused by arthralgia pain,
nourish the liver and kidney, and promote blood circulation; it is
commonly used for the treatment of various diseases, including KOA
(16). It is known that certain
active ingredients of RJNTF are known to act directly on
chondrocytes (17,18), but its mechanism of action has not
yet been thoroughly investigated and explained. The aim of the
present study was to provide a scientific basis for the application
of RJNTF in the treatment of KOA. It is unknown whether RJNTF can
inhibit chondrocyte apoptosis by regulating the PARP1/AIF signaling
pathway. To further clarify the mechanism of action of this drug
and assess its potential as a treatment for KOA, the current study
investigated the inhibition of chondrocyte apoptosis by RJNTF
through modulation of the PARP1/AIF pathway using web-based
pharmacological analytical tools and in vitro experiments.
Finally, the findings of the present study revealed that inhibition
of PARP1 can effectively suppress chondrocyte apoptosis, provide
new insights and understanding of the mechanism of action of folk
medicine, help to promote the clinical translation of folk medicine
in the treatment of KOA and provide important references for
further research and clinical practice.
Materials and methods
Network pharmacology analysis
The data of normal human cartilage and KOA cartilage
were retrieved and downloaded from the Gene Expression Omnibus
database (accession no. GSE75181) (19), and the merged gene expression
matrix was analyzed using the limma package in R (version 4.0.5)
and R Studio (version 4.0.5) (20–22)
to screen the differentially expressed genes (DEGs). The DEGs of
KOA were obtained by screening using the criteria |log2 (fold
change)| ≥1 and Padj<0.05, and a volcano map was
plotted using Hiplot Pro (BGI Group; http://hiplot.com.cn/). The Traditional Chinese
Medicine Systems Pharmacology (TCMSP) database (https://old.tcmsp-e.com/tcmsp.php) was used to
identify the active ingredients of RJNTF, and then the Search Tool
for Interacting Chemicals (http://stitch.embl.de/) database was used to retrieve
the potential targets of each active ingredient. Venn diagrams were
used to visualize the overlapping targets of RJNTF with the DEGs of
KOA. Protein-protein interaction (PPI) analysis was used in the
BisoGenet plug-in of Cytoscape software (version 3.8.2) (23), and the results were analyzed based
on the topological parameters using the CytoNCA plug-in (http://apps.cytoscape.org/apps/cytonca)
to obtain the key targets of RJNTF (24). Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis was performed on the
potential targets of RJNTF that were common DEGs of KOA using the
clusterProfiler package in R (version 4.0.5); the potential targets
related to apoptosis signaling pathways were obtained, and the key
target-signaling pathway map was constructed using Cytoscape
(25,26).
Primary reagents and antibodies
RJNTF is comprised of Achyranthes bidentata,
Angelica sinensis, Angelica biserrata, Hansenia weberbaueriana,
Saposhnikovia divaricata and Glycyrrhiza uralensis at a
ratio of 4:2:3:2:2:2. The decoction protocol of RJNTF included
adding water at a solid:liquid ratio of 1:10, decocting three times
for 1.5 h each time, then filtering the liquid each time to remove
the dregs, before combining and mixing, evaporation of the mixed
liquid to lyophilize into powder, and storing the lyophilized
powder in a vacuum drying oven (preparation method and use of
formula authorized by the National Invention Patent; patent nos.
ZL201710284659.8 and ZL201810899834.9) (27). The Cell Counting Kit-8 (CCK-8)
reagent was purchased from MedChemExpress. DAPI was purchased from
Beijing Solarbio Science & Technology Co., Ltd. The Novolink™
Polymer Detection System was purchased from Fuzhou Maixin Biotech
Co., Ltd. The Annexin V-FITC Apoptosis Detection Kit was purchased
from Nanjing KeyGen Biotech Co., Ltd. L-DMEM was purchased from
Shanghai BasalMedia Technologies Co., Ltd. Hydrogen peroxide
(H2O2) and FBS were purchased from
MilliporeSigma. PARP1-knockdown lentiviral vector was constructed
by Shanghai GeneChem Co., Ltd. The PCR primers and the Nuclear and
Cytoplasmic Protein Extraction Kit used were obtained from Shanghai
Biotechnology Co., Ltd. The PARP1 inhibitor PJ34 was purchased from
MedChemExpress.
Separation and culture of articular
chondrocytes
A total of 30 Male Sprague-Dawley (SD) rats
(4-week-old; 80±10 g) were purchased from SLAC Laboratory Animal
Technology Co., Ltd. [animal license no. SCXK (Hu) 2019-0007] and
were used to obtain chondrocytes from the knee articular cartilage,
which is relatively easy to obtain. The chondrocytes of 4-week-old
rats are in active growth and development, their metabolism is
active and the number of chondrocytes is large so they can better
adapt to the in vitro culture environment (28,29).
Tissues were collected in accordance with the Animal Care and Use
Committee of Fujian University of Traditional Chinese Medicine
(IACUC issue no. FJTCM IACUC 2022044) and the Declaration of
Helsinki. The 4-week-old SPF-grade SD male rats were euthanized by
intraperitoneal injection with 100 mg/kg pentobarbital sodium. The
cessation of breathing and heart rate were used to confirm the
death of the rats. The cartilage of the knee joints was obtained
under aseptic conditions. The cartilage was rinsed with PBS three
times, and then cut into 1×1×1-mm pieces with a scalpel blade,
cleaned with PBS again, and transferred to a 60-mm culture dish; 5
ml 0.2% type II collagenase solution containing 1% penicillin,
streptomycin and amphotericin B triple antibiotic solution, which
was purchased from Shanghai BasalMedia Technologies Co., Ltd., and
placed in a cell culture incubator at 5% CO2 and 37°C
for digestion, collecting the cells every 2 h. The supernatant was
aspirated using a pipette and filtered through a 200-mesh nylon
sieve. The filtrate was transferred to a 15-ml Eppendorf tube,
centrifuged at 1,000 × g for 3 min at room temperature, and the
supernatant was discarded; 4 ml complete medium was used to
resuspend the cell pellet, and then cultured uniformly in T25 cell
culture flasks and incubated for chondrocyte cell culture.
Digestion was carried out by adding 5 ml 0.2% collagenase solution
containing 1% antibiotic to the original 60-mm petri dish for a
total of three times. After 24 h, the culture medium was replaced
when the cells had adhered, and the medium was changed every 2
days. Cell growth was observed using an inverted light microscope.
When the cell confluence reached ~90%, the chondrocytes were
subcultured with 2.5 g/l trypsin (Promega Corporation) and 0.02%
EDTA. Chondrocytes at passage two were used for subsequent
experiments.
Cell viability assay
According to the manufacturer's instructions, a
CCK-8 kit was used to separately evaluate the effects of the
different concentrations of H2O2, PJ34 and
RJNTF on the viability of the chondrocytes. Second-generation
chondrocytes were seeded in 96-well plates (2,000 cells/well), and
then exposed to H2O2 for 4 h (0, 100, 200,
300, 400, 500, 600, 700 or 1,000 µM), PJ34 for 4.5 h (0.001, 0.01,
0.1, 1, 10, 100 or 1,000 µM), and RJNTF (0, 100, 200, 400, 800,
1,600, 3,200, 6,400 or 12,800 µg/ml) for 24, 48 or 72 h,
respectively. Each well was treated with 10% CCK-8 solution and
incubated for further 2 h at 37°C. The absorbance was detected at
450 nm using an EnSpire® multimode plate reader
(PerkinElmer, Inc.).
DAPI staining
Chondrocytes (2×105 cells/well) were
seeded into cell crawls placed in 6-well plates. After 24 h of
incubation, the cell crawls were transferred into a new 6-well
plate and washed twice with PBS. After fixation with 4%
Paraformaldehyde Fix Solution (Beyotime Institute of Biotechnology)
for 30 min and rinsing twice with PBS, the chondrocytes were
stained with 500 µl DAPI for 10 min at room temperature and then
washed twice with PBS for a total of 3 min per wash. The cellular
morphology was observed under a Leica DMi8 inverted fluorescence
microscope (Leica Microsystems, Inc.).
Flow cytometry
The Annexin V-FITC/PI apoptosis kit was used
according to the manufacturer's protocol. Briefly, chondrocytes
were collected and rinsed twice with PBS. Next, chondrocytes were
labeled with 500 µl 1× binding buffer with 5 µl PI and 5 µl Annexin
V-FITC for 15 min in the dark. Apoptosis rates were measured using
a Cyto FLEX flow cytometer (Beckman Coulter Inc.) and CytExpert
(version 2.4; Beckman Coulter, Inc.).
Knockdown of PARP1
Chondrocytes (2×105 cells/well) were
seeded into 96-well plates. A short interfering RNA (si)-negative
control (NC), si-PARP1 #1 (positive virus 114447), si-PARP1 #2
(positive virus 114448) and si-PARP1 #3 (positive virus 114449)
with green fluorescent protein were purchased from GeneChem, Inc.
(Table I). After 24 h, the viruses
were stored at −80°C and taken out when required, placing on ice to
thaw. The four viruses were individually diluted to an multiplicity
of infection (MOI) of 100, 50 and 10 using complete medium. The
culture media was discarded from cells, and using Nucleic acid
transfection kit (Shanghai Genechem, Inc.) with nucleic acid
concentration at 1×108 TU/ml and the effective
transfection enhancers HiTransG A and HiTransG P (GeneChem, Inc.)
were added according to the grouping, and mixed and cultured at
37°C, with 5% CO2, for 16 h. After transfection, the
medium was replaced with 10% FBS DMEM, and cells were incubated at
37°C, with 5% CO2 for 48 h, and the transfection
efficiency was evaluated under a fluorescence microscope. The
transfection efficiency was calculated as follows: Transfection
efficiency=(number of fluorescent cells/total number of cells)
×100. After singling out viruses with high transfection efficiency,
the chondrocyte apoptosis model was replicated.
 | Table I.Sequences of si-PARP1s and si-NC. |
Table I.
Sequences of si-PARP1s and si-NC.
| Gene | Sequences
(5′-3′) |
|---|
|
PARP1-RNAi(114447) |
GCTGATCTGGAATATCAAAGA |
|
PARP1-RNAi(114448) |
GGAGGCAAGTTGACAGGATCT |
|
PARP1-RNAi(114449) |
GCACAGTTATCGGCAGTAACA |
| si-NC |
TTCTCCGAACGTGTCACGT |
Reverse transcription-quantitative PCR
(RT-qPCR)
Chondrocytes (3×106 cells/well) were
seeded in 6-well plates and treated with one of the aforementioned
different siRNA constructs. Total RNA was extracted from cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and the RNA concentration was measured. cDNA was
synthesized using an Evo M-MLV RT kit (Hunan Acre Bioengineering
Co., Ltd.), according to the manufacturer's protocol, and amplified
by qPCR using SYBR Green Pro Tap (Hunan Acre Bioengineering Co.,
Ltd.) on a Bio-Rad CFX96 amplifier. The thermocycling conditions
were 95°C for 3 min; followed by 40 cycles of amplification at 95°C
for 10 sec and 60°C for 30 sec. β-actin was used as the internal
control, and relative gene expression was normalized to β-actin
mRNA expression and analyzed using the 2−ΔΔCq method
(30). The sequences of the
primers are listed in Table
II.
 | Table II.Primer sequences used in reverse
transcription-quantitative PCR. |
Table II.
Primer sequences used in reverse
transcription-quantitative PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| PARP1 |
CTTGGTGGAGTACGAGATTGAC |
GGTGTAGAAGCGATTGGAGAG |
| β-actin |
TCACCCACACTGTGCCCATCTATGA |
CATCGGAACCGCTCATTGCCGATAG |
Western blotting
Cells were washed three times with cold PBS and
blotted dry, and then 120 µl RIPA (Beyotime Institute of
Biotechnology) buffer supplemented with 1 mM PMSF was added for 30
min on ice; the petri dish was shaken once every 10 min, and the
lysate was transferred to a 1.5 ml EP tube which was centrifuged at
4°C for 5 min at 12,000 × g. The supernatant was collected, and a
total of 5 µl supernatant was used for BCA protein quantification,
and 5× SDS-PAGE protein sampling buffer was added to the remaining
supernatant. Nucleus and cytoplasmic proteins were extracted
according to the manufacturer's instructions using Nuclear and
Cytoplasmic Protein Extraction Kit (Beyotime Institute of
Biotechnology; cat. no. P0028), and the buffer was mixed with the
protein supernatant and then heated in a thermostatic metal bath at
99°C for 10 min to denature the proteins sufficiently and stored at
−80°C. A total of 30 µg protein sample was loaded per lane and
separated by 10% or 15% SDS-PAGE at 30 V for 10 min, 80 V for 30
min, and 110 V for 55 min and subsequently transferred to a 0.45-µm
PVDF membrane; 15% PAGE gels were used for caspase-3,
cleaved-caspase-3, AIF, MIF, histone and β-actin, while 10% PAGE
gels were used for PARP1, cleaved-PARP1, PAR and β-actin. Both the
target protein and the corresponding internal reference were run on
the same membrane; this was cut into strips and each was probed
separately to ensure that every protein can be incubated with a
specific antibody. After the membranes were blocked with NcmBlot
blocking buffer for 10 min at 25°C, membranes were incubated with
primary antibodies against PARP1 (1:5,000; Proteintech Group, Inc.;
cat. no. 66520-1-Ig), caspase-3 (1:1,000; Proteintech Group, Inc.;
cat. no. 19677-1AP), cleaved PARP1 (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 94885S), cleaved caspase-3 (1:1,000;
Cell Signaling Technology, Inc.; cat. no. 9661S), PAR (1:200; Enzo
Life Sciences, Inc.; cat. no. ALX-804-220-R100), AIF (1:1,000; cat.
no. 5318S; Cell Signaling Technology, Inc.), MIF (1:1,000; Abcam;
cat. no. Ab175189), β-actin (1:1,000; Cell Signaling Technology,
Inc.; cat. no. 8457S), or histone (1:1,000; Abcam; cat. no. Ab1791)
at 4°C overnight. The following day, membranes were washed with
TBST (TBS with 0.1% Tween-20, 10X) three times and subsequently
incubated with corresponding the HRP-conjugated secondary
antibodies (1:20,000; Cell Signaling Technology, Inc.; cat. no.
7074S) at 25°C for 1 h. The membranes were rinsed three times with
TBST, and signals were visualized with a Bio-Rad ChemiDoc MP
Imaging System (Bio-Rad Laboratories, Inc.), which using enhanced
chemiluminescence western blot substrate. Densitometry analysis was
performed using ImageJ (version win64; National Institutes of
Health) and normalized to the respective β-actin or histone
band.
Co-immunoprecipitation (IP) assay
Diluted antibody (400 µl) against AIF (1:1,000; cat.
no. 5318S; Cell Signaling Technology, Inc.), MIF (1:1,000; cat. no.
Ab175189; Abcam) and β-actin (1:1,000; cat. no. 8457S; Cell
Signaling Technology, Inc.) was aspirated into 50 µl magnetic beads
and mixed well, before incubation at 4°C for 2 h. After magnetic
separation, the magnetic beads were collected, 500 µl
phosphate-buffered saline (Shanghai BasalMedia Technologies Co.,
Ltd.) was added, the magnetic beads were mixed well and the
supernatant was discarded after magnetic separation. Washing was
performed four times. A total of 200 µl lysis buffer (Beyotime
Institute of Biotechnology) was added, followed by lysis at 4°C for
30 min, with agitation of the cells every 10 min to ensure full
contact with the lysis buffer, and then centrifugation at 4°C for
10 min at 12,000 × g. The supernatant was collected and set aside
at 4°C. After BCA quantification, 20 µl SDS-PAGE loading buffer was
added to the magnetic beads and mixed uniformly, after which the
magnetic beads were heated at 99°C for 10 min, the beads were
separated and the supernatant was used for western blotting.
Statistical analysis
Data analysis was performed using GraphPad Prism
(version 8.0; Dotmatics). A unpaired two-sample t-test was used to
compare differences between two groups. For multiple group
comparisons, one-way ANOVA was used, followed by Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Network pharmacology analysis to
determine the key targets and core signaling pathways in the
treatment of KOA with RJNTF
Differential genes of KOA and key targets of
RJNTF
A total of 644 significantly altered and affected
differential genes were obtained including 423 upregulated genes
and 221 downregulated genes (Fig.
1A). A total of 45 active ingredients of RJNTF were obtained by
screening using the TCMSP database, corresponding to 234 targets.
There were 29 common targets between KOA and RJNTF as shown by the
Venn diagram in Fig. 1B; the
number of active ingredients corresponding to each drug was 6 for
Achyranthes bidentata Blume, 10 for Glycyrrhiza
uralensis Fisch, 4 for Hansenia weberbaueriana (Fedde ex
H. Wolff) Pime, 3 for Heracleum hemsleyanum Diels, 11 for
Saposhnikovia divaricata (Turcz.) Schischk and 11 for
Angelica sinensis (Oliv) Diels (Fig. 1C).
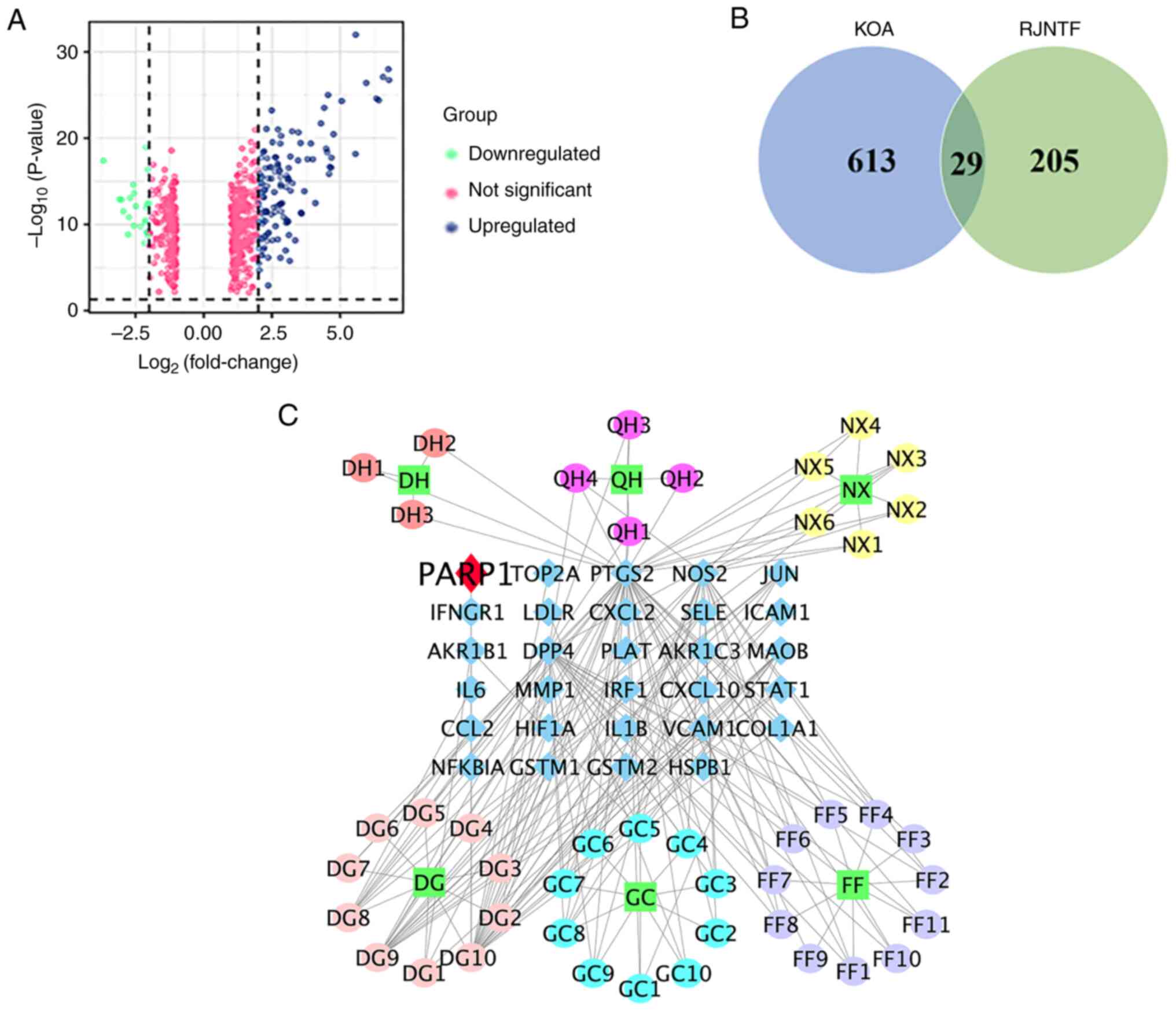 | Figure 1.Identification of targets common to
both RJNTF and KOA. (A) Volcano plot of the differentially
expressed genes in OA. Sample data of the knee joints of patients
with OA from the Gene Expression Omnibus database. Green represents
downregulated genes, blue represents upregulated genes, and pink
represents no significant change. (B) Venn diagram of the genes
common to both KOA and RJNTF. (C) Active ingredients and key
targets of RJNTF. Green squares represent the drug, blue diamonds
represent genes, and circles represent the active ingredient of the
drug; the darker red color represents DH, the rosy red color
represents the active ingredient of QH, the yellow color represents
NX, the pink color represents DG, the azure color represents GC and
the purple color represents FF; RJNTF, Rongjin Niantong Fang; KOA,
knee osteoarthritis; PARP1, poly [ADP-ribose] polymerase-1; DH,
Heracleum hemsleyanum Diels; QH, Hansenia
weberbaueriana (Fedde ex H. Wolff) Pime; NX, Achyranthes
bidentata Blume; DG, Angelica sinensis (Oliv) Diels; GC,
Glycyrrhiza uralensis Fisch; FF, Saposhnikovia
divaricata (Turcz.) Schischk. |
Key targets of RJNTF for the treatment
of KOA
The targets of RJNTF for the treatment of KOA were
imported into Cytoscape to generate the PPI network, and a total of
1,966 related targets and 48,840 target-to-target
interrelationships were obtained; the plugin CytoNCA was further
used to filter the network nodes based on their topological
attributes using the degree centrality value, after which, 531
relevant targets and 2,218 target-target interrelationships were
obtained by excluding irrelevant nodes. Finally, 143 key targets
and 3,600 target-target interrelationships were obtained by
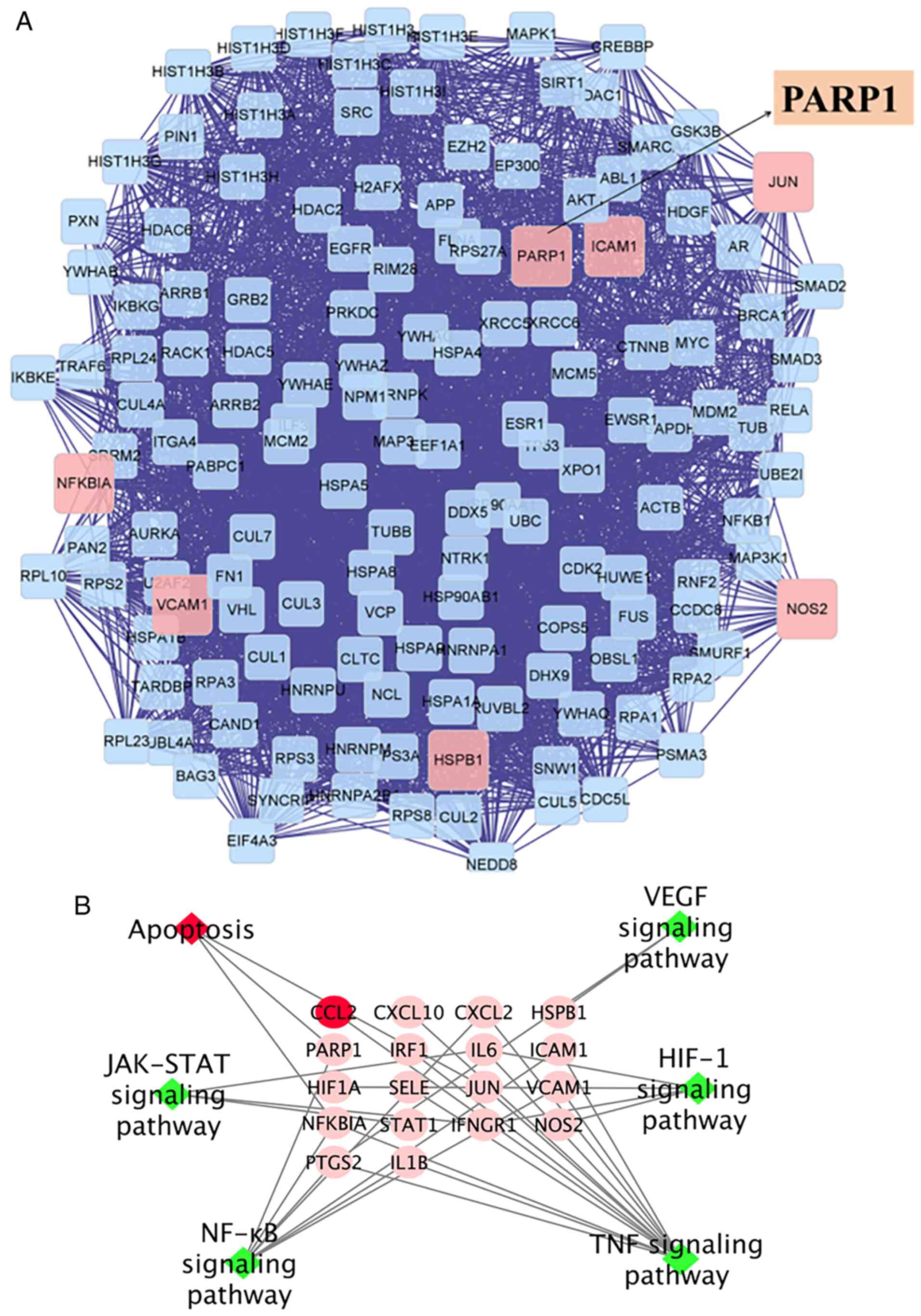
screening with the betweeness centrality value, including STAT3,
PI3K, RAF, RAS, AKT, PARP1 and JALK2, the mechanisms of which
include proliferation, inflammation and apoptosis. After layers of
screening, PARP1, a special apoptosis target, was found among
several targets (Fig. 2A).
Core signaling pathway of RJNTF in
treating KOA
The results of KEGG analysis showed that RJNTF could
treat KOA by interfering with multiple signaling pathways such as
NF-κB, apoptosis, HIF-1 signaling pathway, JAT-STAT signaling
pathway and IL-17 signaling pathway (Fig. 2B) (31–34).
Based on the results of a large number of previous studies showing
that the PARP1/AIF pathway was closely related to apoptosis
(35–37), while the results of network
pharmacology indicated that the treatment of KOA by RJNTF was
related to the PARP1 apoptotic pathway; however, there are no
studies assessing the relationship between the PARP1/AIF pathway
and apoptosis of chondrocytes. Therefore, the subsequent
experiments were designed to ascertain the underlying
mechanism.
Inhibition of chondrocyte apoptosis
through modulation of the PARP1/AIF pathway
Establishment and characterization of an
apoptotic articular chondrocyte cell model
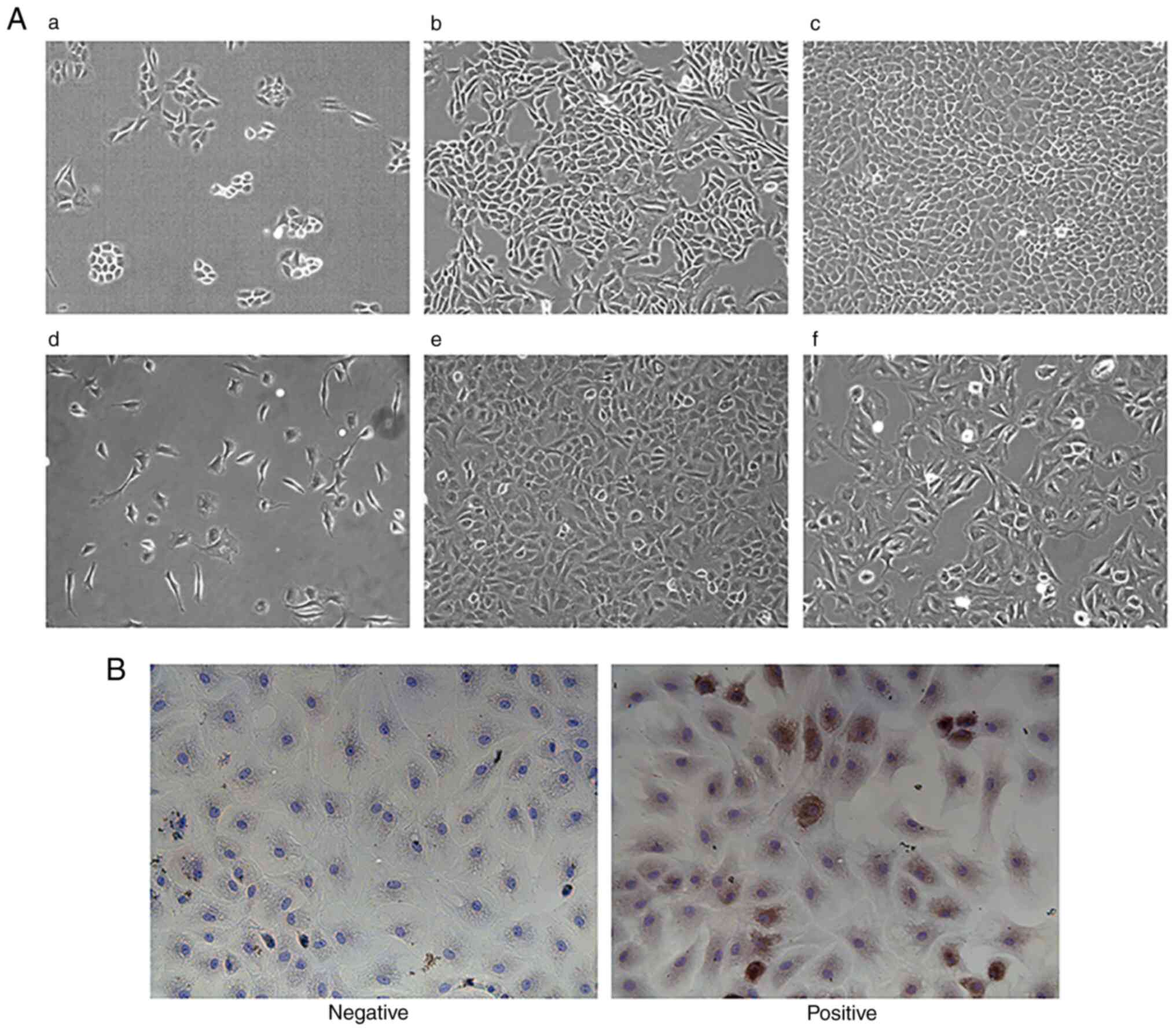
Characterization of normal chondrocytes is shown in
Fig. 3A. In the process of
chondrocyte culture, observed by inverted phase contrast optical
microscope, after 1 day, chondrocytes were completely attached to
the wall and began to divide and grow, and most of the cells were
round or oval. After 7 days of culture, the number of chondrocytes
were markedly increased, and grew in clusters in the shape of
pavements; clusters were connected with each other in a long piece,
and the speed of proliferation was accelerated. When the cells were
grown up to 80% of the density, they could be subcultured to obtain
the F1 chondrocytes. After 3 days of culture, the cytoplasm was
abundant, the nuclei were clearly distinguishable and the edges of
the cells were sharp. After ~4 days of culture, F2 cells could be
obtained by substitution; F2 cells at this time could still
maintain a better morphology, with no obvious tendency of
hypertrophy and degradation, whereas F3 cells appeared to have more
hypertrophic and degraded cells, with overlapping and fuzzy edges,
more pseudopods and a slowed rate of proliferation. Therefore, F2
cells were selected for subsequent cell experiments. In Fig. 3B, Collagen II in the extracellular
matrix of chondrocytes was stained by immunocytochemistry (DAB
method), with the cytoplasm exhibiting a brownish-yellow color and
the nucleus exhibiting a blue color. In the negative group, the
nucleus exhibited a blue color, while no brownish-yellow color was
seen in the cytoplasm. It was concluded that the experimental cells
could synthesize Collagen II and proteoglycan, and had the function
of chondrocytes, so were given this identification. CCK-8 assay
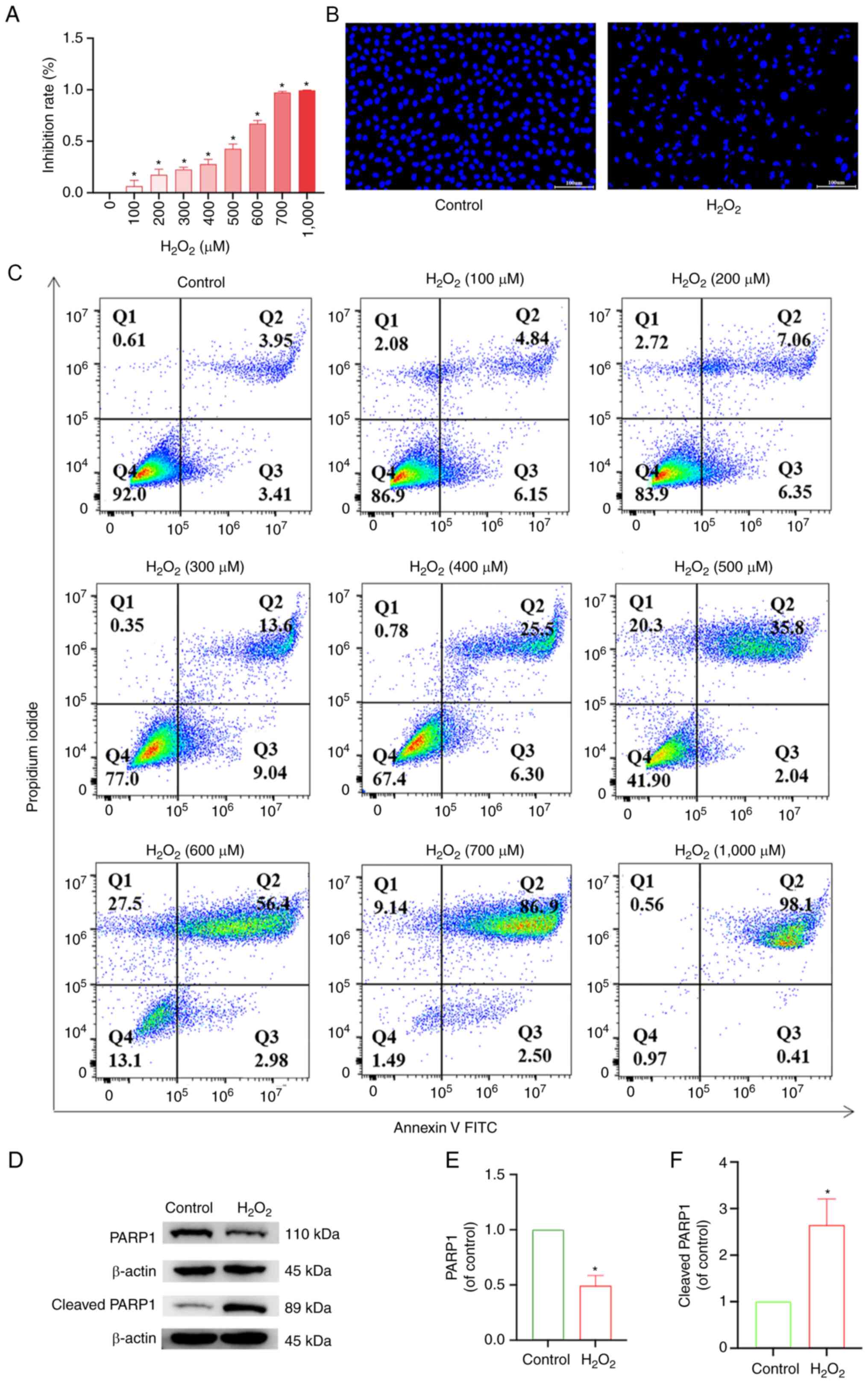
results showed that the inhibition rate of chondrocytes increased
with increasing H2O2 concentration (Fig. 4A). Similarly, flow cytometry
results also showed that the apoptosis rate increased with
increasing H2O2 concentration (Fig. 4C). Subsequently, using DAPI
staining, compared with the control group, the model group treated
with 500 µM H2O2, in the early stage of
apoptosis, exhibited condensation of the nucleus resulting in
uneven DAPI staining of the nucleus due to the aggregation of
chromatin on the side of the nucleus. In the later stages of
apoptosis, the nucleus of the apoptotic cell exhibited round
vesicles of different sizes, which were likely apoptotic vesicles
(Fig. 4B). Western blotting
results showed that compared with the control group, the expression
of PARP1 in the model group was reduced, and the expression of
cleaved PARP1 increased (Fig.
4D-F), which showed that the model of articular chondrocyte
apoptosis was successfully established, and it could be used for
subsequent experiments.
Effect of PARP1 knockdown on the
apoptosis of chondrocytes
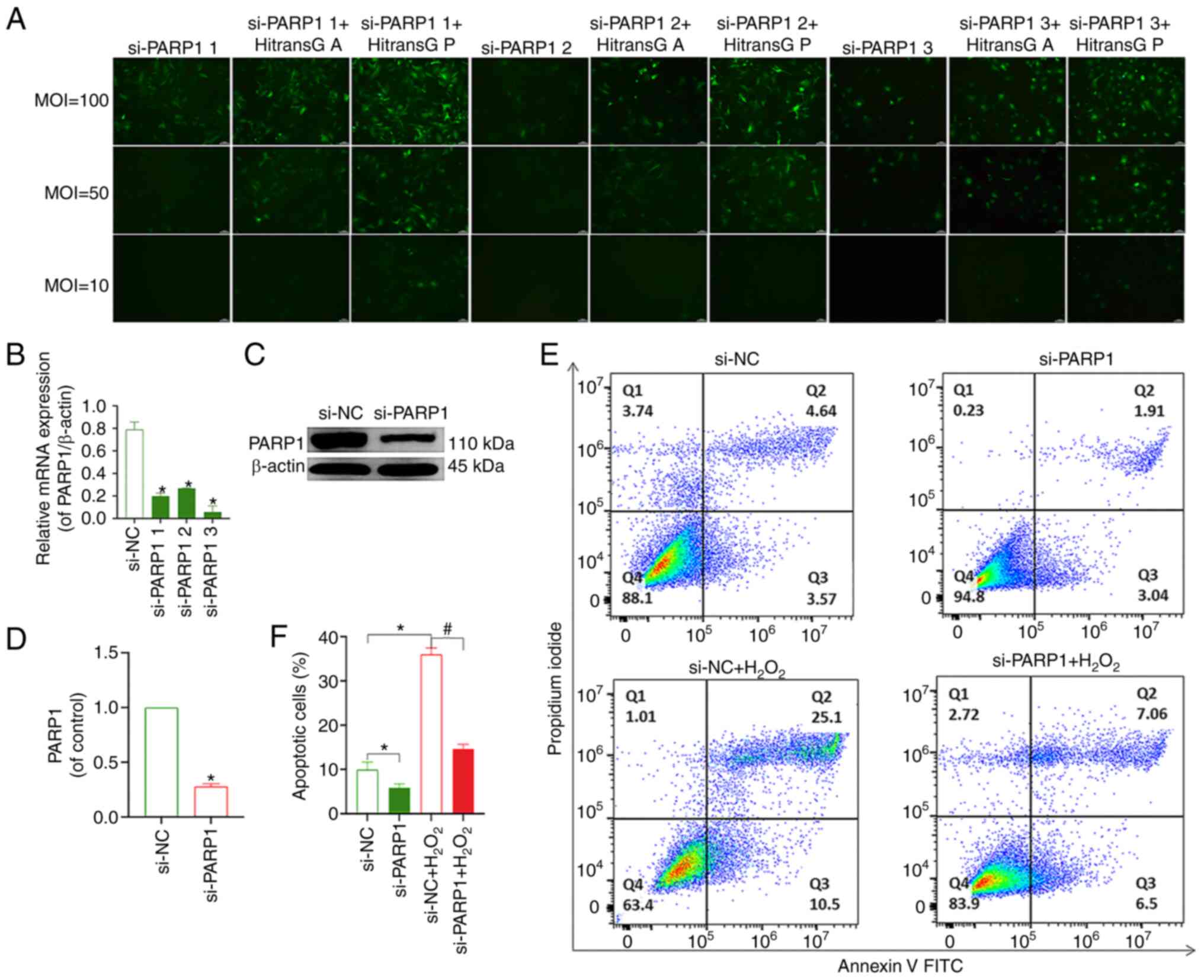
The transfection efficiency of all three viruses
reached >80% when using HiTransG P transfection enhancement
solution and an MOI of 100. The effective transfection conditions
for all three PARP1-knockdown lentiviruses included MOI=100, the
transfection enhancement reagent was HitransG P and the length of
time after which the solution was changed following transfection
was 16 h (Fig. 5A). The results of
RT-qPCR and western blotting showed that PARP1 expression was
lowest in cells transfected with si-PARP1 #3 (Fig. 5B-D); thus, this construct was used
for all subsequent experiments. The flow cytometry results showed
that chondrocyte apoptosis was decreased in the si-PARP1 group
compared with that in the si-NC group. Compared with the si-NC
group, the rate of apoptosis was increased in the si-NC+
H2O2 group. The chondrocyte apoptosis rate
was decreased in the si-PARP1+ H2O2 group
compared with the si-NC+ H2O2 group (Fig. 5E and F). Meanwhile, it was found
that knockdown of PARP1 resulted in a decrease in PARP1, cleaved
PARP1, PAR and nucleus AIF and MIF levels, and an increase in
cytoplasmic AIF and MIF expression (P<0.05; Fig. 6A-I). In conclusion, the knockdown
of PARP1 effectively inhibited chondrocyte apoptosis.
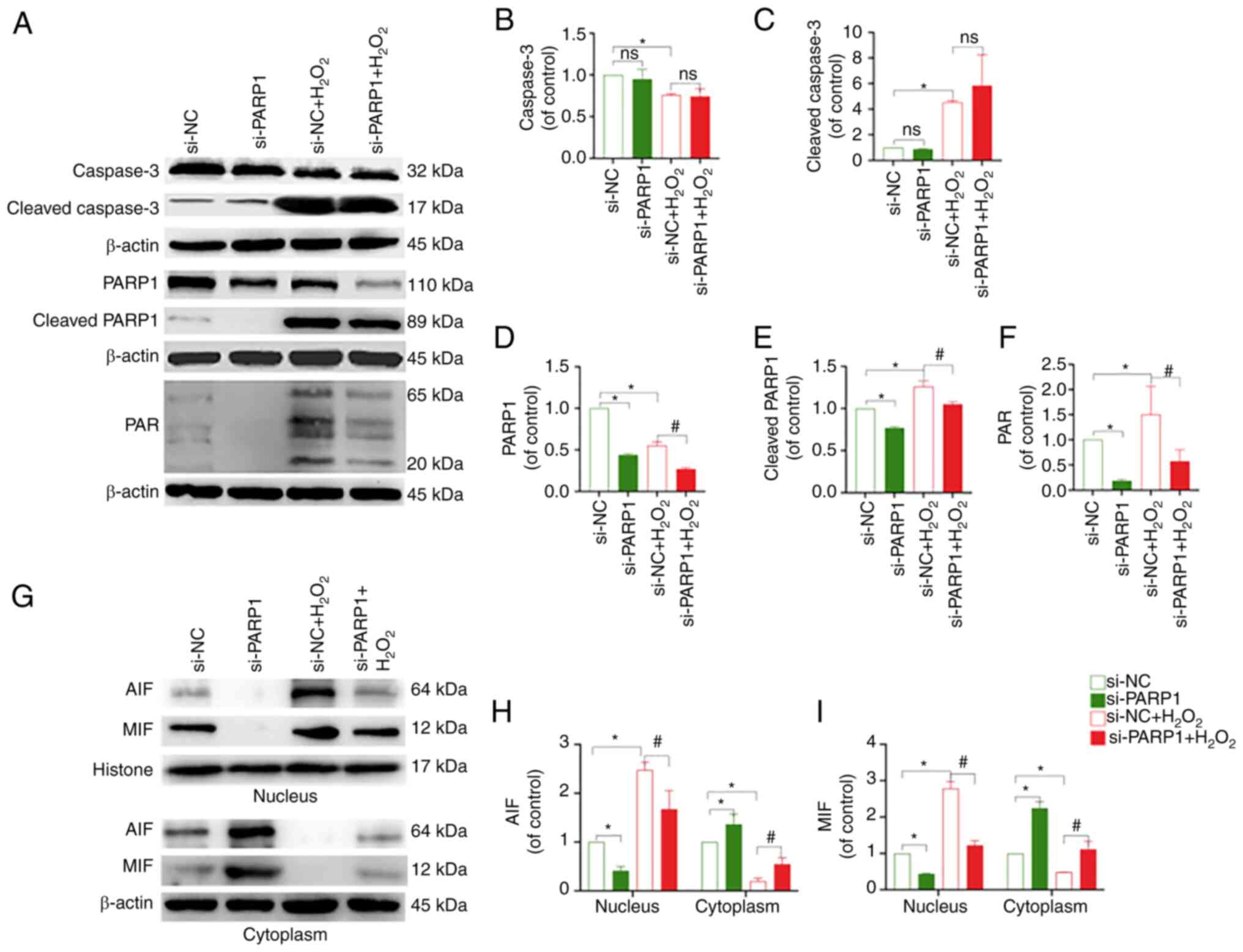 | Figure 6.Effect of PARP1 knockdown on the
expression of PARP1/AIF pathway-related proteins. (A) Protein
expression levels of caspase-3, cleaved caspase-3, PARP1, cleaved
PARP1 and PAR in the chondrocytes in the different treatment
groups. Quantitative analysis of the relative protein expression
levels of (B) caspase-3, (C) cleaved caspase-3, (D) PARP1, (E)
cleaved PARP1 and (F) PAR. *P<0.05 vs. si-NC group;
#P<0.05 vs. si-NC + H2O2 group.
(G) Protein expression of AIF and MIF in the nucleus and cytoplasm
of chondrocytes in the different treatment groups. Quantitative
analysis of the relative protein expression levels of (H) AIF and
(I) MIF. *P<0.05 vs. si-NC group; #P<0.05 vs.
si-NC + H2O2 group. PARP1, poly [ADP-ribose]
polymerase-1; PAR, poly ADP-ribose; AIF, apoptosis inducing factor;
MIF, migration inhibitory factor; ns, not significant; si, small
interfering RNA; NC, negative control; H2O2,
hydrogen peroxide. |
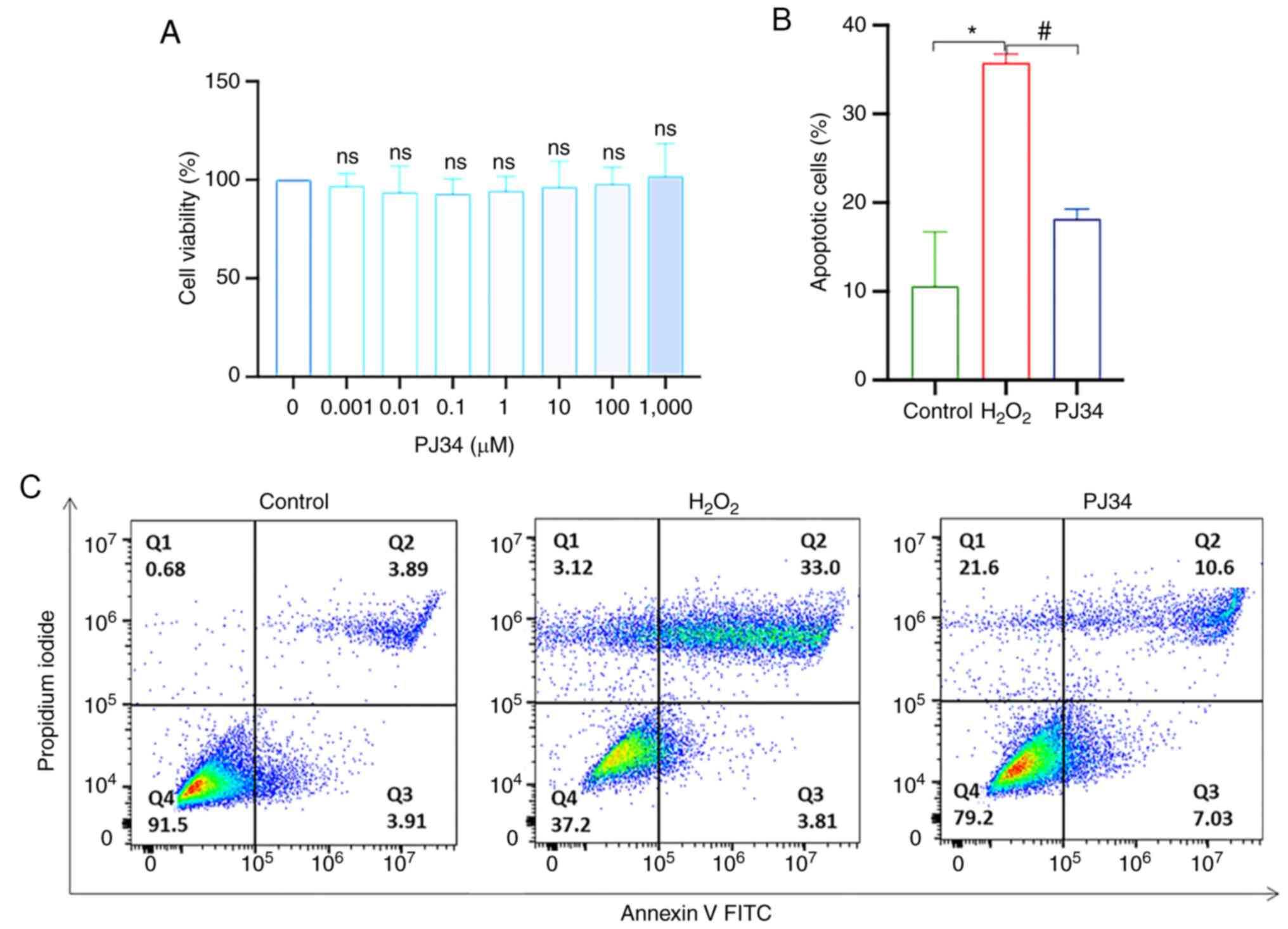
Effect of PJ34-mediated inhibition of
PARP1 on chondrocyte apoptosis
Apoptosis is closely related to the target PARP1,
and although PJ34 is a commonly used PARP1 inhibitor, its
inhibitory effect on chondrocytes is unknown. To determine the
cytotoxic effect of PJ34, chondrocytes were exposed to PJ34 at
different concentrations (0, 0.001, 0.01, 0.1, 1, 10, 100 and 1,000
µM) for 4.5 h. PJ34 had no effect on the activity of chondrocytes
at all tested concentrations (Fig.
7A). Based on a previous study (12), 10 µM PJ34 was selected for
subsequent experiments. Moreover, apoptosis was reduced when cells
were pretreated with PJ34 hydrochloride, suggesting the
PARP1-dependent pathway played a pivotal role in chondrocyte
apoptosis (Fig. 7B and C). To
understand the mechanism of action and determine the impact of PJ34
on chondrocytes, an immunoblot profiling of key factors was
performed. Compared with the model group, the protein expression
levels of PAR and nucleus AIF and MIF were downregulated, while
that of cytoplasmic AIF and MIF was upregulated in the inhibitor
group (P<0.05), and the differences in the protein expression
levels of caspase-3, cleaved caspase-3, PARP1, and cleaved PARP1
were not statistically significant. The results of Co-IP suggested
that AIF interacted with MIF, and that this interaction was reduced
by the addition of PJ34. In summary, inhibition of PARP1
effectively reduced chondrocyte apoptosis (Fig. 8A-K).
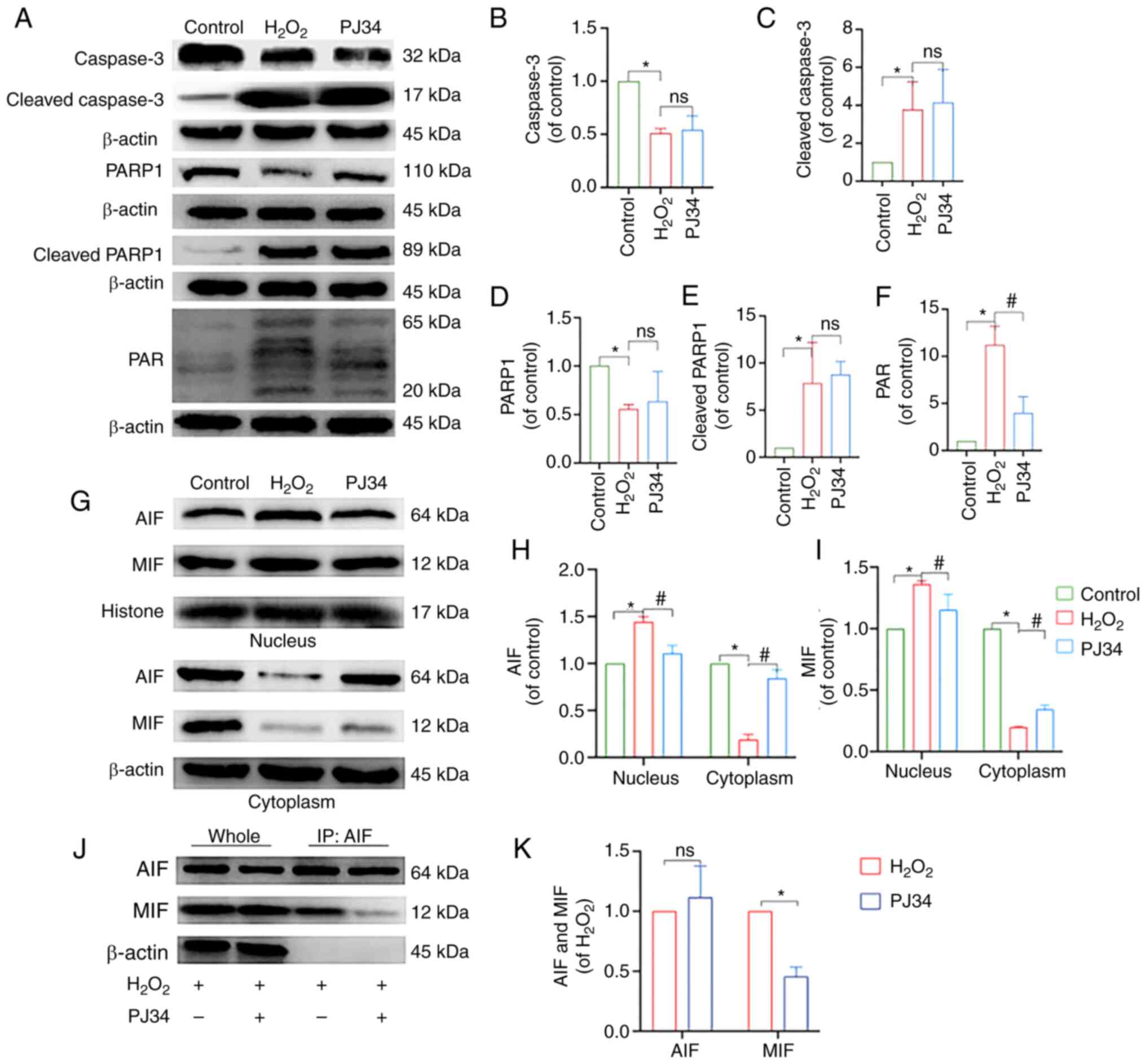 | Figure 8.Effect of PJ34-mediated PARP1
inhibition on the expression of PARP1/AIF pathway-related proteins.
(A) Protein expression levels of caspase-3, cleaved caspase-3,
PARP1, cleaved PARP1 and PAR in chondrocytes in each treatment
group. Quantitative analysis of the relative protein expression
levels of (B) caspase-3, (C) cleaved caspase-3 (D) PARP1, (E)
cleaved PARP1 and (F) PAR. (G) Protein expression levels of AIF and
MIF in the nucleus and cytoplasm of the chondrocytes in the
different treatment groups. Quantitative analysis of the relative
protein expression levels of (H) AIF and (I) MIF. (J) IP analysis
of AIF with MIF, where the immunoprecipitate is AIF. (K) Analysis
of AIF and MIF protein interactions. *P<0.05 vs. control group;
#P<0.05 vs. H2O2 group. PARP1,
poly [ADP-ribose] polymerase-1; PAR, poly ADP-ribose; AIF,
apoptosis inducing factor; MIF, migration inhibitory factor;
H2O2, hydrogen peroxide; ns, not significant;
IP, immunoprecipitation. |
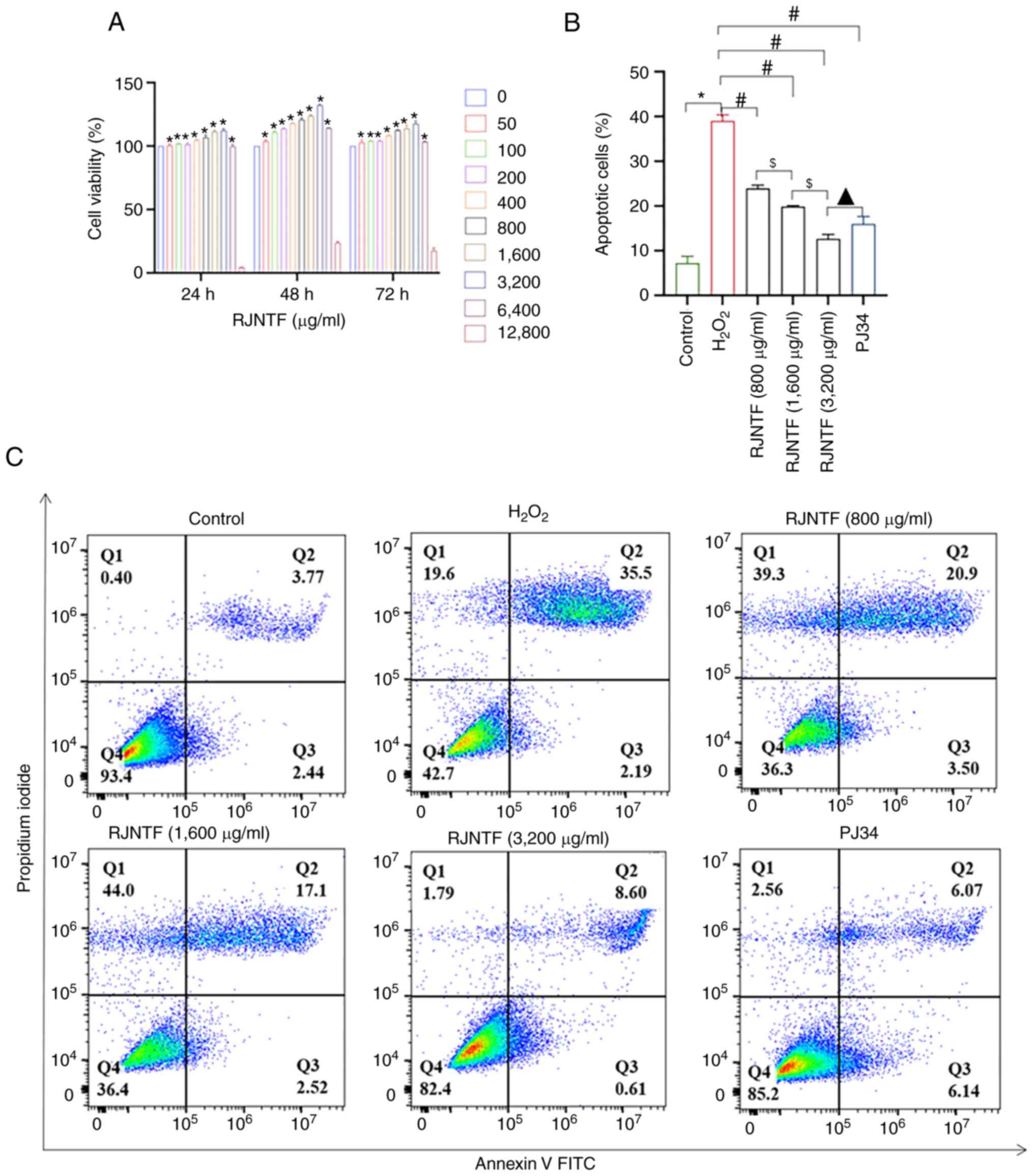
RJNTF inhibits chondrocyte apoptosis
by regulating the PARP1/AIF pathway
As it was aforementioned, chondrocyte apoptosis was
at least partly mediated by the PARP1/AIF pathway. Using CCK-8
assays, 800, 1,600 and 3,200 µg/ml RJNTF were finally selected to
treat cells for 48 h for the subsequent experiments (Fig. 9A). The flow cytometry results
showed that compared with the model group, the apoptotic rate of
chondrocytes was reduced in the RJNTF low, medium and high dose
group, and the apoptotic rate of chondrocytes in the RJNTF high
dose group was lower in the PJ34 treated group (P<0.05; Fig. 9B and C). Western blotting showed
that compared with the model group, the protein expression levels
of cleaved caspase-3, cleaved PARP1, PAR, and nucleus AIF and MIF
were downregulated, and PARP1, caspase-3, and cytoplasmic AIF and
MIF were upregulated in the RJNTF medium and high dose groups
(P<0.05). Compared with the inhibitor group, caspase-3 and PARP1
protein expression levels were upregulated in the RJNTF high dose
groups, and cleaved caspase-3, cleaved PARP1 and protein expression
levels were downregulated in the RJNTF high dose groups (P<0.05;
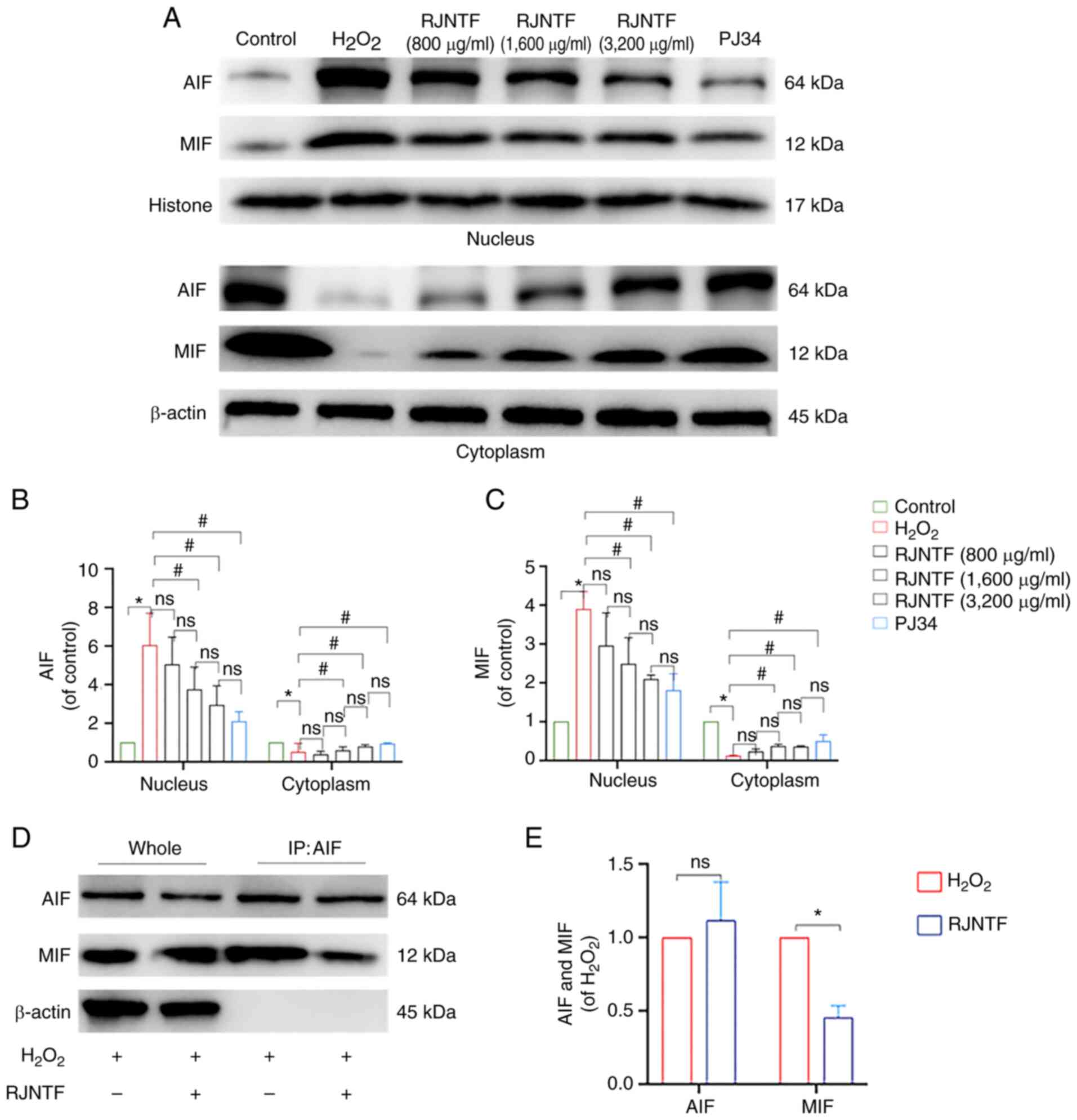
Fig. 10A-F). AIF and MIF protein
expression levels were detected in the precipitates of both the
model group and the RJNTF group, indicating that there was an
interaction between AIF and MIF, and this interaction was reduced
after the addition of RJNTF (Fig.
11A-E). In conclusion, RJNTF could effectively reduce
chondrocyte apoptosis by inhibiting the PARP1/AIF pathway.
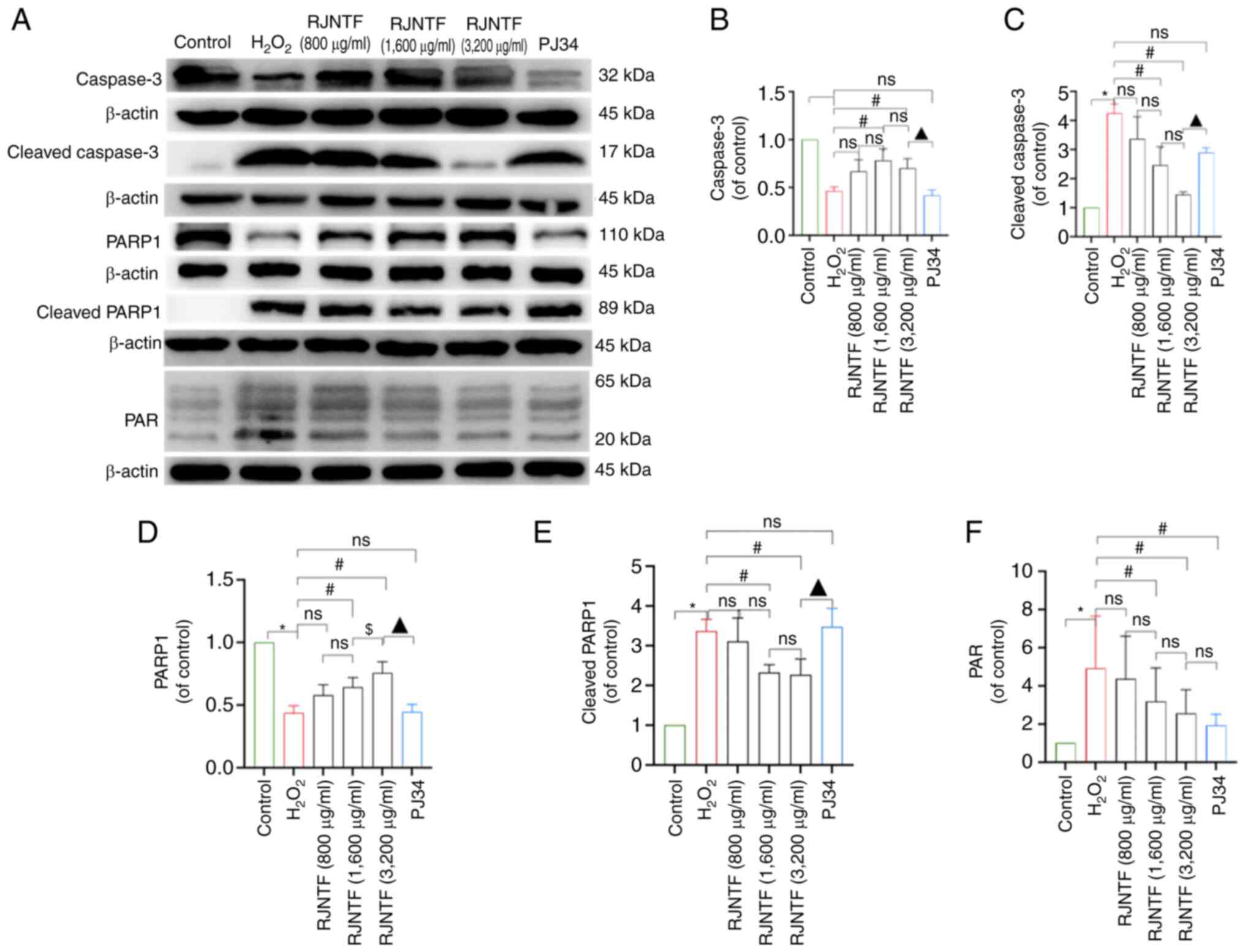 | Figure 10.Effect of RJNTF on the expression of
PARP1/AIF pathway-related proteins. (A) The protein expression
levels of caspase-3, cleaved caspase-3, PARP1, cleaved PARP1, and
PAR in the chondrocytes in the different treatment groups were
determined. Quantitative analysis of the relative protein
expression levels of (B) caspase-3, (C) cleaved caspase-3 (D)
PARP1, (E) cleaved PARP1 and (F) PAR. *P<0.05 vs. control group;
#P<0.05 vs. H2O2 group;
$P<0.05 vs. RJNTF group; ▲P<0.05 vs.
3,200 µg/ml RJNTF. PARP1, poly [ADP-ribose] polymerase-1; PAR, poly
ADP-ribose; AIF, apoptosis inducing factor;
H2O2, hydrogen peroxide. |
Discussion
Network pharmacology analysis of protein
interactions showed that RJNTF exerted its beneficial effects on
KOA via regulation of STAT3, PI3K, RAF, RAS, AKT, PARP1, JALK2 and
PARP1. Sun et al (38)
found that TANK-binding kinase 1 (TBK1) was expressed at high
levels in KOA by constructing an animal model of OA in C57BL/6 J
mice. TBK1 activated the JAK/STAT signaling pathway, and knockdown
of TBK1 inhibited extracellular matrix degradation. The beneficial
effect of TBK1 knockdown was abrogated by transfecting cells with a
STAT3 overexpression plasmid. AKT1 is an important downstream
target kinase in the PI3K/AKT signaling pathway, which can regulate
the mTOR signaling pathway and other pathways, IL-1β-induced
chondrocyte proliferation, and can reduce apoptosis (39). It has been shown that Ras
independently inhibits the activation of Erk1/2. This effect is
associated with reduced activation of transcription factors such as
Elk-1, which inhibits cartilage damage by suppressing major
catabolic factors involved in KOA cartilage degradation processes
(40). The aforementioned results
suggest that the mechanism of KOA action includes proliferation,
inflammation and apoptosis (41–44).
KEGG analysis showed that RJNTF exerts its effects
on KOA by interfering with multiple signaling pathways such as the
NF-κB pathway, apoptotic pathways, the HIF-1 signaling pathway, the
JAT-STAT signaling pathway, the IL-17 signaling pathway and other
signaling pathways to treat knee joints (45–48).
NF-κB is closely related to inflammation, and network pharmacology
combined with in vitro experiments confirmed that the
mechanism of action of Fengshi Gutong Capsule in treating KOA was
associated with the NF-κB signaling pathway (49). It has been noted that maintaining a
hypoxic environment in the subchondral bone alleviates
osteoarthritis progression. HIFs are core regulators that induce
hypoxia genes, repair the cellular oxygen environment and play an
important role in the treatment of OA (50,51).
It has been shown that the IL-17 signaling pathway is highly
regulated in synovial tissues of patients with KOA, suggesting that
the IL-17 signaling pathway has a better transcriptional response
in KOA chondrocytes and synovial tissues, which can induce
osteoclastogenesis and bone erosion in arthritic diseases;
therefore, the IL-17 signaling pathway may be a potential target
gene for the treatment and diagnosis of KOA (52,53).
Several pathways play a regulatory role in KOA; among them, PARP1
is closely related to apoptosis (37,54).
For example, chaperone-mediated autophagy can inhibit cardiomyocyte
apoptosis induced by oxidative stress by inhibiting cleaved PARP1
protein expression (55); aurora
kinase A knockdown promotes apoptosis in Epstein-Barr
virus-infected atypical glandular cells through cleavage of
caspase-3 and −9, and PARP1 (56).
Additionally, the colocalization of PARP-1/AIF in the nucleus
indicates that PARP-1 plays a pivotal role in AIF-mediated carbon
ion pro-apoptosis (14).
Therefore, it is hypothesized that RJNTF may exert an inhibitory
effect on chondrocyte apoptosis through regulation of the PARP1/AIF
pathway, and the subsequent experiments were designed to test this
hypothesis.
PARP1 is a DNA repair enzyme expressed in most
eukaryotic cells, activated by recognizing structurally damaged DNA
fragments and is considered to be a DNA damage, and is also a
cleavage substrate for the apoptotic core member protein caspase
(57). The primary function of
PARP1 is to sense and repair DNA breaks (48). PARP1 binds to DNA strand breaks and
then transfers PAR multimers to PARP1 itself, which recruits the
DNA repair machinery to DNA damage. PARP1 catalyzes the formation
of PAR on histones, which allows it to induce chromatin relaxation
in chromatin to increase the viability of the DNA repair machinery
to DNA breaks. Thus, PARP1 is activated by activated caspase-3 to
inhibit these processes and activate apoptosis when the extent of
damage is too large. In the present study, it was found that in
chondrocytes, after PARP1-bound DNA strand breaks, activated
caspase-3 can cleave PARP1, and the 89-kDa PARP1 fragment is
released from the damaged DNA as it can no longer bind the 24-kDa
PARP1 fragment to DNA. At the same time, initiation of PAR
synthesis results in the release of AIF, translocation to the
nucleus in combination with macrophage MIF, and the action of DNA
endonuclease and nucleic acid exonuclease G results in chromatin
condensation as well as DNA breakage (36). It was found that this type of
apoptosis could be inhibited by the PARP1 inhibitor PJ34, but the
protein expression levels of PARP1 and cleaved PARP1 were not
altered after the addition of PJ34, while PAR expression was
notably downregulated, and the phenomenon of AIF and MIF nuclear
translocation had been suppressed. Thus, PJ34 further inhibited
apoptosis caused by AIF entry into the nucleus by inhibiting the
synthesis of PAR.
The flow cytometry results indicated that RJNTF
could inhibit chondrocyte apoptosis by interfering with the
PARP1/AIF pathway. However, with the addition of RJNTF, although
the expression of cleaved caspase-3, cleaved PARP1 and nucleus AIF
and MIF decreased and that of caspase-3, PARP1 and cytoplasmic AIF
and MIF increased, it did not show a dose-dependent effect with
increasing concentrations of RJNTF. Combining the results of flow
cytometry, the apoptosis rate of the RJNTF high dose group was
lower than that of the PJ34 group; a significant difference which
suggests that the use of H2O2 modeling not
only affected the apoptotic pathway of PARP1/AIF, but also the
involvement of other apoptotic pathways. RJNTF can not only inhibit
apoptosis by interfering with the PARP1/AIF pathway, but also other
pathways to inhibit apoptosis, and thus play a more potent role in
inhibiting apoptosis compared with a single inhibitor. Once again,
this provides a reliable experimental basis for the multi-targeting
and multi-pathway effect of RJNTF in the treatment of KOA.
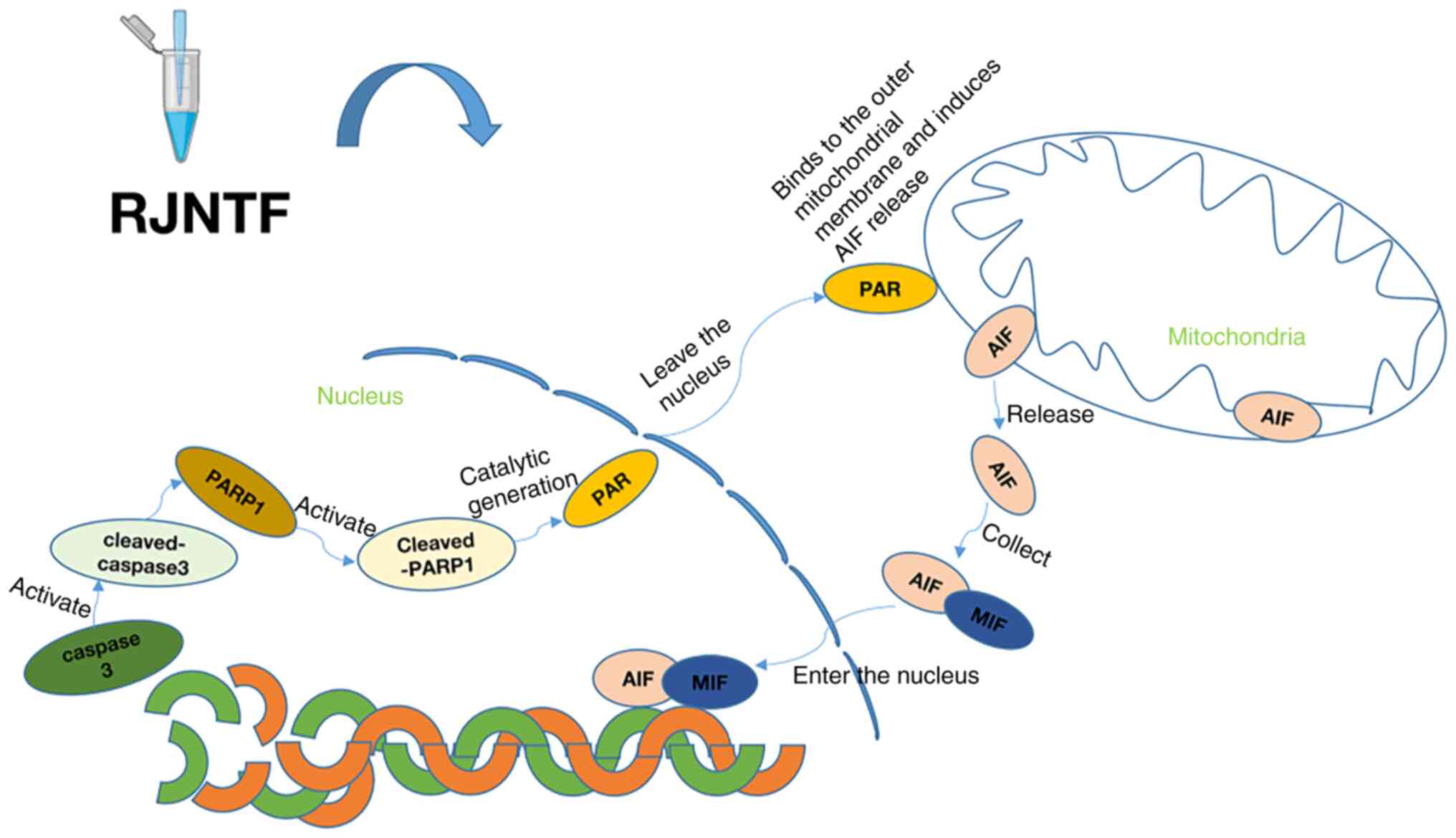
In conclusion, the present study demonstrated that
RJNTF reduces H2O2-induced chondrocyte
apoptosis via the PARP1/AIF signaling pathway (Fig. 12). These findings highlight novel
avenues for understanding the therapeutic mechanism of RJNTF, and
provide a reference for further exploration and clinical
application of RJNTF. However, it is noteworthy that a limitation
of the present study is that it is an in vitro study where
RJNTF is directly contacted with chondrocytes. This may not reflect
the situation in vivo since RJNTF is taken orally and is not
clear if all the components of RJNTF directly contact the
chondrocytes after oral administration; this will be investigated
further in future studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82074465), the Natural Science
Foundation of Fujian Province (grant no. 2022J01843), the Key
Laboratory of Integrative Medicine of Fujian Province University
(Fujian University of Traditional Chinese Medicine; grant nos.
KLIM2022003 and CKJ2022003), the NATCM's Project of High-level
Construction of Key TCM Disciplines (Traditional Chinese
Orthopedics) (grant no. zyyzdxk-2023106) and the Traditional
Chinese Orthopedics Open subject of FJTCM (grant no.
XGS2023004).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JC and TZ conducted the majority of the experiments.
YD and ZC assisted with the immunofluorescence experiments. CZ and
XC performed the flow cytometry. RW and QL performed the
statistical analysis, and wrote, reviewed and edited the original
manuscript. JC and TZ revised the manuscript. JC, TZ, QL and RW
designed the study. GW conceived and designed the study, and wrote,
reviewed and edited the original manuscript. RW, YD and GW confirm
the authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All animals received humane care and the
experimental methods were approved by the Animal Care and Use
Committee of Fujian University of Traditional Chinese Medicine
(Fuzhou, China; approval no. FJTCM IACUC 2022044).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ramírez-Noguera P, Marín IZ, Chavarin BM,
Valderrama ME, López-Barrera LD and Díaz-Torres R: Study of the
early effects of chitosan nanoparticles with glutathione in rats
with osteoarthrosis. Pharmaceutics. 15:21722023. View Article : Google Scholar
|
|
2
|
Rousseau JC, Sornay-Rendu E, Bertholon C,
Garnero P and Chapurlat R: Serum periostin is associated with
prevalent knee osteoarthritis and disease incidence/progression in
women: The OFELY study. Osteoarthritis Cartilage. 23:1736–1742.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
GBD 2015, . Disease and Injury Incidence
and Prevalence Collaborators: Global, regional, and national
incidence, prevalence, and years lived with disability for 310
diseases and injuries, 1990–2015: A systematic analysis for the
global burden of disease study 2015. Lancet. 388:1545–1602. 2016.
View Article : Google Scholar
|
|
4
|
Yu D, Peat G, Bedson J and Jordan KP:
Annual consultation incidence of osteoarthritis estimated from
population-based health care data in England. Rheumatology
(Oxford). 54:2051–2060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rewald S, Lenssen AFT, Emans PJ, de Bie
RA, van Breukelen G and Mesters I: Aquatic cycling improves knee
pain and physical functioning in patients with knee osteoarthritis:
A randomized controlled trial. Arch Phys Med Rehabil.
101:1288–1295. 2020. View Article : Google Scholar
|
|
6
|
Mahmoudian A, Lohmander LS, Mobasheri A,
Englund M and Luyten FP: Early-stage symptomatic osteoarthritis of
the knee-time for action. Nat Rev Rheumatol. 17:621–632. 2021.
View Article : Google Scholar
|
|
7
|
Godziuk K, Prado CM, Woodhouse LJ and
Forhan M: The impact of sarcopenic obesity on knee and hip
osteoarthritis: A scoping review. BMC Musculoskelet Disord.
19:2712018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang G, Wen X, Jiang Z, Du X, Liu R, Zhang
C, Huang G, Liao W and Zhang Z: FUNDC1/PFKP-mediated mitophagy
induced by KD025 ameliorates cartilage degeneration in
osteoarthritis. Mol Ther. 31:3594–3612. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song J, Kim EH, Yang JH, Kim D, Robby AI,
Kim SA, Park SY, Ryu JH and Jin EJ: Upregulated FOXM1 stimulates
chondrocyte senescence in Acot12-/-Nudt7-/- double knockout mice.
Theranostics. 13:5207–5222. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen C, Yin P, Hu S, Sun X and Li B:
Circular RNA-9119 protects IL-1β-treated chondrocytes from
apoptosis in an osteoarthritis cell model by intercepting the
microRNA-26a/PTEN axis. Life Sci. 1:1179242020. View Article : Google Scholar
|
|
11
|
Huang P, Chen G, Jin W, Mao K, Wan H and
He Y: Molecular mechanisms of parthanatos and its role in diverse
diseases. Int J Mol Sci. 23:72922022. View Article : Google Scholar
|
|
12
|
Mashimo M, Onishi M, Uno A, Tanimichi A,
Nobeyama A, Mori M, Yamada S, Negi S, Bu X, Kato J, et al: The
89-kDa PARP1 cleavage fragment serves as a cytoplasmic PAR carrier
to induce AIF-mediated apoptosis. J Biol Chem. 296:1000462021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koehler RC, Dawson VL and Dawson TM:
Targeting parthanatos in ischemic stroke. Front Neurol.
12:6620342021. View Article : Google Scholar
|
|
14
|
Xu X, Sun B and Zhao C: Poly (ADP-Ribose)
polymerase 1 and parthanatos in neurological diseases: From
pathogenesis to therapeutic opportunities. Neurobiol Dis.
15:1063142023. View Article : Google Scholar
|
|
15
|
Zhang Y, Yang X, Ge X and Zhang F:
Puerarin attenuates neurological deficits via Bcl-2/Bax/cleaved
caspase-3 and Sirt3/SOD2 apoptotic pathways in subarachnoid
hemorrhage mice. Biomed Pharmacother. 109:726–733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Q, Zhou X, Yang L, Zhao Y, Chew Z,
Xiao J, Liu C, Zheng X, Zheng Y, Shi Q, et al: The natural compound
notopterol binds and targets JAK2/3 to ameliorate inflammation and
arthritis. Cell Rep. 32:1081582020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zada S, Pham TM, Hwang JS, Ahmed M, Lai
TH, Elashkar O, Kim JH, Kim DH and Kim DR: Chlorogenic acid
protects human chondrocyte C28/I2 cells from oxidative
stress-induced cell death through activation of autophagy. Life
Sci. 285:1199682021. View Article : Google Scholar
|
|
18
|
Jia C, Hu F, Lu D, Jin H, Lu H, Xue E and
Wu D: Formononetin inhibits IL-1β-induced inflammation in human
chondrocytes and slows the progression of osteoarthritis in rat
model via the regulation of PTEN/AKT/NF-κB pathway. Int
Immunopharmacol. 113:1093092022. View Article : Google Scholar
|
|
19
|
Comblain F, Dubuc JE, Lambert C, Sanchez
C, Lesponne I, Serisier S and Henrotin Y: Identification of targets
of a new nutritional mixture for osteoarthritis management composed
by curcuminoids extract, hydrolyzed collagen and green tea extract.
PLoS One. 11:e01569022016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
20:e472015. View Article : Google Scholar
|
|
21
|
R Core Team (2012), . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: ISBN 3-900051-07-0, URL. http://www.R-project.org/
|
|
22
|
RStudio Team (2015), . RStudio. RStudio,
Inc.; Boston, MA: URL. http://www.rstudio.com/
|
|
23
|
Martin A, Ochagavia ME, Rabasa LC, Miranda
J, Fernandez-de-Cossio J and Bringas R: BisoGenet: A new tool for
gene network building, visualization and analysis. BMC
Bioinformatics. 11:912010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M: ‘Post-genome Informatics’.
Oxford University Press; 2000, View Article : Google Scholar
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J, Chen N, Zhang T, Lin J, Huang Y
and Wu G: Rongjin Niantong Fang ameliorates cartilage degeneration
by regulating the SDF-1/CXCR4-p38MAPK signalling pathway. Pharm
Biol. 60:2253–2265. 2022. View Article : Google Scholar
|
|
28
|
Li X, Xu Y, Li H, Jia L, Wang J, Liang S,
Cai A, Tan X, Wang L, Wang X, et al: Verification of pain-related
neuromodulation mechanisms of icariin in knee osteoarthritis.
Biomed Pharmacother. 144:1122592021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin J, Wu G, Chen J, Fu C, Hong X, Li L,
Liu X and Wu M: Electroacupuncture inhibits sodium
nitroprusside-mediated chondrocyte apoptosis through the
mitochondrial pathway. Mol Med Rep. 18:4922–4930. 2018.PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xia GQ, Zhu MP, Li JW and Huang H: An
alkaloid from Menispermum dauricum, dauricine mediates Ca influx
and inhibits NF-κB pathway to protect chondrocytes from
IL-1β-induced inflammation and catabolism. J Ethnopharmacol.
321:1175602024. View Article : Google Scholar
|
|
32
|
Zhang H, Wang L, Cui J, Wang S, Han Y,
Shao H, Wang C, Hu Y, Li X, Zhou Q, et al: Maintaining hypoxia
environment of subchondral bone alleviates osteoarthritis
progression. Sci Adv. 9:eabo78682023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun C, Peng S, Lv Z, Guo T and Zhang L:
Research of STEAP3 interaction with Rab7A and RACK1 to modulate the
MAPK and JAK/STAT signaling in osteoarthritis. Int Immunopharmacol.
124:1110342023. View Article : Google Scholar
|
|
34
|
Xiao J, Zhang P, Cai FL, Luo CG, Pu T, Pan
XL and Tian M: IL-17 in osteoarthritis: A narrative review. Open
Life Sci. 18:202207472023. View Article : Google Scholar
|
|
35
|
Raghavan A and Shah ZA: Withania somnifera
improves ischemic stroke outcomes by attenuating PARP1-AIF-mediated
caspase-independent apoptosis. Mol Neurobiol. 52:1093–1105. 2015.
View Article : Google Scholar
|
|
36
|
Wang Z, Qiu Z, Hua S, Yang W, Chen Y,
Huang F, Fan Y, Tong L, Xu T, Tong X, et al: Nuclear Tkt promotes
ischemic heart failure via the cleaved Parp1/Aif axis. Basic Res
Cardiol. 117:182022. View Article : Google Scholar
|
|
37
|
Abulikemu A, Zhao X, Qi Y, Liu Y, Wang J,
Zhou W, Duan H, Li Y, Sun Z and Guo C: Lysosomal
impairment-mediated autophagy dysfunction responsible for the
vascular endothelial apoptosis caused by silica nanoparticle via
ROS/PARP1/AIF signaling pathway. Environ Pollut. 304:1192022022.
View Article : Google Scholar
|
|
38
|
Sun P and Xue Y: Silence of TANK-binding
kinase 1 (TBK1) regulates extracellular matrix degradation of
chondrocyte in osteoarthritis by janus kinase (JAK)-signal
transducer of activators of transcription (STAT) signaling.
Bioengineered. 13:1872–1879. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang Z, Shi X, Li X, Zhang L, Wu P, Mao
J, Xing R, Zhang N and Wang P: Network pharmacology approach to
uncover the mechanism governing the effect of simiao powder on knee
osteoarthritis. Biomed Res Int. 2020:69715032020. View Article : Google Scholar
|
|
40
|
Boileau C, Martel-Pelletier J, Brunet J,
Schrier D, Flory C, Boily M and Pelletier JP: PD-0200347, an
alpha2delta ligand of the voltage gated calcium channel, inhibits
in vivo activation of the Erk1/2 pathway in osteoarthritic
chondrocytes: A PKCalpha dependent effect. Ann Rheum Dis.
65:573–580. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xue JF, Shi ZM, Zou J and Li XL:
Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of
articular chondrocytes and attenuates inflammatory response in rats
with osteoarthritis. Biomed Pharmacother. 89:1252–1261. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu K, He Y, Moqbel SAA, Zhou X, Wu L and
Bao J: SIRT3 ameliorates osteoarthritis via regulating chondrocyte
autophagy and apoptosis through the PI3K/Akt/mTOR pathway. Int J
Biol Macromol. 175:351–360. 2021. View Article : Google Scholar
|
|
43
|
Chevalley T, Brandi ML, Cashman KD,
Cavalier E, Harvey NC, Maggi S, Cooper C, Al-Daghri N, Bock O,
Bruyèreet O, et al: Role of vitamin D supplementation in the
management of musculoskeletal diseases: Update from an European
society of clinical and economical aspects of osteoporosis,
osteoarthritis and musculoskeletal diseases (ESCEO) working group.
Aging Clin Exp Res. 34:2603–2623. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hou SM, Chen PC, Lin CM, Fang ML, Chi MC
and Liu JF: CXCL1 contributes to IL-6 expression in osteoarthritis
and rheumatoid arthritis synovial fibroblasts by CXCR2, c-Raf,
MAPK, and AP-1 pathway. Arthritis Res Ther. 22:2512020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang XA and Kong H: Mechanism of HIFs in
osteoarthritis. Front Immunol. 14:11687992023. View Article : Google Scholar
|
|
46
|
Bauer C, Moser LB, Kern D, Jeyakumar V and
Nehreret S: The combination of glucocorticoids and hyaluronic acid
enhances efficacy in IL-1β/IL-17-treated bovine osteochondral
grafts compared with individual application. Int J Mol Sci.
24:143882023. View Article : Google Scholar
|
|
47
|
Luo P, Zhao T and He H: IL-38-mediated
NLRP3/caspase-1 inhibition is a disease-modifying treatment for TMJ
inflammation. Ann N Y Acad Sci. 1508:92–104. 2022. View Article : Google Scholar
|
|
48
|
Lepetsos P, Papavassiliou KA and
Papavassiliou AG: Redox and NF-κB signaling in osteoarthritis. Free
Radic Biol Med. 132:90–100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun Y, Liu J, Wang J, He M, Chen X and
Chen L: Network pharmacology integrated with experimental
validation revealed the mechanism of Fengshi Gutong capsule in the
treatment of osteoarthritis. J Ethnopharmacol. 319:1172612024.
View Article : Google Scholar
|
|
50
|
Li X, Mei W, Huang Z, Zhang L, Zhang L, Xu
B, Shi X, Xiao Y, Ma Z, Liao T, et al: Casticin suppresses
monoiodoacetic acid-induced knee osteoarthritis through inhibiting
HIF-1α/NLRP3 inflammasome signaling. Int Immunopharmacol.
86:1067452020. View Article : Google Scholar
|
|
51
|
Zhang H, Wang L, Cui J, Wang S, Han Y,
Shao H, Wang C, Hu Y, Li X, Zhou Q, et al: Maintaining hypoxia
environment of subchondral bone alleviates osteoarthritis
progression. Sci Adv. 9:eabo78682023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mimpen JY, Baldwin MJ, Cribbs AP, Philpott
M, Carr AJ, Dakin SG and Snelling SJB: Interleukin-17A causes
osteoarthritis-like transcriptional changes in human
osteoarthritis-derived chondrocytes and synovial fibroblasts in
vitro. Front Immunol. 12:6761732021. View Article : Google Scholar
|
|
53
|
Liu SC, Hsieh HL, Tsai CH, Fong YC, Ko CY,
Wu HC, Chang SL, Hsu CJ and Tang CH: CCN2 facilitates IL-17
production and osteoclastogenesis in human osteoarthritis synovial
fibroblasts by inhibiting miR-655 expression. J Bone Miner Res.
37:1944–1955. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alemasova EE and Lavrik OI: Poly
(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory
proteins. Nucleic Acids Res. 47:3811–3827. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang D, Lai W, Liu Y, Wan R and Shen Y:
Chaperone-mediated autophagy attenuates
H2O2-induced cardiomyocyte apoptosis by
targeting poly (ADP-ribose) polymerase 1 (PARP1) for lysosomal
degradation. Cell Biol Int. 46:1915–1926. 2022. View Article : Google Scholar
|
|
56
|
Varshney N, Murmu S, Baral B, Kashyap D,
Singh S, Kandpal M, Bhandari V, Chaurasia A, Kumar S and Jha HC:
Unraveling the Aurora kinase A and Epstein-Barr nuclear antigen 1
axis in Epstein Barr virus associated gastric cancer. Virology.
588:1099012023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sonar SA, Meitei HT, Karmakar S, Mishra A,
Inamdar S, Lenka N and Lal G: Th17 cell promotes apoptosis of
IL-23R neurons in experimental autoimmune encephalomyelitis. Clin
Immunol. 259:1098982024. View Article : Google Scholar
|