Introduction
During hematopoiesis, hematopoietic stem cells
(HSCs) self-renew and give rise to progenitors and their mature
lymphoid and myeloid linages. This maturation process is controlled
by internal and external cues and mediated by specific
transcription factors in blood progenitor cells (1,2).
Extensive studies have been carried out to uncover the function of
hematopoietic transcription factors and their impact on normal
development and disease (1,2).
Red blood cells and megakaryocytes arise from a
common precursor, designated megakaryocyte-erythroid progenitor
(MEP) (3). Commitment of MEPs to
the erythroid/megakaryocytic lineage takes place within the bone
marrow microenvironment under the influence of multiple regulatory
factors. Protein-protein interaction of heptad transcription
factors (TFs) friend leukemia integration factor 1 (Fli-1), GATA
binding protein (GATA)1, GATA2, runt-related transcription factor 1
(RUNX1), T-Cell acute lymphocytic leukemia 1 (TAL1) in MEP cells
controls linage specific erythroid and megakaryocytic
differentiation (4). In the
combinatorial binding of heptad factors in bulk human hematopoietic
stem progenitor cells (HSPCs), individual progenitors and cell
lines reveal cell-specific changes in the regulatory architecture
of these transcription factors during lineage specific
differentiation (5–7).
Fli-1 was first identified as the integration sites
of provirus, involved in transformation of erythroid cells by
friend murine leukemia virus (F-MuLV) (8,9).
Fli-1 downregulation in erythroleukemic cells promotes erythroid
differentiation (10–12), whereas overexpression or
drug-mediated activation of Fli-1 induces differentiation of MEP to
megakaryocytic cells (13,14). This is consistent with the fact
that in zebra fish FLI1 acts at the top of the transcriptional
network driving both blood and endothelial development (15). In the present study, FLI1 was found
upstream of Gata2, Stem cell leukemia/Tal1), LIM-onlyprotein2
(Lmo2) and Zebrafish ets-related protein (Etsrp). The expression of
the Fli-1 related erythroblast transformation-specific-related gene
(ERG) has also been implicated in erythroid and
megakaryocytic differentiation (16). However, the association between
these two genes during the entire maturation process of MEP has not
yet been investigated. Studies also implicate the LIM domain
binding 1 (LDB1) in the regulation of erythroid differentiation
(17–23). Chromatin immunoprecipitation
analysis reveals that most DNA bound murine Gata1 and Tal1 proteins
are contained within higher order complexes (Ldb1-complexes) that
include the nuclear adapters Ldb1 and Lmo2 (17). Notably, FLI1 in complex with LDB1
was recently shown to regulate megakaryocytic gene expression
through interaction with GATA1 in murine erythroleukemic cells
(24). LDB1 is deemed to act as a
scaffold protein that brings Fli-1 to proximity of Gata1 though DNA
looping, leading to activation of megakaryocytic genes (24).
Previous studies identified GATA1 as a direct target
of FLI1 and its expression is suppressed by this TF (12,25).
However, the role of GATA2 in erythroid or megakaryocytic
differentiation is not fully understood. FLI1 was shown in the
present study to bind the GATA2 promoter and activates its
transcription. By contrast, ERG expression was shown to block
GATA2 transcription leading to suppression of megakaryocytic
differentiation. Knockdown studies revealed that while GATA1 is
critical for erythroid differentiation, GATA2 is mainly required
for megakaryocytic differentiation. Moreover, the regulation of
LDB1 by FLI1, which is mediated through direct regulation of GATA1,
played a critical role in its ability to control erythroid and
megakaryocytic lineages. The present study provided new insights
into the intricate regulatory circuits between FLI1 and other
factors such as GATA1, GATA2, ERG and LDB1 that govern erythroid
and megakaryocytic differentiation.
Materials and methods
Cell lines and vector
The human erythroleukemia cell lines HEL (cat. no.
ATCC-TIB-180) and 293T (cat. no. ATCC-CRL3216) [a derivative of the
293T (cat. no. 293tsA1609neo) cell line (cat. no. ATCC CRL-11268)],
were previously obtained from ATCC. These cells were cultured and
maintained in Dulbecco's Modified Eagle Medium supplemented with 5%
fetal bovine serum (HyClone; Cytiva). The luciferase reporter
vector PGL3 (PGL3-basic) was purchased from Promega Corporation
(cat. no. U47295). The vector PCDNA3 Flag Erg (Addgene, Inc.) was
transfected into HEL cells by Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions.
Gene cloning, transfections and
luciferase activity
GATA2 promoter sequences were downloaded from
the ensemble genome browser (https:/www.ensembl.org/index.html) to download. To
clone the GATA2 promoter, the upstream region of the
promoter (position −560 to +10; see Fig. 1F and Table SI), containing a potent FLI1
binding site, was cloned into the luciferase reporter vector PGL3
(Promega Corporation), as previously described (26,27).
The promotor cloning was performed by (GenScript Biotech, Cn). The
GATA2 promoter and negative control PGL3 vector DNAs (1.25
µg) with either MigR1 (1.25 µg) or MigR1-FLI1 (1.25 µg) were mixed
well, incubated 12 min at room temperature and transfected into
293T cells at 37°C using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer protocol. Renilla luciferase was used in transfection
as internal control to test the transfection efficiency, according
to manufacturer recommendations (Promega Corporation). The
transiently transfected cells were then plated into 96 well plates
and luciferase activity were determined 48 h later, as previously
mentioned (27).
Chromatin immunoprecipitation (ChIP)
assay
In brief, growing HEL cells (4×106 cells
per reaction) were washed, crosslinked with formaldehyde and then
resuspended in 500 µl of lysis buffer (Enzymatic Chromatin IP Kit;
Cell Signaling Technology, Inc.). Micrococcal Nuclease (0.5 µl) was
added to the fixed cells and incubated at 37°C for 20 min with
frequent mixing in order to digest DNA to length of approximately
150–900 bp. The cell lysates were sonicated in three sets of 20-sec
pulses using the Sonics Vibra VCX150 (Ningbo Scientz Biotechnology
Co., Ltd.) to break the nuclear membrane. As a control, a portion
of chromatin aliquot (20 µl) was removed as input DNA.
Immunoprecipitations were performed overnight at 4°C with 100 µl
process solution and 5 µl of ChIP specific FLI1 antibody (Abcam)]
or 1 µl of nonspecific normal rabbit immunoglobulin G (IgG)
antibodies (CST). To each IP reaction then added 30 µl of Protein G
Magnetic Beads and incubated for 2 h at 4°C with rotation.
Precipitates were washed with buffer provided with the kit and
reverse crosslinked, using the instructions provided for company's
Enzymatic Chromatin IP Kit (CST Biological Reagents Co., Ltd.).
Precipitated chromatin was incubated with proteinase K at 65°C for
2 h and used for DNA purification using spin columns from company's
Enzymatic Chromatin IP Kit (CST). Quantitative (RT-q) PCR was
performed to amplify the indicated promoter regions containing FLI1
binding sites. The sequences of the ChIP primers were GATA2L sense:
CGAGTTGCATCTGATTGTATGG and antisense: GCTCCTCTGTCTTCAACCCA. The
percentage of input was calculated by qPCR based upon the intensity
of the amplified FLI1 DNA divided by the amplified input DNA.
Amplified DNA was also resolved in 2% agarose gel as shown in
Fig. 1I.
RT-qPCR
Total RNA was extracted from culture of HEL cells
(4×105) using TRIzol® (Thermo Fisher
Scientific, Inc.) by using the manufacturer's recommended protocol.
RNA concentrations were measured using a NanoDrop 2000
spectrophotometer (Thermo Scientific Fisher, Inc.). To generate
cDNA, reverse transcription reaction was performed using the
PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.).
RT-qPCR was performed using FastStart Universal SYBR Green Master
(Roche Diagnostics GmbH) and the Step One Plus Real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
expression was normalized to GAPDH. The 2−ΔΔCq method
was used for relative quantification (28). RNA extraction, cDNA synthesis and
RT-qPCR were all performed according to the manufacturer protocols.
The primer sequences are in Table
SII. A total of three biological triplicates were used for all
RT-qPCRs, each in triplicates (n=3).
Heatmap analysis
The RNA sequencing for short hairpin (sh)FLI1 in HEL
cells has been published previously (29). TBtools software (TBtoolsV1.098) was
used for Heatmap analysis (30).
The original contributions presented in the study are publicly
available and found at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA682304.
STRING, ENCODE and ENSEMBLE database
analysis
STRING database (www.string-db.org) was used for protein protein
interaction data analysis. The ENCODE database (encodeproject.org)
was used to determine DNA-protein interaction for binding of FLI1
to GATA2. The ensemble genome browser (https://www.ensembl.org/index.html) was used to
extract promoter sequences shown in Fig. 1E.
shRNA and short interfering (si)RNA
expression
The construction of sh-FLI1 expression construct
(shFLI1) has been previously described (29,31).
In brief, the shRNA expression plasmid (12 mg) and packaging
plasmid psPAX2 (6 mg), pMD2. G (12 mg) (Didier Trono, Addgene
plasmid # 12259 and # 12260) were mixed and transfected into
HEK293T cells, using Lipofectamine®2000 48 h after
transfection, the cell supernatant was collected for transduction
of HEL (1×106) cells. The positive cells after
transduction were selected and cultured for 24 h using RPMI-1640
medium containing puromycin (5 mg/ml; Solarbio, China]. Other
shRNAs such as shGATA1, shGATA2 and shLDB1 as well as their control
scrambled plasmids were generated in a similar fashion. The
sequences are shown in Table
SIII. Transfection of siRNAs into HEL cells was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions, as
previously described (27). In
brief, Lipofectamine 2000 was used to transfect siRNA (15 uL of 20
uM) into HEL cells according to manufacturer's instructions
(Invitrogen; Thermo Fisher Scientific). After 48 hours, cells were
harvested for subsequent assays.
Overexpression of ERG in HEL
cells
DNA vectors (1.25 µg) containing PCDNA3.1(−) (1.25
µg; cat. no. V79520; Invitrogen; Thermo Fisher Scientific, Inc.) or
PCDNA3 Flag Erg (1.25 µg; cat. no. 66977; Addgene, Inc.) were
transfected into HEL cells according to the manufacturer's
protocol. The transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The medium was changed 48 h post transduction
and positive cells were selected for using medium containing G418
(250 µg/ml; Beijing Solarbio Science & Technology Co.,
Ltd.).
Western blotting
Western blotting was performed as described
elsewhere (14). In brief, cells
were collected and protein was extracted using RIPA Lysis Buffer
(Beyotime Institute of Biotechnology). After lysis and protein
density determination using BCA protein assay kit, 50 ug samples
were loaded on 10% acrylamide gels and transferred onto the PVDF
membrane. Blocked with TBS buffer containing 5% skimmed milk for
1.5 h at room temperature. Polyclonal rabbit antibodies for FLI1
(cat. no. ab133485) [dilution 1:1,000], GATA1 (ab181544) [dilution
1:2,000], LDB1 (cat. no. ab96799) [dilution 1:1,000] and ERG (cat.
no. ab92513) [dilution 1:1,000] were purchased from Abcam; GATA2
(cat. no. 4595S) [dilution 1:500] from CST Biological Reagents Co.,
Ltd.; GAPDH (cat. no. AB-P-R 001) [dilution 1:1000] from Hangzhou
Goodhere Biotechnology Co., Ltd.; secondary antibodies (Anti-rabbit
IgG (H+L) (DyLight™ 800 4X PEG Conjugate)) from CST Biological
Reagents Co., Ltd. (cat. no. 5151S) [dilution 1:30,000]. The
antibodies were diluted with TBS buffer containing 3%BSA, the
primary antibodies were incubated overnight at 4°C, and the
secondary antibodies were incubated for 1.5 h at room temperature.
The Odyssey Imaging System (LI-COR Biosciences) is used for western
blot protein imaging, and the protein density is determined using
the software (Odyssey CLX Image Studio 3.1) that comes with the
system.
Flow cytometry
Immunofluorescence staining was conducted to detect
erythroid and megakaryocytic cells, as previously described
(14,29). In brief, 1×105 cells
were stained with APC-conjugated antibodies for 40 min at 4°C.
Cells were then washed twice and resuspended in 200 µl PBS and used
for flow analysis. The following primary antibodies were used:
Human cluster of differentiation CD41a-APC (cat. no. 559777), human
CD61-APC (cat. no. 564174), human CD71-APC (cat. no. 551374) and
human CD235a-APC (cat. no. 551336; all purchased from BD
Biosciences). Flow cytometry was performed using a NovoCyte flow
cytometer and Novo-express software (ACEC Biosciences Inc.).
The gating strategies were used as followed:
FSC-A/SSC-A plots were used to separate live cells from debris.
Erythroid cells were differentiated using a scatter/anti-CD71+ and
a scatter/anti-CD235a+ gate from unstained control, respectively.
Megakaryocytes were differentiated using a scatter/anti-CD41a+ and
a scatter/anti-CD61+ gate from unstained control, respectively.
Count/anti-CD71+, count/ anti-CD235a+, count/anti-CD41a+ and count
anti-CD61+ in histograms present the expression of these
markers.
Statistical analysis
Statistical analysis was carried out using the
two-tailed Student t-test or using Welch's ANOVA followed by
Tamhane's T2 post hoc test, using Prism 8 software (GraphPad;
Dotmatics). Results were expressed as mean ± standard deviation
from at least three independent experiments. P<0.05 was
considered to indicate a statistically significant difference.
Results
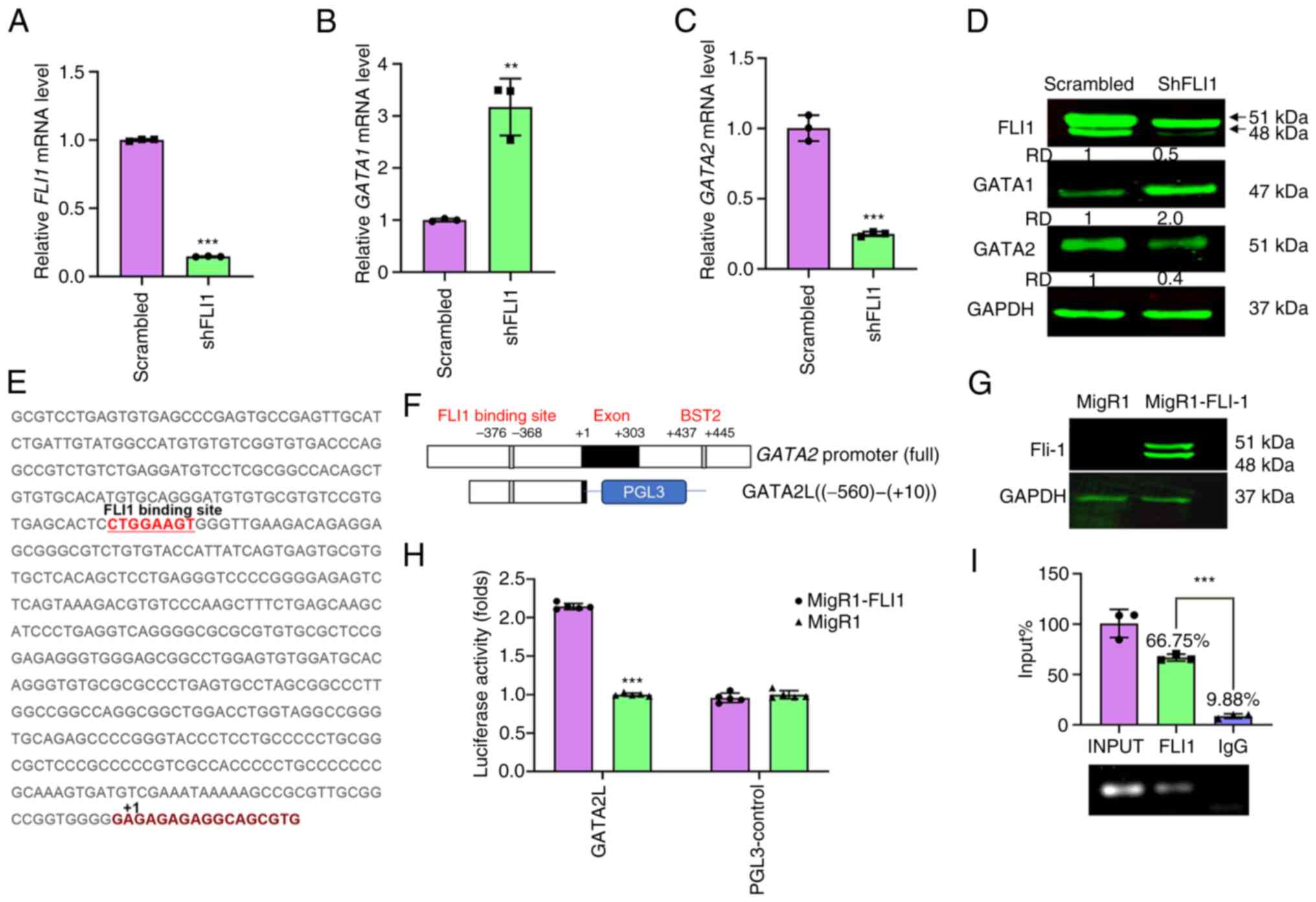
FLI1 directly binds the promoter of
GATA2 and regulates its transcription
The authors have previously reported RNAseq data
following lentivirus-shRNA-mediated FLI1 knockout in
erythroleukemia cell line HEL (shFLI1), which displays biopotential
megakaryopoiesis and erythroid progenitor capacity (29,31).
This RNAseq analysis identified a cluster of genes associated with
megakaryopoiesis, whose expression is altered following FLI1
depletion (Fig. S1A) (29). A heatmap of the erythroid
differentiation expressed genes (EDEGs), shown in Fig. S1B, identified 52 genes whose
expressions was altered in lentivirus-mediated FLI1 knockout
(shFLI1) relative to control (Scrambled) cells.
Among DEGs associated with erythroid and
megakaryocytic differentiation, GATA1 and GATA2 expression has been
previously shown to play pivotal roles in these blood maturation
processes, although the underlying mechanism remains to be
elucidated (16). GATA1 is
negatively regulated by FLI1 (12), as is shown in the present study by
both RT-qPCR and western blot analysis (Fig. 1A, B and D). This is consistent with
a previous report demonstrating binding of FLI1 to a putative site
within the promoter of GATA1 (12). By contrast, GATA2 expression was
notably reduced in shFLI1 compared with scrambled control cells
(Fig. 1C and D), suggesting that
GATA2 expression may be directly regulated by FLI1. Indeed, the
present study identified a putative FLI1 binding site in the
promoter of GATA2 at position −376 to −368 (Fig. 1E). To determine whether FLI1 is
recruited to GATA2 through this site, the promoter of
GATA2 was cloned into the luciferase reporter gene PGL3
(designated GATA2L; Fig. 1F).
Transfection of the GATA2L plasmid together with a FLI1 expression
vector (MigR1-FLI1) into 293T cells significantly increased
luciferase activity (Fig. 1H). The
expression of FLI1 in 293T cells was verified via western blotting
(Fig. 1G). ChIP analysis of HEL
cells using primers that flank the putative FLI1-binding site
within the GATA2 promoter detected a band that was
immunoprecipitated with FLI1 but not control IgG antibodies
(Fig. 1I). These results for the
first time point to the human GATA2 promoter as a direct target of
FLI1.
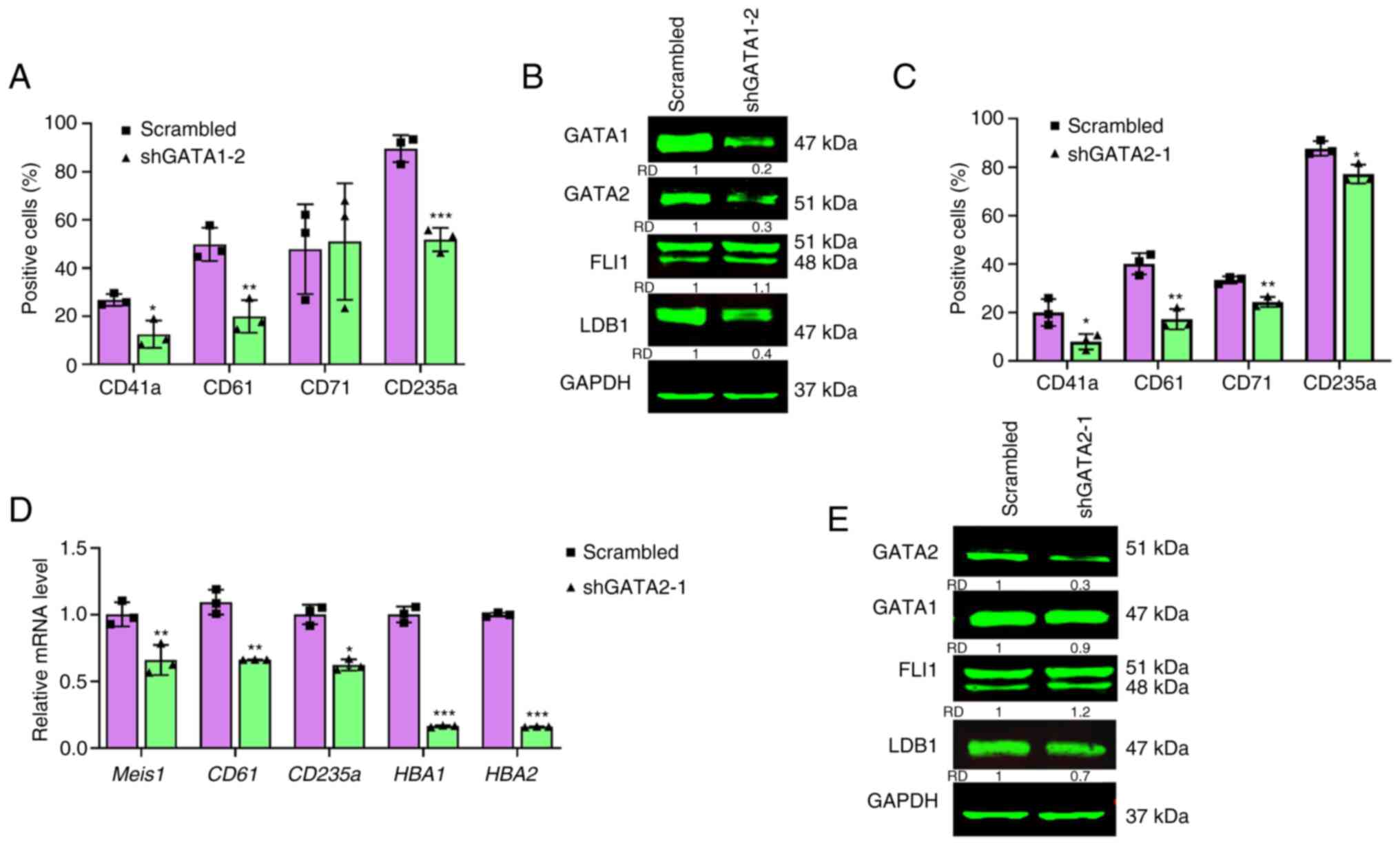
Consequences of FLI1 regulation of
GATA1 and GATA2 in erythroleukemic cell proliferation
To understand the consequences of FLI1 regulation of
GATA1 and GATA2 in cell proliferation and differentiation,
lentivirus shRNAs were used to knockdown these genes in HEL cells.
Significant knockdown was obtained in shGATA1-1 and shGATA1-2 cells
at both mRNA (Fig. 2A) and protein
(Fig. 2B) levels. Similarly,
silencing of GATA2 in HEL cells via shGATA2-1, shGATA-2 and
shGATA2-3 vectors blocked both transcription abundance (Fig. 2C) and protein (Fig. 2D) expression. While knockdown of
GATA2 did not alter cell proliferation (Fig. 2E), loss of GATA1 significantly
reduced proliferation rate in culture compared to scrambled control
(Fig. 2F). Knockdown of GATA1 and
GATA2 slightly, but significantly increased FLI1 expression
(Fig. 2G). Accordingly, multiple
GATA recognition sites were found within the Fli-1 promoter and
GATA1 is shown to bind these sequences (32). The expression analysis is also
predicted binding of GATA2 to FLI1 (Fig. 3E) that may need further analysis in
future studies.
Effect of GATA1 and GATA2 silencing in
erythroid and megakaryocytic differentiation
To determine the effect of GATA1/GATA2 on
erythroid/megakaryocytic lineage development, the present study
examined the effect of silencing these genes in HEL cells, using
the erythroid (CD71 and CD235a) and megakaryocytic (CD41a and CD61)
markers (33,34). It has previously been shown that
overexpression of Fli-1in erythroblasts blocks erythroid
differentiation (10,11). Indeed, FLI1 silencing in HEL cells
reduced percentage of megakaryocytic CD41a and CD61 positive cells
and increased percentage of late erythroid CD235a cells (Fig. S2A and B). FLI1 knockdown had no
effect on expression of early erythroid markers CD71 (Fig. S2A and B).
Knockdown of GATA1 (shGATA1) resulted in a dramatic
reduction in expression of the erythroid differentiation marker
CD235a, but negligible changes in CD71 levels (Figs. 3A and S3). GATA1 knockdown also suppressed
megakaryocytic differentiation as both CD41a and CD61 levels were
significantly reduced in shGATA1-2 cells (Figs. 3A and S3). Ablation of GATA1 in HEL cells
resulted in downregulation of GATA2 and LDB1, but slightly higher
expression of FLI1, as determined by western blotting (Fig. 3B).
GATA2 silencing in shGATA2-1 cells revealed a
significant lower percentage of megakaryocytic CD41a and CD61
expressing cells. This analysis also found significant reduction in
the percentage of CD71 and CD235a expression in shGATA2-1 cells
(Figs. 3C and S4). Indeed, lower expression of
megakaryocytic markers CD61 and Meis1, as well as
reduced levels of the erythroid CD235a marker and the globin
genes HBA1 and HBA2, was observed in shGATA2-1 cells
(Fig. 3D). Knockdown of GATA2 in
HEL cells resulted in downregulation of LDB1, but slightly higher
expression of FLI1, as determined by western blotting (Fig. 3E). The expression of GATA1 was
slightly reduced in shGATA2-1 cells, probably through increased
expression of FLI1 (Fig. 3E).
These results suggested a commitment role for GATA2 in
megakaryocytic differentiation and some involvement in erythroid
maturation via FLI1. GATA1 is probably involved in megakaryocytic
differentiation through regulation of GATA2 (Fig. 3B).
Using a protein-protein interaction database
(STRING), it was found that FLI1 binds GATA2 as well as other
factors including GATA1, RUNX1/2, MYB, SPI1, TAL1, known to play
critical roles in erythroid and megakaryocytic differentiation
(Fig. S5A). GATA2 is also
predicted to interact with LDB1 partner LMO2 that participates in
both erythroid and megakaryocytic maturation (17,18).
As FLI1 ablation suppresses the expression of ITGAB3 (CD61)
(Fig. S2A and B), knockdown of
GATA2 causes similar downregulation (Fig. 3D). Accordingly, the present study
found that the promoter of CD61 has a putative GATA2 binding
site at position −102 to −96 (Fig.
S5B). In the ENCODE database (35), GATA2 strongly binds (Affinity 7.93)
to this region of promoter (Fig.
S5C). These results confirmed a critical role for GATA2 as a
limiting factor in megakaryocytic differentiation.
Overexpression of ERG in HEL cells
suppresses megakaryocytic differentiation
While GATA1 has been shown to control megakaryocytic
differentiation, the underlying mechanism remains to be elucidated.
In contrast to overexpression of FLI1 in HEL cells, the level of
another FLI1-related ETS gene, ERG, is negligible (29). These two genes are known to have
distinct functions in hematopoiesis (16). The present study found that in both
GATA1 and GATA2 knockdown cells, the ERG expression was
significantly induced, suggesting a negative regulation of ERG via
these GATA genes in HEL cells (Fig. 4A
and B). In common with FLI1 (32), the ERG promoter also
contains multiple GATA-binding sites (Fig. S6A). ENCODE analysis indeed found
binding of both GATA1 and GATA2 to the ERG promoter
(Fig. S6B). Thus, GATA1 and GATA2
negatively regulate expression of ERG and FLI1 in HEL cells. To
further investigate the role of ERG in erythroid and megakaryocytic
differentiation, ERG was overexpressed in HEL cells (Fig. 4C). Higher expression of ERG in HEL
cells had no significant effect on cell proliferation (Fig. 4D) but resulted in significant
downregulation of FLI1 and GATA2 (Fig.
4E) and upregulation of GATA1 (Fig. 4E). Higher ERG expression also
resulted in strong downregulation of CD41a/CD61 level, indicating
that, in contrast to FLI1, ERG is a robust suppressor of
megakaryocytic differentiation (Figs.
4F and S7). Higher ERG
expression slightly but significantly induced erythroid CD235a
expression, consistent with the higher GATA1 level in these cells.
These results for the first time suggest that GATA1/2 control
megakaryocytic differentiation through suppression of ERG.
FLI1 negatively regulates LDB1 through
GATA1 to control cell differentiation
The LDB1 gene has also implicated in both erythroid
and megakaryocytic differentiation (17,18,24).
The present study found that LDB1 is negatively regulated by FLI1
(Fig. S1B), using both RT-qPCR
and western blot analysis (Fig. 5A and
B). Notably, the LDB1 partner Lmo2 was also negatively
regulated by FLI1, and its upregulation in shFLI1 cells was
associated with erythroid differentiation (Fig. S1B). The LDB1 promoter does
not have a canonical FLI1 binding site (data not shown) and
therefore its induction in shFLI1 cells is likely through GATA1
and/or GATA2 (Fig. 3B and E).
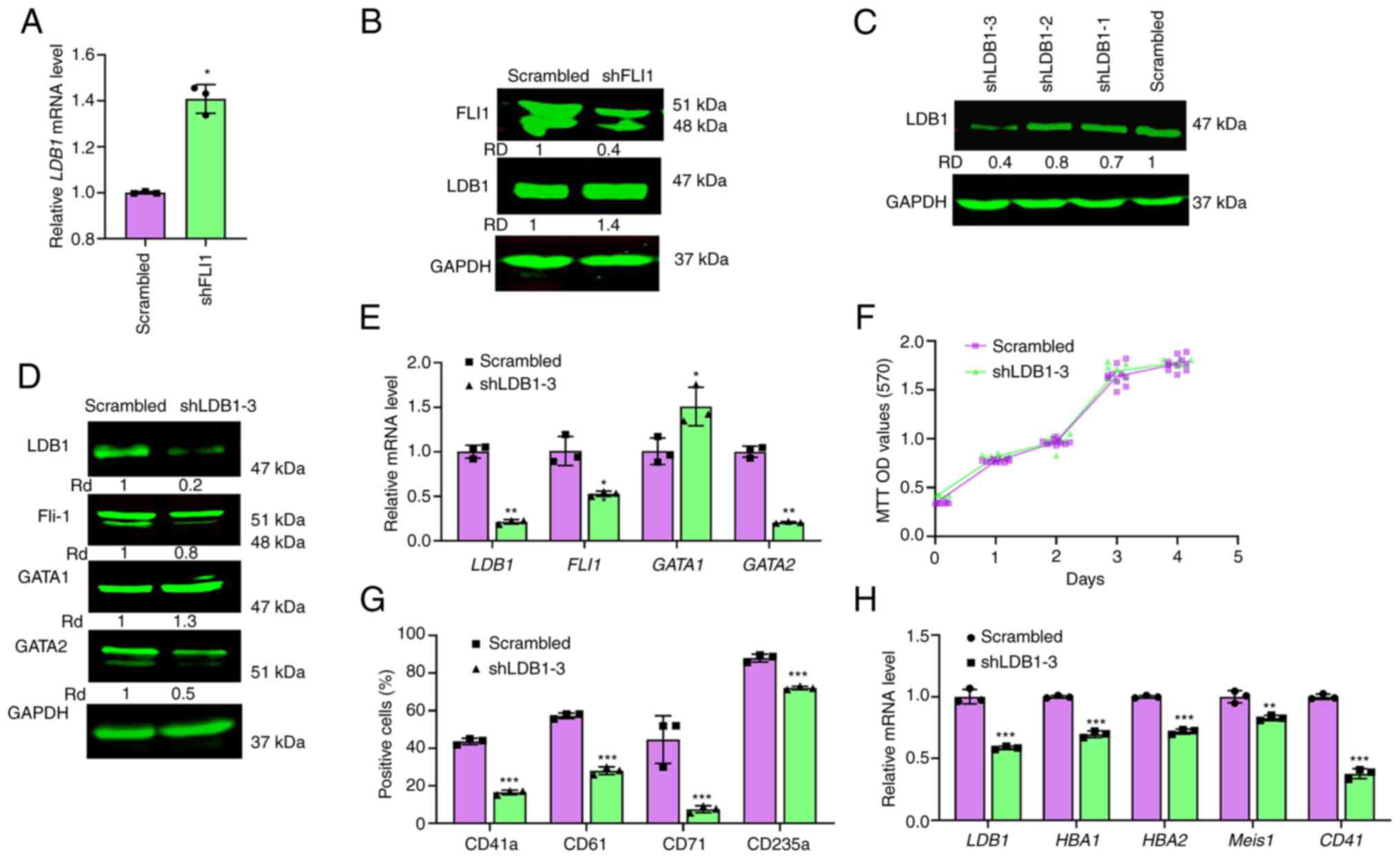 | Figure 5.Regulation of erythroid and
megakaryocytic differentiation by LDB1. The expression of LDB1 in
shFLI1 cells as detected by (A) RT-qPCR or (B) western blotting.
(C) Expression of LDB1 protein in HEL cells transfected with shRNA
lentiviruses (shLDB1-1, shLDB1-2 and shLDB1-3), as determined by
western blotting. (D) The expression of FLI1, LDB1 GATA1 and GATA2
in shLDB1-3 cells by western blotting. GAPDH was used as the
loading control. (E) The expression of genes in shLDB1-3 relative
to scrambled control cells, as determined by RT-qPCR. (F)
Proliferation rate of shLDB1-3 and scrambled control cells, as
determined by MTT. (G) The flow cytometry analysis for expression
of megakaryocytic (CD41a/CD61) and erythroid (CD71/CD235a) markers
in shLDB1-3 cells versus scrambled control cells. Average of three
experiments was indicated. (H) The relative expression of indicated
genes was determined by RT-qPCR. *P<0.05, **P<0.01 and
***P<0.001. LDB1, LIM domain binding 1; sh, short hairpin;
RT-qPCR, reverse transcription-quantitative PCR; GATA, GATA binding
protein; FLI1, friend leukemia integration 1; CD, cluster of
differentiation; RD, relative density. |
To uncover the role of LDB1 in erythroid and
megakaryocytic differentiation, its expression was silenced using
lentivirus-shRNA (Fig. 5C).
shLDB1-3 cells with efficient LDB1 knockdown exhibited strong
reduction in FLI1 and GATA2 and robust induction of GATA1
expression (Fig. 5D and E).
Nonetheless, LDB1 silencing in HEL cells had no effect on cell
proliferation in culture (Fig.
5F).
Flow cytometry showed that knockdown of LDB1
significantly reduced the percentage of cells expressing
megakaryocytic markers CD41a and CD61. LDB1 ablation resulted in a
lower percentage of erythroid CD71 and CD235a expressing cells
(Figs. 5G and S8). These results were further supported
by lower expression of erythroid hemoglobin genes HBA1, HBA2
and lower megakaryocytic markers CD41 and Meis1, by
RT-qPCR (Fig. 5H). Notably, while
FLI1 knockdown in HEL cells accelerated erythroid and moderately
decelerated megakaryocytic differentiation (Fig. S2), LDB1 knockdown suppressed FLI1
(Fig. 5D), induced GATA1
expression (Fig. 5D) and
unexpectedly inhibited erythroid maturation (Fig. 5G).
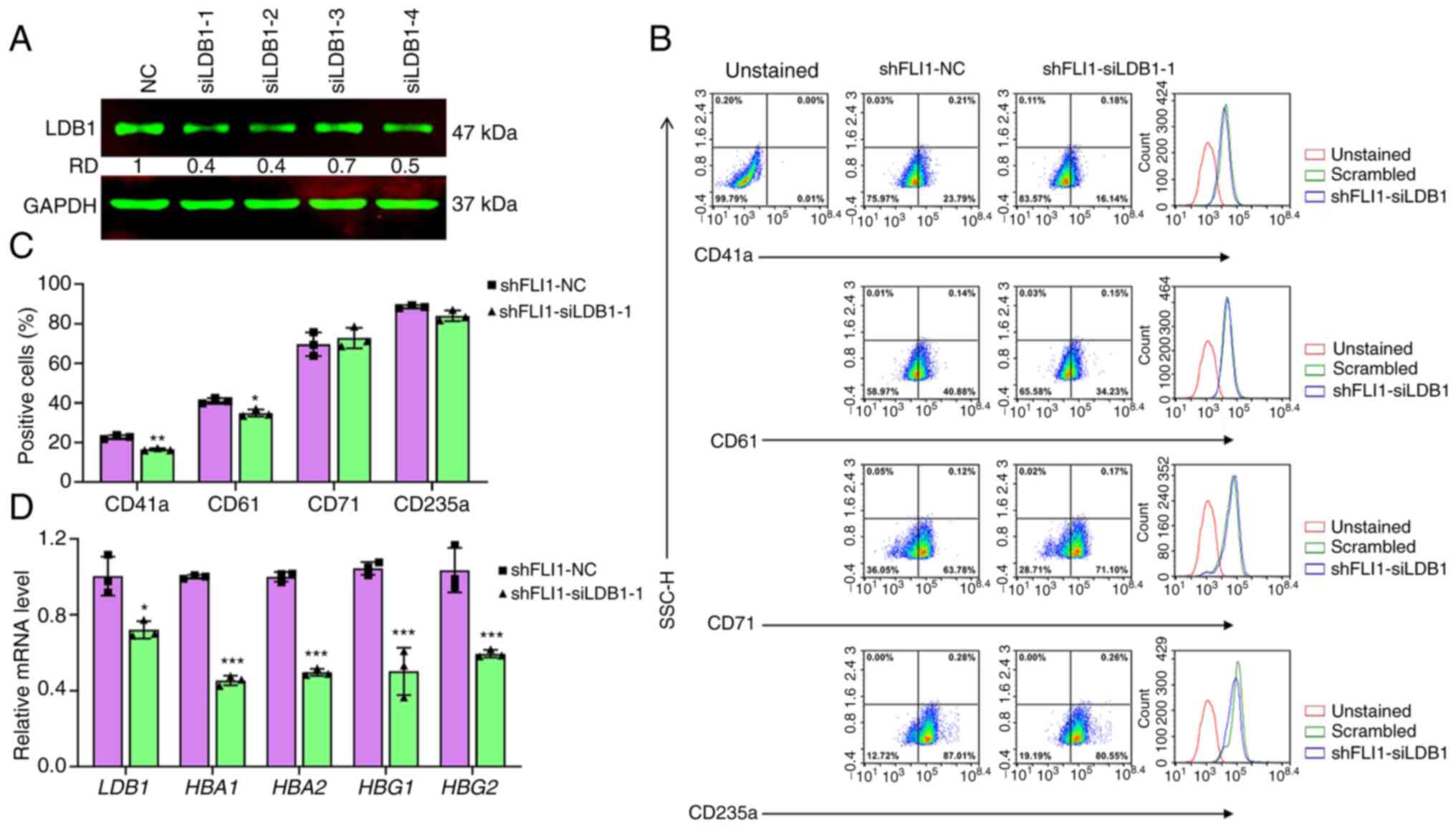
The above results suggested that a certain threshold
level of LDB1 expression may be necessary to enable FLI1 to block
erythroid differentiation. To test this possibility, LDB1
expression was knocked down using siRNAs (siLDB1-1-siLDB1-4) in
shFLI1 cells (Fig. 6A).
Downregulation of LDB1 in shFLI1 by siLDB1-1 resulted in a lower
percentage of CD41a and CD61, but slight and insignificant
reduction of CD235a expressing cells (Fig. 6B and C). However, in RT-qPCR,
siLDB1-1 significantly inhibited expression of erythroid markers
HBA1, HBA2, HBG1 and HBG2 (Fig. 6D). These results showed the
essential role of LDB1 in controlling erythroid and megakaryocytic
commitments via FLI1.
Interplay between FLI1 and KLF1 during
erythroid differentiation
Finally, the KLF1 (EKLF) transcription factor
controls erythroid differentiation through binding to CACCC motifs
within various globin gene promoters (36). KLF1 and FLI1 were both previously
reported to negatively regulate each other (37,38).
KLF1 is also known to positively regulate GATA1, leading to
erythroid differentiation (37–39).
In GATA1 knockdown cells (Fig.
7A), KLF1 expression was considerably suppressed
(Fig. 7B), supporting a direct
regulation of KLF1 by GATA1. Notably, a negligible change in
KLF1 expression was detected in the LDB1 knockdown cells
(Fig. 7C). Consistent with
inhibition of megakaryopoiesis, FLI1 expression was reduced
(Fig. 5D), whereas ERG
expression was strongly elevated in shLDB1-3 cells (Fig. 7D). These results further confirmed
the important role of LDB1 in controlling erythroid and
megakaryocytic differentiation in cooperation with FLI1 and ERG. As
depicted in Fig. 7E,
transcriptional regulation of GATA1, GATA2, LDB1 and ERG by FLI1 is
critical for commitment to either erythroid or megakaryocytic
differentiation. In HEL cells, LDB1 silencing decreased FLI1 and
increased GATA1 levels (Fig. 5D).
This increase in GATA1 expression unexpectedly failed to induce
erythroid differentiation. Conversely, reduced LDB1 expression led
to GATA2 downregulation, resulting in lower megakaryocytic
differentiation. Thus, cooperation between FLI1 and LDB1 is
necessary for proper regulation of erythroid differentiation.
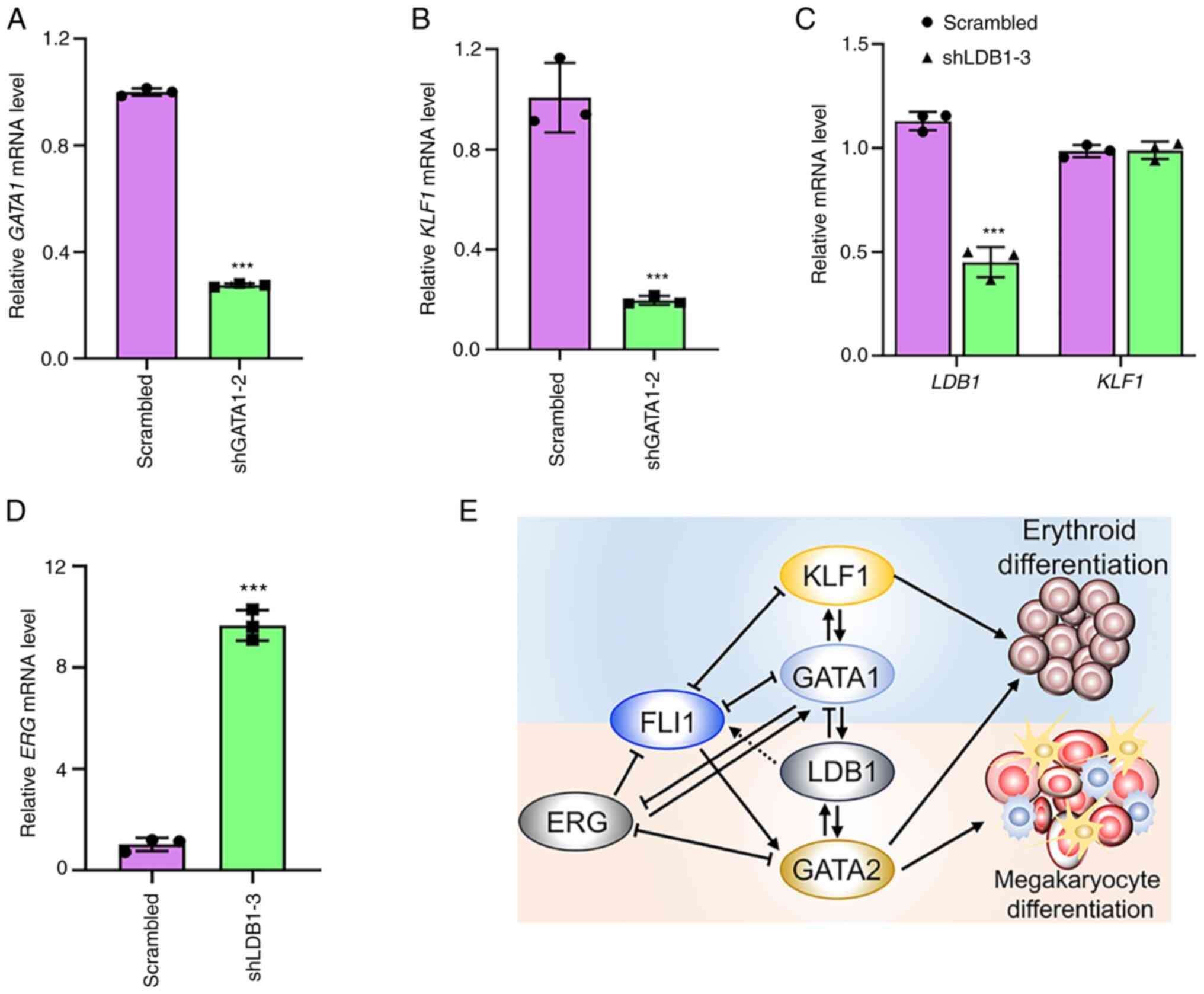 | Figure 7.KLF1 transcription regulated by FLI1
controls erythroid differentiation. (A,B) The expression of (A)
GATA1 and KLF1 (B) in shGATA1-2 cells as determined by RT-qPCR.
(C,D) The expression of (C) KLF1 and (D) ERG in shLDB1-2 cells, as
determined by RT-qPCR. (E) The intricate regulatory circuit of FLI1
and other transcription factors leading to erythroid and
megakaryocytic differentiation in erythroleukemia HEL cells.
Depicted model shows that FLI1 loss through activation of GATA1
induces erythroid differentiation. FLI1 loss suppresses
GATA2 transcription, leading to reduce megakaryocytic
differentiation. Loss of GATA1 and GATA2 activates ERG which in
contrast to FLI1, blocks megakaryocytic differentiation. LDB1,
through negative regulation by FLI1, plays a critical during
erythroid or megakaryocytic differentiation. Dotted line shows
indirect regulation. ***P<0.001. KLF1, KLF transcription factor
1; FLI1, friend leukemia integration 1; GATA, GATA binding protein;
sh, short hairpin; RT-qPCR, reverse transcription-quantitative PCR;
ERG, ETS transcription factor ERG; ETS, E26
transformation-specific; CD, cluster of differentiation. |
Discussion
The present study showed that FLI1 controls the
transcription of GATA1, GATA2 and LDB1, thereby coordinating the
erythroid versus megakaryocytic cell differentiation in HEL cells.
While FLI1 negatively controls GATA1 to block erythroid
differentiation, the present study showed that direct GATA2
transcriptional regulation by FLI1 is essential in promoting the
differentiation of the MEP-like erythroleukemia cell line HEL
toward megakaryocytic lineage maturation. LDB1 plays a broader role
in commitment of progenitors to both erythroid and megakaryocytic
differentiation via FLI1. These results provided novel insights
into an intra-regulatory role of FLI1 and its accessories during
erythroid and megakaryocytic differentiation. However, further
studies on animal models may be necessary to confirm the function
of these TFs in vivo.
The association between FLI1 and GATA2 was
previously observed, but it is not known whether this regulation is
direct or indirect (15). The
present study showed that regulation of GATA2 by FLI1 is critical
for megakaryocytic commitment. RNAseq analysis of FLI1 knockdown
cells indeed identified at least 47 genes associated with
megakaryocytic differentiation, among them ITGB3 (CD61), ITGA2B
(CD41) and Meis1 (Fig.
S1A). The human ITGAB3 promoter contains binding site
for GATA2, further confirming a role for GATA2 in megakaryocytic
differentiation. Indeed, in high-risk acute myeloid leukemia,
chromosomal rearrangements between 3q21 and 3q26 is often
associated with elevated platelet and megakaryocyte numbers
(40). The 3q rearrangements
reposition a GATA2 enhancer near the EVI1 (or MECOM) locus, which
results in both EVI1 and GATA2 overexpression leading to higher
number of megakaryocytic cells. Deleting GATA2 enhancer in mice
also results in significant reduction in differentiation of
progenitors to megakaryocytes and erythrocytes (41). Moreover, in inv(16) leukemia, the
CBFβ-MYH11 fusion inhibits megakaryopoiesis by blocking the
expression of GATA2/KLF1 and interfering with a balanced
transcriptional program involving these two factors (42). Although K562 erythroleukemic cells
do not express FLI1 (14),
overexpression of GATA2 in these cells induces megakaryocytic
differentiation and blocks erythroid maturation (43). Overall, the results of the present
study indicated that FLI1 regulation of GATA2 is essential for
megakaryocytic differentiation.
During differentiation, progenitor cells undergo
maturation processes to become mature blood cells. This process
involves several cell divisions that are probably controlled by
TFs. GATA1 expression controls erythroid differentiation. When
GATA1 is knocked down in HEL cells, there was a reduction in
erythroid differentiation that is also associated with a
significant reduction in the rate of cell proliferation (Fig. 2F). As Fli-1 is an oncogene, its
activation negatively regulates GATA1 expression resulting in
downregulation of this transcription factor and blockage of
erythroid differentiation that eventually leads to the development
of erythroleukemia. Indeed, several studies have reported a role
for GATA1 in cell proliferation and differentiation in various
types of cancer involving several growth promoting genes including
PI3K (44,45). While the present study provided a
correlation between differentiation and proliferation, at least for
GATA1, the other transcription factors that showed no change in the
rate of proliferation may require additional events to control cell
division.
FLI1 homologue gene ERG is also implicated in both
hematopoietic stem cell expansion and hematopoiesis (16). However, the expression of FLI1 and
ERG varies in different hematopoietic cells, suggesting distinct or
overlapping function (16).
Similar to FLI1, overexpression of ERG in hematopoietic cells
affected both erythroid and megakaryocytic differentiation
(46). Notably, ERG expression is
negligible in erythroleukemic cells overexpressing FLI1 (29). This raises the possibility that
FLI1 and ERG may exert opposite functions during erythroid
differentiation and transformation. The present study showed that,
in contrast to FLI1, ERG blocks the expression of GATA2 and its
overexpression in erythroleukemic cells suppressed megakaryocytic
differentiation. It also showed that FLI1 suppressed ERG expression
through downregulation of GATA1. These results pointed to the
opposite roles of FLI1 and ERG in erythroid and megakaryocytic
differentiation. The present study, for the first time to the best
of the authors' knowledge, suggested that GATA1/2 may control
megakaryocytic differentiation through suppression of ERG.
In functional ablation studies in mice, LDB1 has
been shown essential for embryonic erythropoiesis and blood island
formation (47). LDB1 facilitates
nuclear organization of its erythroid partners on the
β-globin gene promoter to initiate transcription during
erythroid differentiation (48).
In contrast to these reports, LDB1 and LMO2 were shown in the
present study to function as negative regulators of erythroid
differentiation in erythroleukemic cells (49). The present study also showed that
FLI1 knockdown in erythroleukemic cells induced higher expression
of both human LDB1 and LMO2, supporting a role for these genes in
promoting erythroid differentiation. Accordingly, shRNA-mediated
downregulation of LDB1 resulted in reduced expression of erythroid
differentiation markers, suggesting a positive role for LDB1, and
probably LMO2, in erythroid differentiation. A study by Giraud
et al (24) reveals that
interaction between FLI1 and LDB1 is critical to activate
megakaryocytic genes in erythroleukemic cells. Notably, in the
erythroleukemic cells of the present study, LDB1 knockdown induced
both downregulation of FLI1 and its target GATA2, two essential
factors for megakaryocytic differentiation. Accordingly, LDB1
ablation resulted in suppression of megakaryopoiesis associated
with downregulation of CD41a/CD61 markers and expression of MEIS1.
This result points to LDB1 and GATA2 as positive regulators of
megakaryocytic differentiation, controlled by FLI1. The LDB1
ablation experiments also revealed that a certain threshold level
of LDB1 expression enables FLI1 to block erythroid differentiation.
While LDB1 itself is not a transcription factor, it may affect
FLI1, ERG and other factors through protein-protein interactions.
Indeed, LDB1 has been reported to interact with FLI1 in
erythroleukemic cells (24).
Moreover, FLI1 inhibition is reported to regulate its own
transcription (50). Overall,
combination of transcription factors and LDB1 can affect the fate
of MEP towards either erythroid or megakaryocytic differentiation.
These factors together create a complex network due to
protein-protein interaction that can affect the fate of MEP that
may need further analysis in future studies.
Finally, the transcription factor KLF1, a master
regulator of erythroid differentiation (36–39),
is confirmed in the present study to be negatively regulated by
FLI1 and positively by GATA1. KLF1 and GATA1 expression both
cooperate to actuate erythroid gene expression in erythroid cells
(39). Notably, in LDB1 knockdown
cells, while the levels of GATA1 were significantly high, these
cells lost commitment to erythroid differentiation. As FLI1complex
with LDB1 is critical for megakaryopoiesis (24), this interaction may also control
commitment of progenitor cells to the erythroid linage, a notion
that should be addressed in future studies. While different complex
binding of these factors is known to be critical for commitment of
MEPs to different lineages, the present study provided a new
perception into the regulatory circuit that fine tuning the level
of these transcription factors required during erythroid and
megakaryocytic differentiation.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by research grants from the
Natural National Science Foundation of China (grant nos. U1812403
and 82260040) to YBD, the Science and Technology Department of
Guizhou Province [grant nos. QKHJC-ZK(2022)YB297,
QKHJC-ZK(2023)YB240 and QKHJC-ZK(2021)YB569] and the Key Laboratory
of Chemistry for Natural Products of Guizhou Province and Chinese
Academic of Sciences Research Grant [grant no. TCZJZ(2022)03] to XX
and CW and the Guizhou Medical University Research Grant (grant no.
RN21025) to BG.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CW, MH, WL, YK, AH, KY, LH and XX contributed to
the conception, design of the study as well as data acquisition and
interpretation. CW performed the experiments and wrote the
manuscript. CW, MH were involved in data analysis and statistics.
YB, EZ, XX and BG contributed to the conception, design of the
study as well as reviewing the manuscript critically. XX and YB
confirm the authenticity of all the raw data. YB supervised and
designed the study. All authors contributed to interpretation of
findings, reviewed and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ATCC
|
American Type culture collection
|
|
ChIP
|
Chromatin immunoprecipitation
|
|
ERG
|
ETS transcription factor ERG
|
|
ETS
|
E26 transformation-specific
|
|
FLI1
|
friend leukemia integration 1
|
|
GATA1
|
GATA binding protein 1
|
|
GATA2
|
GATA binding protein 2
|
|
HSC
|
hematopoietic stem cell
|
|
ITGA2B
|
integrin subunit alpha 2b
|
|
ITGB3
|
integrin subunit beta 3
|
|
KLF1
|
KLF transcription factor 1
|
|
LDB1
|
LIM domain binding 1
|
|
LMO2
|
LIM domain only 2
|
|
MEP
|
megakaryocytic erythroid
progenitors
|
|
TAL1
|
T-Cell acute lymphocytic leukemia
1.
|
References
|
1
|
Orkin SH and Zon LI: Hematopoiesis: An
evolving paradigm for stem cell biology. Cell. 132:631–644. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Teitell MA and Mikkola HK: Transcriptional
activators, repressors, and epigenetic modifiers controlling
hematopoietic stem cell development. Pediatr Res. 59:33R–39R. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wickrema A and Crispino JD: Erythroid and
megakaryocytic transformation. Oncogene. 26:6803–6815. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tijssen MR, Cvejic A, Joshi A, Hannah RL,
Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson
NK, et al: Genome-wide analysis of simultaneous GATA1/2, RUNX1,
FLI1, and SCL binding in megakaryocytes identifies hematopoietic
regulators. Dev Cell. 20:597–609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Subramanian S, Thoms JAI, Huang Y,
Cornejo-Páramo P, Koch FC, Jacquelin S, Shen S, Song E, Joshi S,
Brownlee C, et al: Genome-wide transcription factor binding maps
reveal cell-specific changes in the regulatory architecture of
human HSPC. Blood. 142:1448–1462. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beck D, Thoms JA, Perera D, Schütte J,
Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O'Brien TA,
Göttgens B, et al: Genome-wide analysis of transcriptional
regulators in human HSPCs reveals a densely interconnected network
of coding and noncoding genes. Blood. 122:e12–e22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Diffner E, Beck D, Gudgin E, Thoms JA,
Knezevic K, Pridans C, Foster S, Goode D, Lim WK, Boelen L, et al:
Activity of a heptad of transcription factors is associated with
stem cell programs and clinical outcome in acute myeloid leukemia.
Blood. 121:2289–2300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ben-David Y, Giddens EB and Bernstein A:
Identification and mapping of a common proviral integration site
Fli-1 in erythroleukemia cells induced by friend murine leukemia
virus. Proc Natl Acad Sci USA. 87:1332–1336. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ben-David Y, Giddens EB, Letwin K and
Bernstein A: Erythroleukemia induction by Friend murine leukemia
virus: Insertional activation of a new member of the ets gene
family, Fli-1, closely linked to c-ets-1. Genes Dev. 5:908–918.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pereira R, Quang CT, Lesault I, Dolznig H,
Beug H and Ghysdael J: FLI-1 inhibits differentiation and induces
proliferation of primary erythroblasts. Oncogene. 18:1597–1608.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tamir A, Howard J, Higgins RR, Li YJ,
Berger L, Zacksenhaus E, Reis M and Ben-David Y: Fli-1, an
Ets-related transcription factor, regulates erythropoietin-induced
erythroid proliferation and differentiation: Evidence for direct
transcriptional repression of the Rb gene during differentiation.
Mol Cell Biol. 19:4452–4464. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Athanasiou M, Mavrothalassitis G,
Sun-Hoffman L and Blair DG: FLI-1 is a suppressor of erythroid
differentiation in human hematopoietic cells. Leukemia. 14:439–445.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Starck J, Weiss-Gayet M, Gonnet C, Guyot
B, Vicat JM and Morlé F: Inducible Fli-1 gene deletion in adult
mice modifies several myeloid lineage commitment decisions and
accelerates proliferation arrest and terminal erythrocytic
differentiation. Blood. 116:4795–4805. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu T, Yao Y, Zhang G, Wang Y, Deng B,
Song J, Li X, Han F, Xiao X, Yang J, et al: A screen for Fli-1
transcriptional modulators identifies PKC agonists that induce
erythroid to megakaryocytic differentiation and suppress
leukemogenesis. Oncotarget. 8:16728–16743. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu F, Walmsley M, Rodaway A and Patient
R: Fli1 acts at the top of the transcriptional network driving
blood and endothelial development. Curr Biol. 18:1234–1240. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ben-David Y, Gajendran B, Sample KM and
Zacksenhaus E: Current insights into the role of Fli-1 in
hematopoiesis and malignant transformation. Cell Mol Life Sci.
79:1632022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Love PE, Warzecha C and Li L: Ldb1
complexes: The new master regulators of erythroid gene
transcription. Trends Genet. 30:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Lee JY, Gross J, Song SH, Dean A and
Love PE: A requirement for Lim domain binding protein 1 in
erythropoiesis. J Exp Med. 207:2543–2550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee J, Krivega I, Dale RK and Dean A: The
LDB1 complex Co-opts CTCF for erythroid lineage-specific long-range
enhancer interactions. Cell Rep. 19:2490–2502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Soler E, Andrieu-Soler C, de Boer E, Bryne
JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van
Ijcken W, et al: The genome-wide dynamics of the binding of Ldb1
complexes during erythroid differentiation. Genes Dev. 24:277–289.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krivega I, Dale RK and Dean A: Role of
LDB1 in the transition from chromatin looping to transcription
activation. Genes Dev. 28:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng W, Lee J, Wang H, Miller J, Reik A,
Gregory PD, Dean A and Blobel GA: Controlling long-range genomic
interactions at a native locus by targeted tethering of a looping
factor. Cell. 149:1233–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stadhouders R, Cico A, Stephen T,
Thongjuea S, Kolovos P, Baymaz HI, Yu X, Demmers J, Bezstarosti K,
Maas A, et al: Control of developmentally primed erythroid genes by
combinatorial co-repressor actions. Nat Commun. 6:88932015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Giraud G, Kolovos P, Boltsis I, van
Staalduinen J, Guyot B, Weiss-Gayet M, IJcken WV, Morlé F and
Grosveld F: Interplay between FLI-1 and the LDB1 complex in murine
erythroleukemia cells and during megakaryopoiesis. iScience.
24:1022102021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eisbacher M, Holmes ML, Newton A, Hogg PJ,
Khachigian LM, Crossley M and Chong BH: Protein-protein interaction
between Fli-1 and GATA-1 mediates synergistic expression of
megakaryocyte-specific genes through cooperative DNA binding. Mol
Cell Biol. 23:3427–3441. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li YJ, Zhao X, Vecchiarelli-Federico LM,
Li Y, Datti A, Cheng Y and Ben-David Y: Drug-mediated inhibition of
Fli-1 for the treatment of leukemia. Blood Cancer J. 2:e542012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Song J, Liu W, Yao Y, Kapranov P,
Sample KM, Gajendran B, Zacksenhaus E, Hao X and Ben-David Y: FLI1
promotes protein translation via the transcriptional regulation of
MKNK1 expression. Int J Oncol. 56:430–438. 2020.PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang C, Sample KM, Gajendran B, Kapranov
P, Liu W, Hu A, Zacksenhaus E, Li Y, Hao X and Ben-David Y: FLI1
induces megakaryopoiesis gene expression through WAS/WIP-dependent
and independent mechanisms; Implications for wiskott-aldrich
syndrome. Front Immunol. 12:6078362021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen C, Chen H, Zhang Y, Thomas HR, Frank
MH, He Y and Xia R: TBtools: An integrative toolkit developed for
interactive analyses of big biological data. Mol Plant.
13:1194–1202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song J, Yuan C, Yang J, Liu T, Yao Y, Xiao
X, Gajendran B, Xu D, Li YJ, Wang C, et al: Novel flavagline-like
compounds with potent Fli-1 inhibitory activity suppress diverse
types of leukemia. FEBS J. 285:4631–4645. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barbeau B, Barat C, Bergeron D and Rassart
E: The GATA-1 and Spi-1 transcriptional factors bind to a GATA/EBS
dual element in the Fli-1 exon 1. Oncogene. 18:5535–5545. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
An X and Chen L: Flow cytometry (FCM)
analysis and fluorescence-activated cell sorting (FACS) of
erythroid cells. Methods Mol Biol. 1698:153–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakahata T and Okumura N: Cell surface
antigen expression in human erythroid progenitors: Erythroid and
megakaryocytic markers. Leuk Lymphoma. 13:401–409. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang P, Basu P, Redmond LC, Morris PE,
Rupon JW, Ginder GD and Lloyd JA: A functional screen for
Krüppel-like factors that regulate the human gamma-globin gene
through the CACCC promoter element. Blood Cells Mol Dis.
35:227–235. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Starck J, Cohet N, Gonnet C, Sarrazin S,
Doubeikovskaia Z, Doubeikovski A, Verger A, Duterque-Coquillaud M
and Morle F: Functional cross-antagonism between transcription
factors FLI-1 and EKLF. Mol Cell Biol. 23:1390–1402. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Neuwirtova R, Fuchs O, Holicka M, Vostry
M, Kostecka A, Hajkova H, Jonasova A, Cermak J, Cmejla R,
Pospisilova D, et al: Transcription factors Fli1 and EKLF in the
differentiation of megakaryocytic and erythroid progenitor in
5q-syndrome and in Diamond-Blackfan anemia. Ann Hematol. 92:11–18.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Merika M and Orkin SH: Functional synergy
and physical interactions of the erythroid transcription factor
GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell
Biol. 15:2437–2447. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamaoka A, Suzuki M, Katayama S, Orihara
D, Engel JD and Yamamoto M: EVI1 and GATA2 misexpression induced by
inv(3)(q21q26) contribute to megakaryocyte-lineage skewing and
leukemogenesis. Blood Adv. 4:1722–1736. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Johnson KD, Conn DJ, Shishkova E,
Katsumura KR, Liu P, Shen S, Ranheim EA, Kraus SG, Wang W, Calvo
KR, et al: Constructing and deconstructing GATA2-regulated cell
fate programs to establish developmental trajectories. J Exp Med.
217:e201915262020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yi G, Mandoli A, Jussen L, Tijchon E, van
Bergen MGJM, Cordonnier G, Hansen M, Kim B, Nguyen LN, Jansen PWTC,
et al: CBFβ-MYH11 interferes with megakaryocyte differentiation via
modulating a gene program that includes GATA2 and KLF1. Blood
Cancer J. 9:332019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ikonomi P, Rivera CE, Riordan M,
Washington G, Schechter AN and Noguchi CT: Overexpression of GATA-2
inhibits erythroid and promotes megakaryocyte differentiation. Exp
Hematol. 28:1423–1431. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Y, Ke Q, Shao Y, Zhu G, Li Y, Geng N,
Jin F and Li F: GATA1 induces epithelial-mesenchymal transition in
breast cancer cells through PAK5 oncogenic signaling. Oncotarget.
6:4345–4356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu J, Liu M, Liu H and Zhou L: GATA1
promotes colorectal cancer cell proliferation, migration and
invasion via activating AKT signaling pathway. Mol Cell Biochem.
457:191–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Carmichael CL, Metcalf D, Henley KJ, Kruse
EA, Di Rago L, Mifsud S, Alexander WS and Kile BT: Hematopoietic
overexpression of the transcription factor Erg induces lymphoid and
erythro-megakaryocytic leukemia. Proc Natl Acad Sci USA.
109:15437–15442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mukhopadhyay M, Teufel A, Yamashita T,
Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP,
Dorward D and Westphal H: Functional ablation of the mouse Ldb1
gene results in severe patterning defects during gastrulation.
Development. 130:495–505. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song SH, Kim A, Ragoczy T, Bender MA,
Groudine M and Dean A: Multiple functions of Ldb1 required for
beta-globin activation during erythroid differentiation. Blood.
116:2356–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Visvader JE, Mao X, Fujiwara Y, Hahm K and
Orkin SH: The LIM-domain binding protein Ldb1 and its partner LMO2
act as negative regulators of erythroid differentiation. Proc Natl
Acad Sci USA. 94:13707–13712. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu T, Xia L, Yao Y, Yan C, Fan Y,
Gajendran B, Yang J, Li YJ, Chen J, Filmus J, et al: Identification
of diterpenoid compounds that interfere with Fli-1 DNA binding to
suppress leukemogenesis. Cell Death Dis. 10:1172019. View Article : Google Scholar : PubMed/NCBI
|