Introduction
Obesity has become a marked global public health
issue that remains challenging to overcome (1,2). The
number of individuals who are overweight or obese is notably
increasing, particularly in China, due to the rapid progress in
socioeconomic development. According to recent studies (3), it has been revealed that >50% of
the adult population in China is suffering from obesity or being
overweight due to changes in the dietary structure of the Chinese
society and the decline in the amount of physical labor hours
(4), in addition to genetic,
psychosocial and other factors (5).
Obesity is a persistent, asymptomatic inflammatory
state characterized by elevated levels of inflammatory markers
within the body (6). The
uncontrolled growth of the adipose tissue of the body severely
affects blood supply; hypoxia resulting from insufficient blood
supply can stress and destroy adipocytes, further recruiting
macrophages to phagocytose the dead adipocytes (7). The liberation of unbound fatty acids
from adipocytes triggers the activation of proinflammatory genes,
and the excessive production of inflammatory substances such as
interleukin (IL)-6, tumour necrosis factor α (TNF-α) and IL-1β
induces a condition of persistent, mild inflammation in the body
(8,9). Adipose tissue is an endocrine organ
that regulates energy balance in the body, and persistent chronic
inflammation interferes with this activity, leading to metabolic
disorders in the body (10).
Researchers have recently discovered a connection between
obesity-induced chronic low-grade inflammation and various
cancerous conditions, indicating that the risks of obesity extends
beyond this (11,12).
Current research suggests that dysregulation of the
microbiota-host balance may be responsible for obesity (13). Hundreds of millions of bacteria
reside in the human gut, microorganisms that colonize the human gut
shortly after birth and work in coordination with the immune system
to counter the effects of changes in the external environment
(14). It has been demonstrated
that alterations in the intestinal flora and their metabolites
trigger multiple inflammatory pathways, and that dysregulation of
bile acid metabolism, as well as endotoxemia of intestinal origin,
promotes metabolic dysfunction in the body, leading to insulin
resistance (15,16). The structure of the gut flora can
be influenced by diet, which is considered a notable environmental
factor. Diets that are high in saturated fatty acids have the
potential to disrupt the structure of the gut flora and affect the
barrier protection of the gut (17). In recent years, researchers have
been working to find solutions to obesity by targeting the gut
flora to improve metabolic disorders (18).
Lactoferrin (LF) is a multifunctional transferrin
glycoprotein that is widely distributed in the exocrine fluids of
the human body (19). Due to its
strong iron chelating ability, LF can compete with harmful
intestinal bacteria to grab iron ions thus achieving an
antibacterial effect; it has also been shown to have antiviral,
anticancer and anti-inflammatory effects and a function in
regulating of the immune system (20,21).
A previous study demonstrated that LF improves lipid metabolic
disorders in obese mice in conjunction with a high-fat diet
(22). In the present experiment,
the action of LF was delayed and the detection of blood
inflammatory factors and the expression of intestinal tight
junction proteins was increased.
Materials and methods
Materials and animals
LF was acquired from The Tatua Co-operative Dairy
Company Ltd. The purity of LF was 95% with an iron saturation of
15%. The high-fat diet (energy ratio: Protein 18.14%, fat 60.65%,
carbohydrate 21.22%) and normal diet (protein 27.5%, fat 11.1%,
carbohydrate 67.4%) was obtained from Jiangsu Xietong
Pharmaceutical Bio-engineering Co., Ltd.
A total of 30 male 3-week-old C57BL/6 mice (average
weight 17±1 g) were obtained from SPF (Beijing) Biotechnology Co.,
Ltd. All mice were housed at a temperature of 25±1°C and 60±5%
humidity with 12 h light/dark cycle, and unrestricted availability
to food and water. Food and water were changed daily, while animal
health and behavior are monitored once a day. All procedures were
carried out following strict ethical guidelines, and every effort
was made to minimize animal suffering.
Euthanasia
The experimental animals exhibiting refusal to
consume food or water, lack of physical activity, rapid weight loss
within a short timeframe or any signs indicating infection will be
subjected to euthanasia. However, no such instances were observed
in this experiment.
Experimental design
After the C57BL/6J mice were acclimatized and fed a
regular diet for 1 week, 10 mice were chosen for the control group
(Group K) and continued to be fed a regular diet for 12 weeks. The
remaining mice were randomly divided into two groups of 10 mice
each; the model group drank purified water and was fed a high-fat
diet (Group M), while for the other group, purified water was
replaced with 2% LF (23) water
after 2 weeks (Group Z2). A brief 2-week high-fat dietary
intervention before LF consumption can cause a hepato-intestinal
inflammatory state and alterations in bacterial populations in the
host (24). The rest of the
conditions for Group Z2 were the same as for Group M, and mice were
fed until the end of week 12.
During the animal experiments, body weight changes
were recorded weekly. At the end of the animal experiment, the
experimental animals were subjected to cervical dislocation under
anesthesia. Mice were anesthetized with 40 mg/kg pentobarbital
sodium solution by intraperitoneal injection. The death of the
experimental animal was confirmed by the cessation of breathing,
disappearance of reflexes and dilation of pupils. After the animal
experiments were completed, the mouse lipid levels, circulating
lipopolysaccharide (LPS) levels, serum inflammatory factor levels,
intestinal tight junction proteins relative expression were
measured.
The analysis of fecal samples was conducted using
16S ribosomal (r)RNA sequencing.
Sample collection
On the day prior to concluding the experiment, the
second new fecal pellet of a stressed (pressure exerted on the
abdomen) defecation was obtained on a sterilized table and promptly
moved to a freezing tube that was sterile and frozen at −80°C.
Following collection of fecal samples, the animals underwent a 12-h
fasting period, and their body weight and length were measured in
order to calculate Lee's index (25). Mice were anesthetized with 40 mg/kg
pentobarbital sodium solution by intraperitoneal injection, ~0.8 ml
blood was collected under anesthesia through heart puncture and
then the mice were euthanized by cervical dislocation, and the
collected samples were subsequently preserved at −80°C for future
use. The collected blood was centrifuged at 4°C for 10 min at 1,200
× g for serum separation. Animal tissue samples were stored at
−80°C.
Serum chemistry analysis
The biochemical indices of total cholesterol (TC;
cat. no. ADS-W-ZF014) triglycerides (TG; cat. no. ADS-W-ZF013),
low-density lipoprotein (LDL; cat. no. ADS-0428M1) and high-density
lipoprotein (HDL; cat. no. ADS-0459M1) in mouse serum were analyzed
using commercial enzyme assay kits (Jiangsu Aidisheng
Biotechnology). Serum LPS (cat. no. MM-0634M1), interleukin (IL)-6
(cat. no. MM-0163M1), IL-1β (cat. no. MM-0040M1) and tumor necrosis
factor-α (TNF-α; cat. no. MM-0132M1) levels were determined using
enzyme-linked immunosorbent assay kits (Jiangsu Meimian Industrial
Co., Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Briefly, total RNA from mouse intestine tissue was
extracted with TransZol (TransGen Biotech Co., Ltd.). The
first-strand cDNA was synthesized with a cDNA synthesis kit (Takara
Bio, Inc.) through RT kit was used according to the manufacturer's
protocol. RT-qPCR was subsequently performed using a
SYBR® Green Real-time PCR Master Mix (Toyobo Life
Science), qPCR was performed using a 7300 real-time PCR instrument
system (Thermo Fisher Scientific, Inc.). PCR was performed in
duplicate at 95°C for 3 min and subjected to 40 cycles at 95°C for
5 sec, 60°C for 5 sec and 72°C for 15 sec. To standardize the mRNA
levels of all genes, β-actin was employed as an internal control.
The relative expression of each gene was determined by the
2−ΔΔCq method (26).
The primers used for qPCR are presented in Table SI.
Western blotting
Mouse small intestines tissue were lysed with
ice-cold lysis buffer containing RIPA buffer (Beyotime Institute of
Biotechnology) and a proteinase inhibitor cocktail (Baiaolaibo
Technology, Inc.). The lysates were sonicated with an oscillation
frequency of 20–25 kHz at 4°C for 15 sec, followed by
centrifugation at 12,000 × g for 15 min at 4°C and the supernatants
were retained. Total protein concentration was quantified using a
BCA protein assay kit (Epizyme Biomedical Technology, Inc.).
Following the total of 30 µg protein/lane were separation of
protein samples on a 10% SDS-PAGE gel, an electrophoretic transfer
was undertaken to a PVDF membrane. This membrane was subsequently
incubated in TBS containing 0.1% Tween-20 and 5% non-fat dry milk
powder for a duration of 1 h at ambient temperature, followed by an
overnight incubation period at 4°C along with the selected primary
antibodies. The primary antibodies included zonula occludens-1
(ZO-1; 1:500; cat. no. bs-1329R; BIOSS), Occludin (1:500; cat. no.
bs-10011R; BIOSS) and β-actin (1:10,000; cat. no. AB0035; Shanghai
Abways Biotechnology Co., Ltd.). After several washes, the
membranes were incubated with an appropriate horseradish peroxidase
(HRP)-conjugated secondary antibody (1:1,000; cat. no. A0208;
Beyotime Institute of Biotechnology) for 1 h at room temperature.
The proteins bands were visualized using enhanced chemiluminescence
kits (Sangon Biotech Co., Inc.), and the optical density of the
protein bands was quantified using the ImageJ v1.8.0 software
(National Institutes of Health), using β-actin as an internal
control.
Gut microbiota analysis
Genomic DNA was extracted from mouse feces using
hexadecyltrimethylammonium bromide, and its purity and
concentration were assessed through agarose gel electrophoresis. A
suitable quantity of genetic material was placed into a tube
designed for centrifugation, then it was diluted with sterile water
until the concentration reached 1 ng/µl. The diluted DNA sample
served as a template, and based on the attributes of the sequencing
region, specific primers were chosen for barcode primers. To
guarantee both efficiency and precision in amplification,
Phusion® High-Fidelity PCR primers (New England BioLabs,
Inc.) were used to amplify the marker gene for 16S rRNA V4. Primers
515F (5′-GTTTCGGTGCCAGCMGCCGCGGTAA-3′) and 806R
(5′-CAGATCGGACTACHVGGGTWTCTAAT-3′) in the 16S V4 region were
selected to amplify the DNA samples of each bacterial 16S rRNA
gene, 470 bp for length and ‘paired end’ for direction of
sequencing. After the completion of amplification, the target bands
were thoroughly examined and purified. The assembly of the library
was accomplished using the TruSeq® DNA PCR-Free Sample
Preparation Kit (New England BioLabs, Inc.). After quantification
using Qubit 2.0 (Thermo Fisher Scientific, Inc.) and qPCR, the
constructed library underwent qualification before being subjected
to sequencing with NovaSeq6000 (Illumina, Inc.).
FLASH (version 1.2.11; http://ccb.jhu.edu/software/FLASH/) was used to splice
the measured sequences, and the initial tags underwent quality
filtering with specific conditions to eliminate chimeras and obtain
the ultimate valid data. Following the identification of chimera,
high-quality sequences with up to 97% sequence similarity were
clustered into operational taxonomic units (OTUs) and assigned
species annotations (27). Every
OTU represents the taxonomic levels of phylum, order, family, genus
and species. QIIME (version 2) (28) underwent executions of α diversity
and β diversity investigations as a means to dissect the intricacy
within the samples. Comparing the intricacy of species diversity
among samples, α diversity (Chao 1 and Simpson's metric) and β
diversity [weighted UniFrac β metrics and unweighted pair-group
method with arithmetic mean (UPGMA) clustering] assess the
complexity across various samples. The linear discriminant analysis
(LDA) effect size (LEfSe) software was used to perform LEfSe
(version 1.1.2) analyses, with the LDA score screening value set at
4 (29). Statistical analyses were
performed using R statistical software (version 3.0.3) (30). The extraction, detection, and
quantitative analysis of gut microbiota in the samples were
performed by Wuhan Metware Biotechnology Co., Ltd. (www.metware.cn).
Statistical analysis
The experiments were repeated three times.
Statistical analyses were conducted using SPSS (version 26.0; IBM
Corp.). The significance of variations among groups was assessed
using one-way analysis of variance followed by the LSD test. The
Kruskal-Wallis test was used when the data were not able to be
properly transformed. Plotting bar graphs were created using
GraphPad Prism (version 9.0; Dotmatics). Data are presented as mean
± standard deviation. P≤0.05 was considered to indicate a
statistically significant difference.
Results
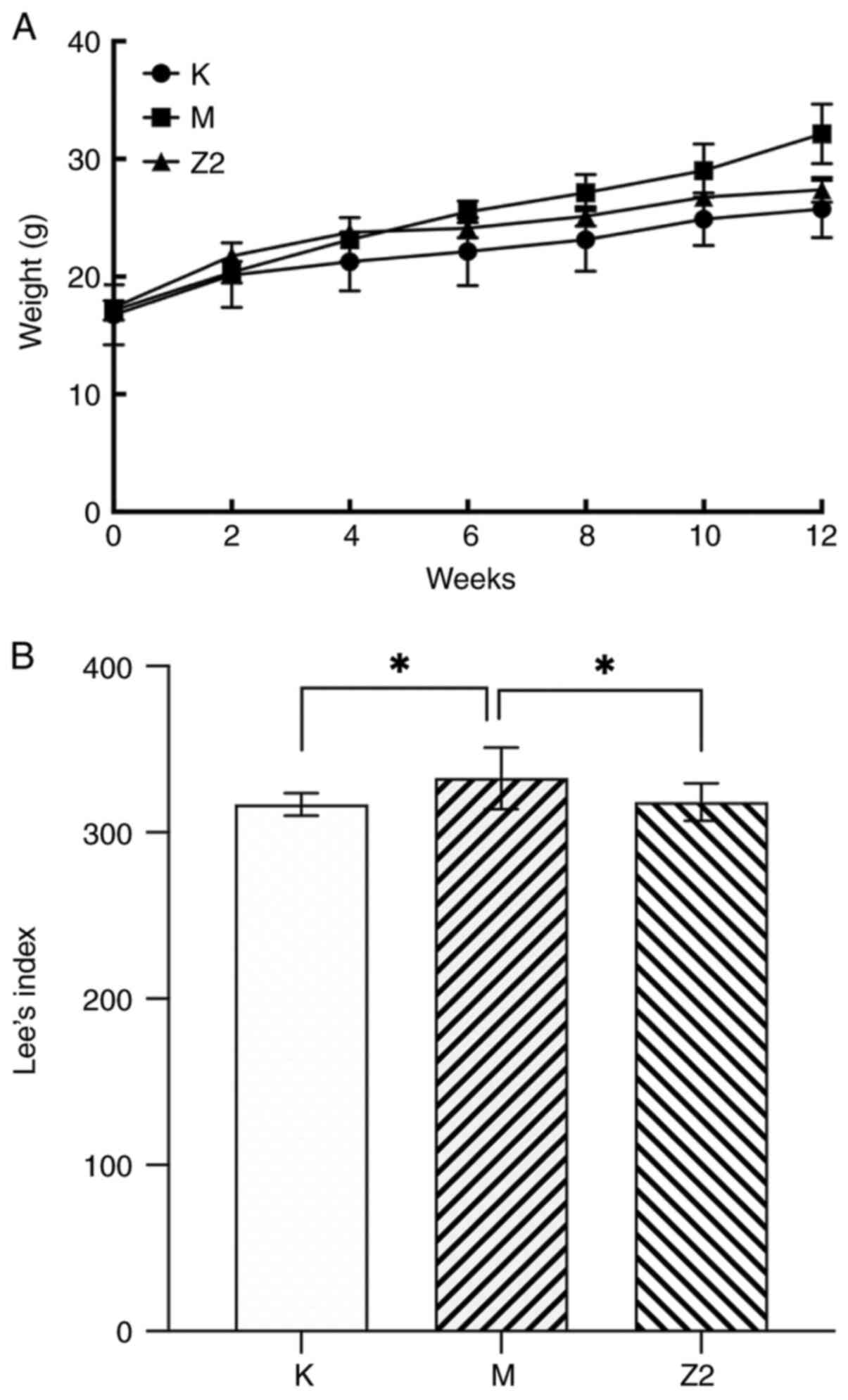
Impact of LF on body weight and the
Lee's index in mice
As the mice aged, their food and water consumption
increased. The Z2 group exhibited an increase in their daily water
intake. The experiment started with ~8 mm per mouse and ended with
~12 m per mouse. As depicted in Fig.
1A, throughout the entire duration of the animal experiment,
mice in the M group exhibited a greater weight gain compared with
those in the K group, while the Z2 group, also subjected to a
high-fat diet, demonstrated a slower rate of weight gain with LF
intervention. Lee's index is a reliable indicator of the
correlation between the mass of body fat and the weight of the
body. As shown in Fig. 1B, the
Lee's index of Group M was significantly higher (P<0.05)
compared with that of Group K, indicating that the mouse obesity
model was successfully constructed. Compared with group M, Lee's
index of Group Z2 exhibited a significant decrease (P<0.05),
suggesting that LF has a therapeutic effect on mouse obesity. The
Lee's index of mice in Group Z2 was closer to that of mice in Group
K and was significantly different from that of mice in Group M
(P<0.05).
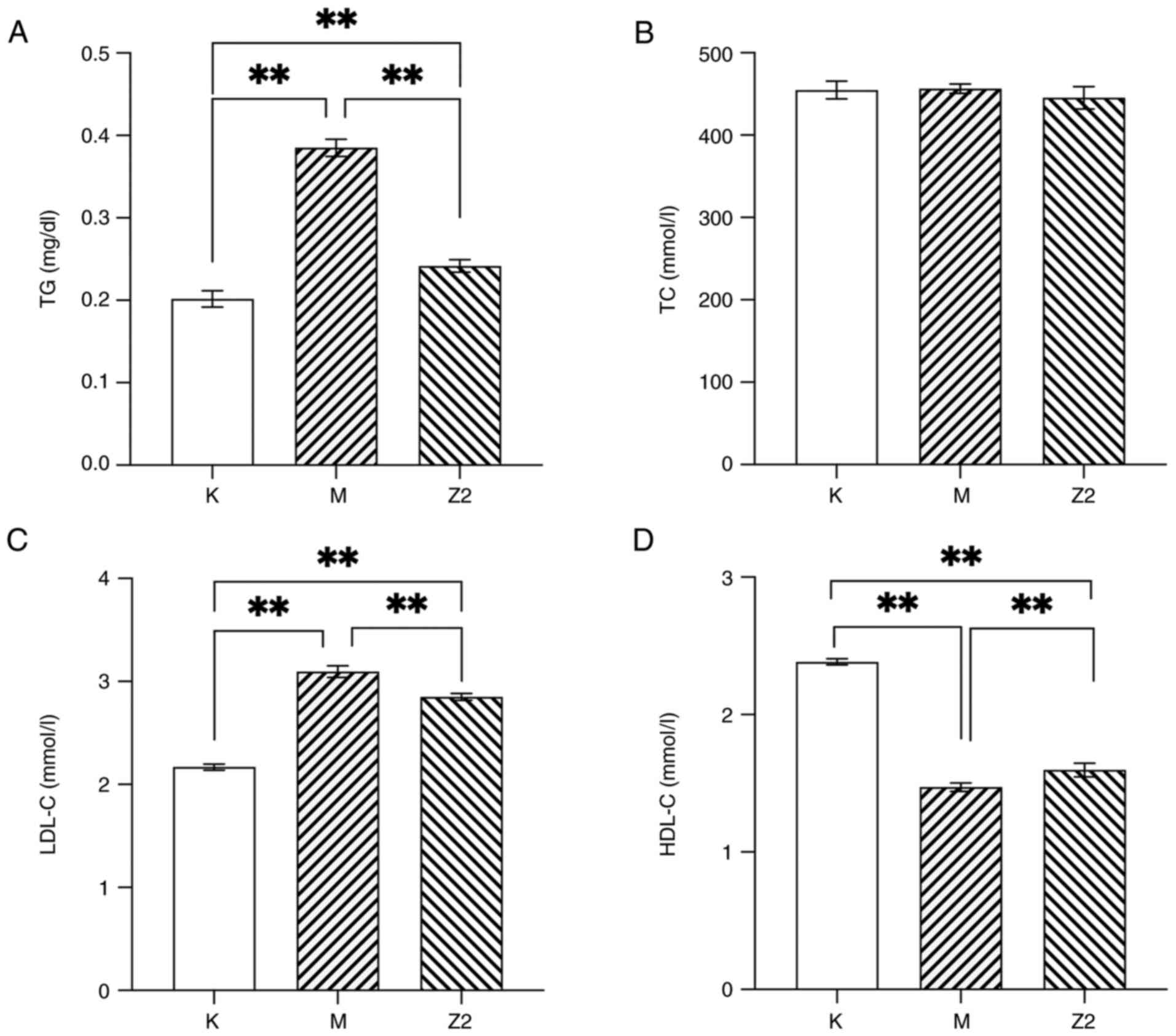
Effect of LF on blood lipids in mice
fed a high-fat diet
A high-fat diet resulted in a significant increase
in TG and LDL-C levels (P<0.01) as well as a decrease in HDL-C
levels (P<0.01) in Group M compared with Group K, suggesting
that abnormalities of lipid metabolism existed in the mice that had
been reared on a high-fat diet (Fig.
2). After the intervention, the results showed that LF was able
to significantly reduce TG and LDL-C levels in obese mice
(P<0.01; Fig. 2A and C), but
there was no effect on TC concentration (Fig. 2B). In Fig. 2D, the HDL-C concentration was
significantly higher in both Group K and Z2 compared with that in
Group M (P<0.01). It can be observed that the intervention of LF
can improve dyslipidemia in obese mice.
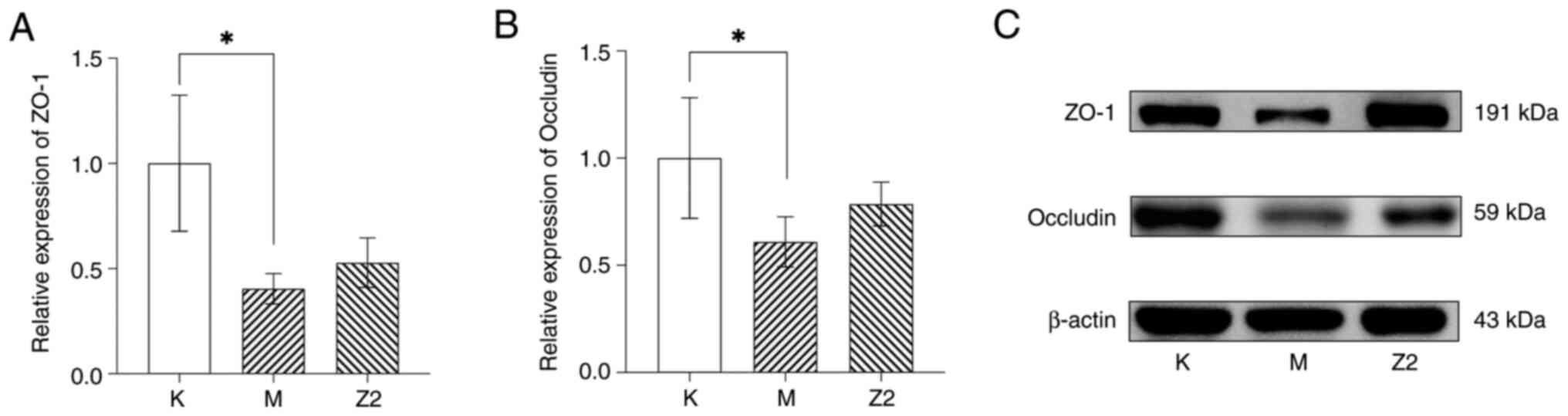
Effect of LF on intestinal tight
junction proteins
In the obese mouse model, the levels of ZO-1 and
occludin expression levels were shown to be reduced compared with
those in Group K (P<0.05; Fig.
3). After the intervention of LF, it could be observed that the
expression of ZO-1 and occludin in Group Z2 gradually recovered,
but the difference was not statistically significant.
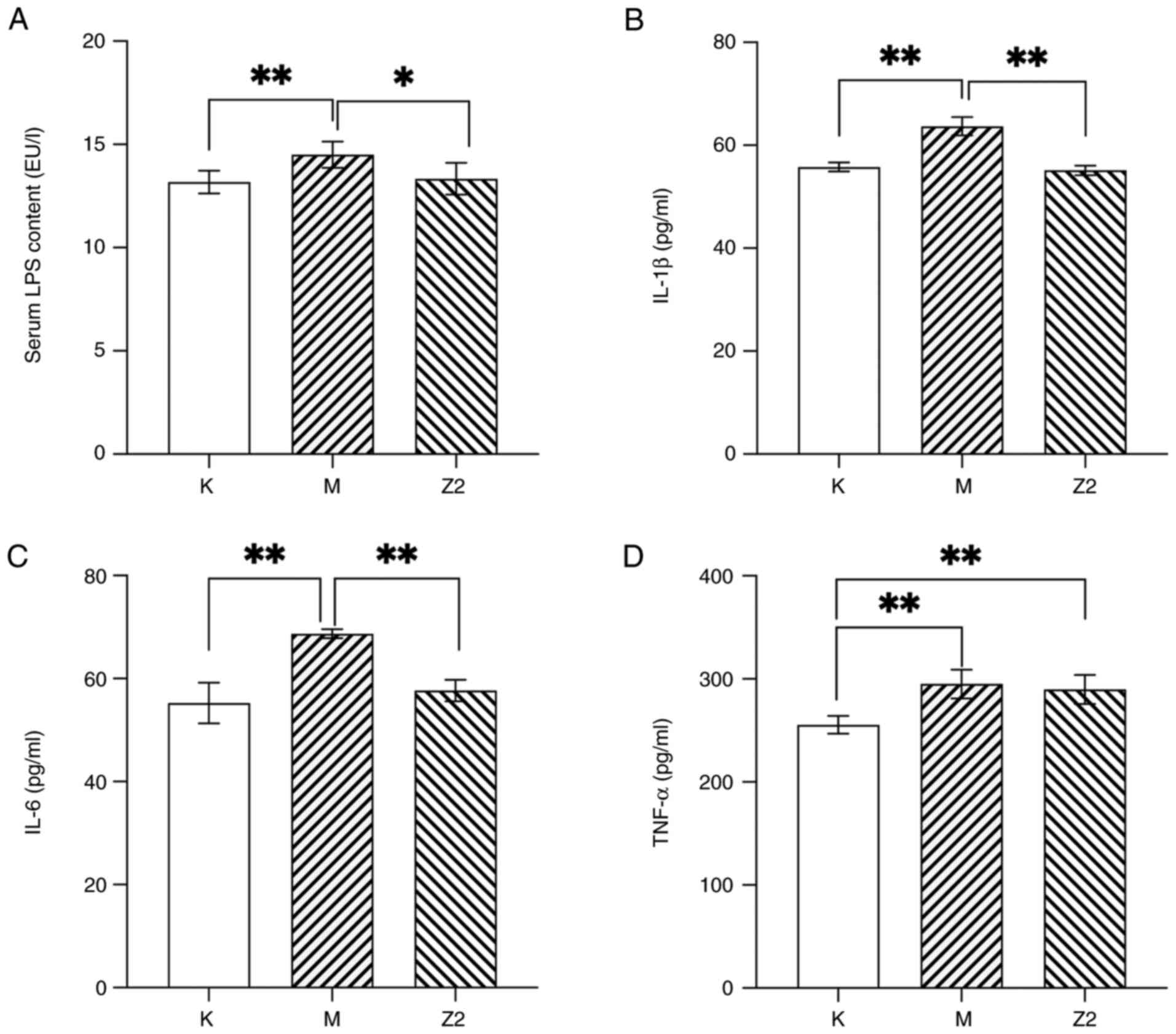
Effect of LF on serum levels of LPS as
well as inflammatory factors
The levels of LPS in the three groups of mice were
compared to the potential relationship between chronic low-grade
inflammation in obesity and LPS stimulation (Fig. 4A). Compared with Group K, LPS was
significantly increased in Group M (P<0.01). However, in Group
Z2, there was a decrease in LPS concentration compared with that in
Group M (P<0.05). This indicates that LF can reduce elevated LPS
levels caused by a high-fat diet.
Blood LPS can activate the relevant inflammatory
pathways and promote the release of inflammatory factors (31); therefore, the effect of LF on the
serum inflammatory factor mass concentration in experimental mice
was further investigated (Fig. 4).
Compared with Group K, the mass concentrations of IL-1β, IL-6 and
TNF-α were elevated in Group M, and the difference was significant
(P<0.01). Compared with Group M, the mass concentrations of
IL-1β, IL-6 and TNF-α were reduced in Group Z2, with significant
differences in the mass concentrations of IL-1β and IL-6
(P<0.01). These results indicate that consuming a diet rich in
fats results in a rise in inflammatory factors within the
bloodstream, and that LF reduces the blood mass concentration of
inflammatory factors, especially that of IL-1β and IL-6.
Effect of LF on the intestinal flora
of mice on a high-fat diet
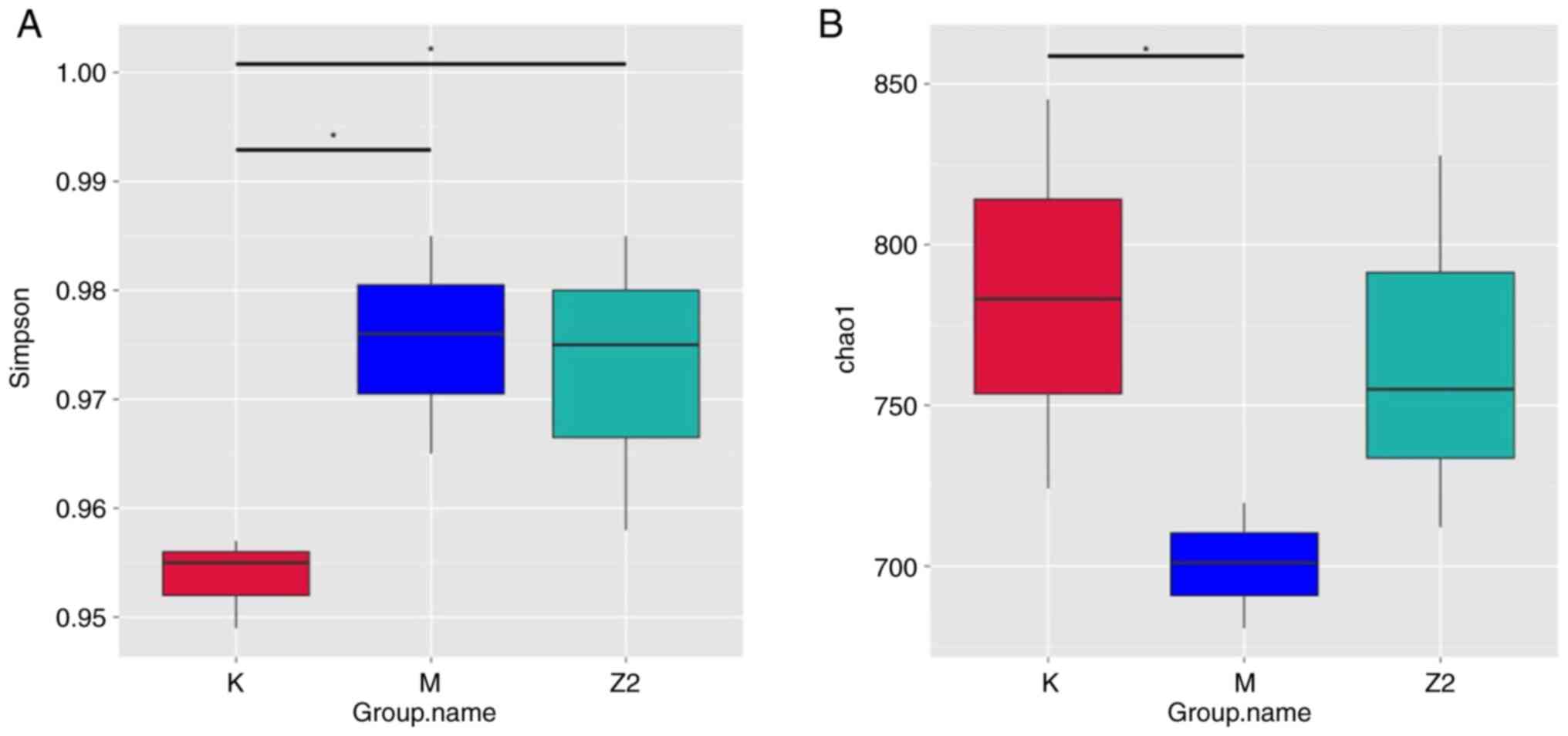
Analysis of differences between groups in the α
diversity index
α diversity is used to indicate the abundance and
variety of organisms in a given specimen (32). Chao 1 and Simpson's metrics are
shown in Fig. 5; in the Chao 1
index, the difference between group K and group M was significant,
and in Simpson's index, group K differed significantly from groups
M and Z2. However, the Chao 1 index plot shows that the total
number of species in the intestinal flora of mice in Group M was
significantly lower compared with that of Group K, and the total
number of species in the intestinal flora of mice in Group Z2 was
closer to that of the K group. From the Simpson's index plot, it
can be observed that the diversity and evenness of species
distribution within the bacterial community of Group M were
significantly increased, and the diversity and evenness of species
distribution within the community of Group Z2 were reduced, which
differed significantly from those of Group K. The diversity and
evenness of species distribution within the bacterial community of
Group Z2 were significantly increased.
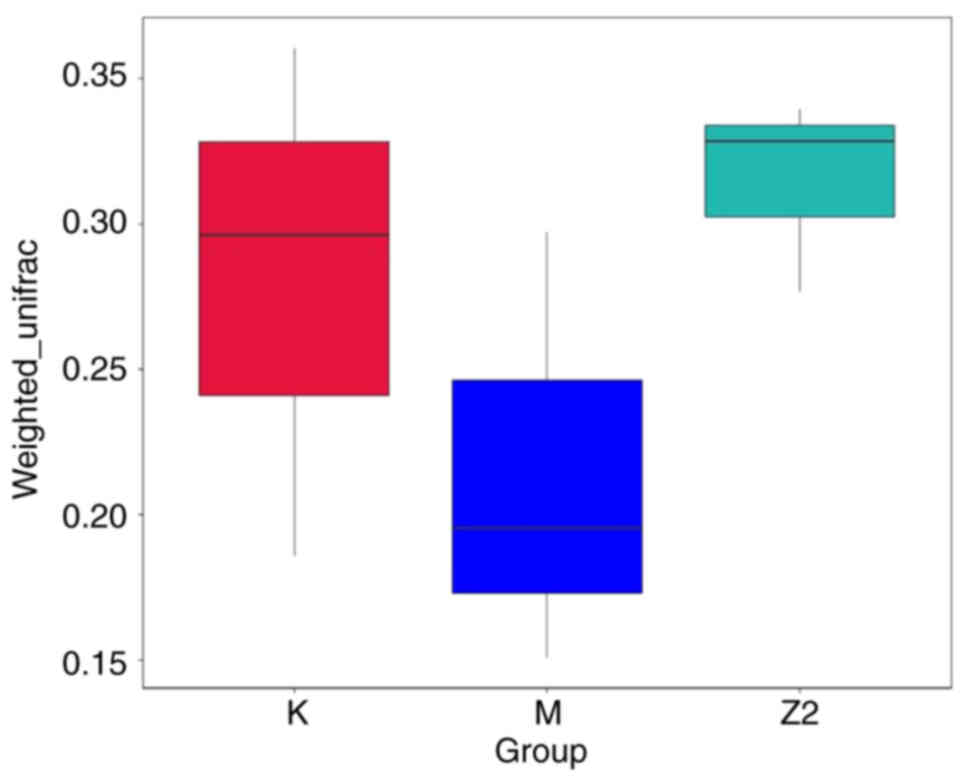
β diversity index intergroup
difference analysis
Beta diversity is used to compare differences in
species composition between communities. Fig. 6 displays the boxplot representation
of the analysis of intergroup differences in β diversity. It was
revealed that the intestinal flora composition of Group M differed
from that of Group K, while the intestinal flora composition of
Group Z2 was more similar to that of Group K, but the differences
between groups was not significant.
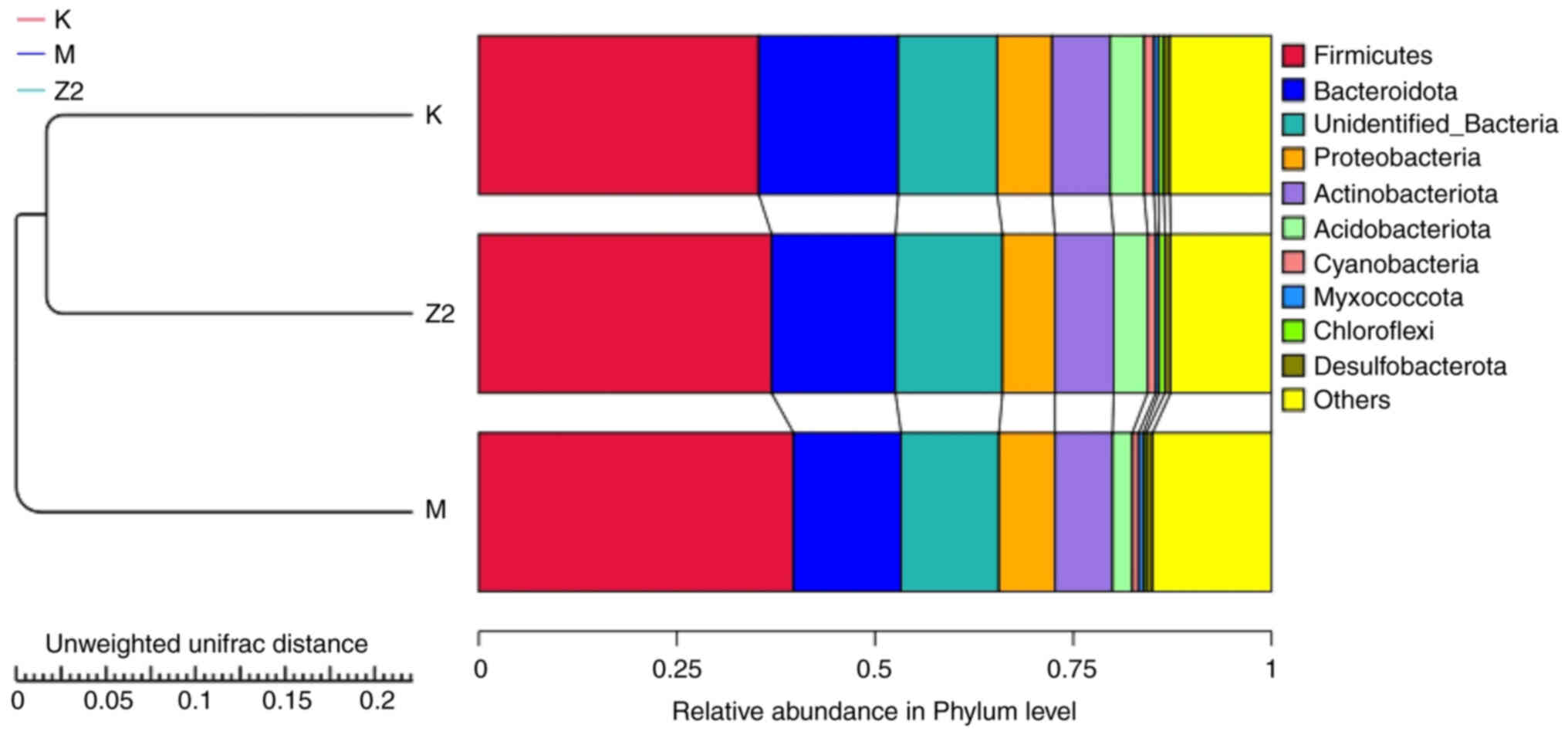
The execution of the UPGMA clustering analysis was
facilitated through an unweighted UniFrac distance matrix. The
assimilation of the clustering consequences coincided with the
proportional prevalence of species at the gateway echelon for every
specimen (Fig. 7). Group K and Z2
exhibited a closer clustering, revealing a higher abundance of
Firmicutes in Group M compared with Group K and Z2. By
contrast, the proportion of Bacteroidota was relatively low.
Following the administration of LF, Group Z2 exhibited a
comparatively reduced ratio of Firmicutes and an elevated
ratio of Bacteroidota, with the
Firmicutes/Bacteroidota ratio resembling that of Group
K.
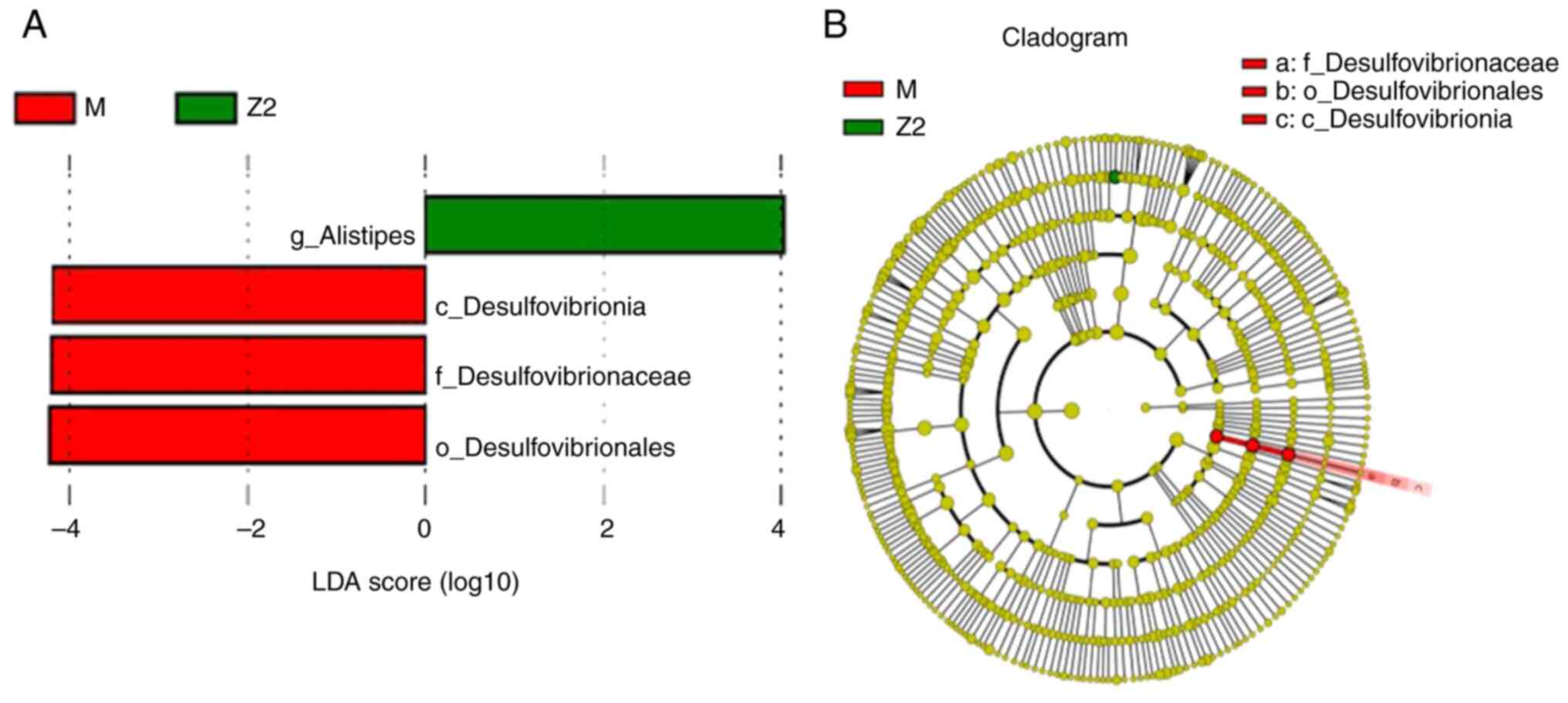
Association study of three groups of
intestinal flora compartments
Biomarkers that were statistically different between
the analyzed groups were investigated using the LEfSe method. The
results of the LEfSe histogram and evolutionary branching diagram
are shown in Fig. 8, where the
abundance of the four species of enterobacteria in the intestinal
flora of the two groups was compared, and the difference was
statistically significant (P<0.05). Among them, Alistipes
was significantly higher in Group Z2 compared with Group M
(P<0.05), while Desulfovibrionia, Desulfovibrioniaceae
and Desulfovibrionales were more abundant in Group M
compared with in Group Z2 (P<0.05).
Analysis of species differences
between groups
To look for species that differed between groups at
different levels of classification, a t-test test was completed
between groups (Fig. 9). At the
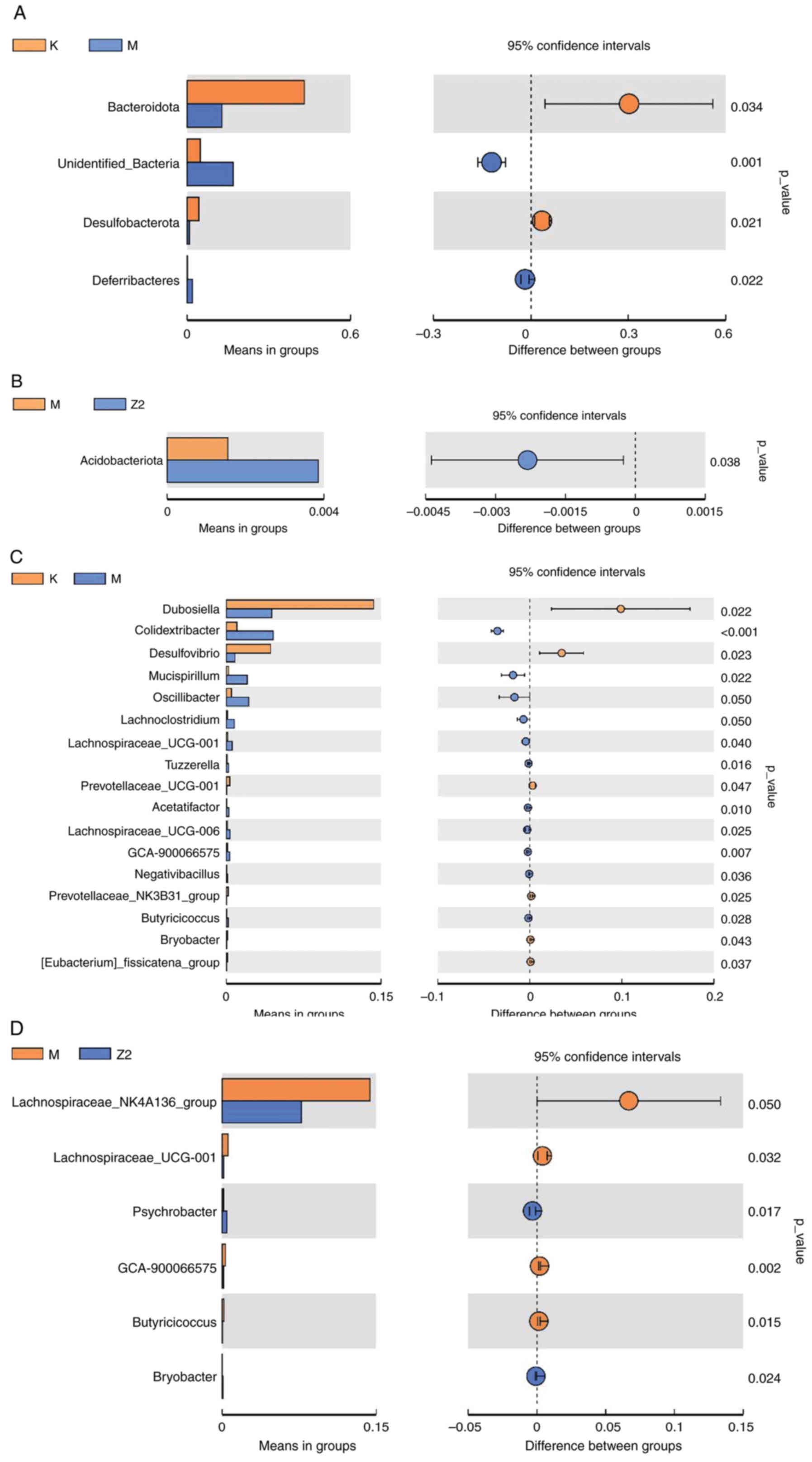
gate level, the abundance of Deferribacteres was increased,
and the abundance of Bacteroidata and
Desulfobacterota was decreased in Group M compared with
Group K (Fig. 9A); the abundance
of Acidobacteriota was significantly higher in Group Z2
compared with Group M (Fig. 9B).
At the genus level, the abundance of Colldextribacter,
Mucispirillum, Oscillibacter, Lachnoclostridium and
Lachnospiraceae-UCG-001 was significantly higher in Group M
compared with Group K, while the abundance of Dubosiella and
Desulfovibrio was decreased (Fig. 9C); Group Z2 showed relatively
higher abundance of Psychrobacter and Bryobacter
compared with Group M (Fig.
9D).
Discussion
The present study investigated the impact of LF on
lipid metabolism and the composition of intestinal flora in mice
that were fed a high-fat diet. Obese mice exposed to a long-term
high-fat diet exhibited severe disruption in lipid metabolism and a
persistent low-level inflammatory reaction. Following
administration of LF, a substantial reduction in Lee's index and
serum TC, LDL and levels of inflammatory factors were observed in
obese mice; at the same time, serum HDL levels increased.
LF-treated mice have improved gut flora structure with weight loss.
The results of the current study indicated that LF has a beneficial
impact on lipid metabolism and chronic low-grade inflammation in
obese mice. Furthermore, the mechanism of action appears to be
strongly linked to gut microbiota.
The perception of obesity has shifted, and obesity
is not only seen as a change in physical appearance but it is
considered a disease. Obesity is accompanied not only by elevated
inflammatory markers, but also by a variety of metabolic diseases,
including type 2 diabetes, hypertension and dyslipidemia. Central
obesity with increased abdominal fat is the main type of obesity
leading to metabolic disorders caused by increased inflammatory
factors in the body (33). On one
hand, macrophage activation in this obese population transforms
into a proinflammatory M1 phenotype and infiltrates target organs
in large numbers, producing inflammatory factors such as IL-1β,
IL-6, TNF-α and others that can affect insulin receptor signaling
and generate insulin resistance, which is a key link in the
development of metabolic diseases (34–36).
On the other hand, elevated concentrations of intestinal-derived
LPS in the circulatory system are an independent risk factor for
chronic low-grade inflammation, and LPS can stimulate TLR4 to
elicit a series of inflammatory cascade responses (37,38).
After intervening with LF in mice fed a high-fat diet, a decrease
in the levels of LPS, IL-1β and IL-6 was observed in the present
study. It can be seen that LF can improve the type of central
obesity that is closely associated with increased inflammatory
factors, and the role of LF in other metabolic diseases needs to be
further explored.
The intestinal tract is exposed to a variety of
stimuli from the external environment daily despite the presence of
a mucus layer as well as a barrier layer made up of intestinal
epithelial cells that help limit the entry of intestinal contents
into the body. If the integrity of the intestinal barrier is
compromised, intestinal immune homeostasis is also implicated
(39). Tight junction proteins are
intercellular junction complexes (40) that seal the paracellular space and
act as a restriction on the exchange of substances between the gut
and the body. The results of the current study showed that the
expression of ZO-1 and occludin was significantly reduced in obese
mice fed a high-fat diet. This result is detrimental to the
integrity of the intestinal barrier and may lead to large amounts
of LPS in the intestine entering the circulation with intestinal
leakage, triggering an excessive immune response in the intestine
and promoting the expression of inflammatory factors (41). The expression of both compact
proteins increased in the LF intervention group relative to the
high-fat group, although the difference was not significant. It can
be argued that LF intervention can increase the expression of
intestinal tight junction proteins as a means of limiting
intestinal-sourced LPS entry into the bloodstream and attenuating
the chronic low-grade inflammatory response at source in obese
mice.
The variations in the composition of the gut
microbiota among the three mouse groups were also examined in the
present study. At the gate level, the findings indicated that the
proportion of Firmicutes/Bacteroidota in the gut of mice in
the model group was greater compared with Group K and Z2. Although
the latter two ratios are more similar, this outcome aligns with
the findings of previous studies (42–44).
Research on overweight mice has consistently indicated that obesity
is frequently accompanied by a higher
Firmicutes/Bacteroidota ratio. Additionally, there are
chronic low-grade inflammatory response in the intestines and
disorders of glucose and lipid metabolism in the body. After a more
in-depth analysis of the differences between the intestinal flora
compartments of the three groups of mice, the present study
revealed that at the phylum-to-genera level, Group M exhibited
higher levels of Deferribacteres, Colldextribacter,
Mucispirillum, Oscillibacter and Lachnoclostridium
compared with Group K. The percentage of these enteric bacteria is
elevated in patients with both obesity and intestinal inflammation,
and is strongly associated with intestinal infections (45–47).
LF intervention in mice fed a high-fat diet was followed by changes
in the structure of the intestinal flora, as shown by the LEfSe
analysis. Acidobacteriota and Alistipes were elevated
in abundance in Group Z2 compared with Group M.
Acidobacteriota are bacteria enriched in the gut of healthy
individuals (48).
Alistipes is a genus in the order Bacteroidetes, an
anaerobic bacterium present in the intestinal tract of healthy
humans (49). A previous study has
shown that the abundance of Alistipes is reduced in the gut
of patients with non-alcoholic fatty liver disease with liver
fibrosis (50). Alistipes
is also a producer of acetic and propionic acids (51), which belong to a group of
short-chain fatty acids (SCFAs), a class of metabolites of
intestinal flora with anti-inflammatory properties (52). Other findings have shown positive
effects of SCFAs on both intestinal barrier integrity and metabolic
diseases as well (53–55).
These changes in intestinal flora may be closely
related to the role played by LF. In the intestinal ecosystem, LF
can achieve bacteriostatic effects through its iron chelating
effect to rob iron ions from intestinal pathogenic bacteria and
inhibit the formation of pathogenic bacterial biofilm (56), which can help host intestinal
microorganisms to increase the resistance of colonization to reduce
the threat of potential pathogens (57). Meanwhile, the low-iron environment
can promote the growth of probiotics (58), activate intestinal immune cells and
maintain the integrity of the intestinal barrier (59). All of these experimental results
suggest that LF attenuates the inflammatory response in obese mice,
possibly by modulating the structure and relative abundance of the
intestinal flora, including a reduction in the abundance of
inflammation-associated flora as well as an increase in the
abundance of short-chain fatty acid-producing flora.
In conclusion, the present study revealed that LF
attenuated the inflammatory response and reduced body weight and
lipid levels in HFD-fed mice, and that the underlying mechanisms
may be through an increase in intestinal tight junction proteins as
well as modulation of the structure and relative abundance of
intestinal flora. The current study provides a theoretical basis
for LF to be used as a safe, anti-obesity treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Inner Mongolia Health Commission
Project (grant no. 202202144), the Inner Mongolia Medical
University Research Project (grant no. YKD2022MS078) and the
Natural Science Foundation of Inner Mongolia Autonomous Region
(grant no. 2023LHMS03065).
Availability of data and materials
High-throughput sequencing data have been uploaded
to NCBI's SRA database (ID, PRJNA1085847). The other data generated
in the present study may be requested from the corresponding
author.
Authors' contributions
The animal experiments were conceptualized and
overseen by LL. WW was responsible for writing the manuscript as
well as data analysis and interpretation. JZ and YL were
responsible for acquiring data and writing the manuscript. SS, LW
and RH were responsible for analyzing the data. WW and LL confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Experimental animal use license no. SYXK-2020-0003.
The procedures received approval from the Medical Ethics Committee
of Inner Mongolia Medical University (approval no.
YKD202301158).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mohammed MS, Sendra S, Lloret J and Bosch
I: Systems and WBANs for controlling obesity. J Healthc Eng.
2018:15647482018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Caballero B: Humans against obesity: Who
will win? Adv Nutr. 10:S4–S9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pan XF, Wang L and Pan A: Epidemiology and
determinants of obesity in China. Lancet Diabetes Endocrinol.
9:373–392. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Du SF, Wang HJ, Zhang B, Zhai FY and
Popkin BM: China in the period of transition from scarcity and
extensive undernutrition to emerging nutrition-related
non-communicable diseases, 1949–1992. Obes Rev. 15 (Suppl
1):S8–S15. 2014. View Article : Google Scholar
|
|
5
|
Meldrum DR, Morris MA and Gambone JC:
Obesity pandemic: Causes, consequences, and solutions-but do we
have the will? Fertil Steril. 107:833–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esser N, Legrand-Poels S, Piette J, Scheen
AJ and Paquot N: Inflammation as a link between obesity, metabolic
syndrome and type 2 diabetes. Diabetes Res Clin Pract. 105:141–150.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Longo M, Zatterale F, Naderi J, Parrillo
L, Formisano P, Raciti GA, Beguinot F and Miele C: Adipose tissue
dysfunction as determinant of obesity-associated metabolic
complications. Int J Mol Sci. 20:23582019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Meng Y, He S, Tan X, Zhang Y, Zhang
X, Wang L and Zheng W: Macrophages, chronic inflammation, and
insulin resistance. Cells. 11:30012022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leuti A, Fazio D, Fava M, Piccoli A, Oddi
S and Maccarrone M: Bioactive lipids, inflammation and chronic
diseases. Adv Drug Deliv Rev. 159:133–169. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiurchiu V, Leuti A and Maccarrone M:
Bioactive lipids and chronic inflammation: Managing the fire
within. Front Immunol. 9:382018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iyengar NM, Gucalp A, Dannenberg AJ and
Hudis CA: Obesity and cancer mechanisms: Tumor microenvironment and
inflammation. J Clin Oncol. 34:4270–4276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kolb R, Sutterwala FS and Zhang W: Obesity
and cancer: Inflammation bridges the two. Curr Opin Pharmacol.
29:77–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clemente JC, Ursell LK, Parfrey LW and
Knight R: The impact of the gut microbiota on human health: An
integrative view. Cell. 148:1258–1270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Cui W, Li X and Yang H:
Interaction between commensal bacteria, immune response and the
intestinal barrier in inflammatory bowel disease. Front Immunol.
12:7619812021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patterson E, Ryan PM, Cryan JF, Dinan TG,
Ross RP, Fitzgerald GF and Stanton C: Gut microbiota, obesity and
diabetes. Postgrad Med J. 92:286–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dabke K, Hendrick G and Devkota S: The gut
microbiome and metabolic syndrome. J Clin Invest. 129:4050–4057.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Usuda H, Okamoto T and Wada K: Leaky gut:
Effect of dietary fiber and fats on microbiome and intestinal
barrier. Int J Mol Sci. 22:76132021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Z, Jiang W, Huang W, Lin Y, Chan FKL
and Ng SC: Gut microbiota in patients with obesity and metabolic
disorders-a systematic review. Genes Nutr. 17:22022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ramirez-Rico G, Drago-Serrano ME,
Leon-Sicairos N and de la Garza M: Lactoferrin: A nutraceutical
with activity against colorectal cancer. Front Pharmacol.
13:8558522022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antoshin AA, Shpichka AI, Huang G, Chen K,
Lu P, Svistunov AA, Lychagin AV, Lipina MM, Sinelnikov MY, Reshetov
IV and Timashev PS: Lactoferrin as a regenerative agent: The
old-new panacea? Pharmacol Res. 167:1055642021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vega-Bautista A, de la Garza M, Carrero
JC, Campos-Rodriguez R, Godinez-Victoria M and Drago-Serrano ME:
The impact of lactoferrin on the growth of intestinal inhabitant
bacteria. Int J Mol Sci. 20:47072019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li L, Ma C, Hurilebagen, Yuan H, Hu R and
Wang W: Weilisi: Effects of lactoferrin on intestinal flora of
metabolic disorder mice. BMC Microbiol. 22:1812022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Min QQ, Qin LQ, Sun ZZ, Zuo WT, Zhao L and
Xu JY: Effects of metformin combined with lactoferrin on lipid
accumulation and metabolism in mice fed with high-fat diet.
Nutrients. 10:16282018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yildirim A, Tamer SA, Sahin D, Bagriacik
F, Kahraman MM, Onur ND, Cayirli YB, Kaya ÖT, Aksu B, Akdeniz E, et
al: The effects of antibiotics and melatonin on hepato-intestinal
inflammation and gut microbial dysbiosis induced by a short-term
high-fat diet consumption in rats. Br J Nutr. 122:841–855. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan E, Duan X, Xiang L, Ren J, Lai X, Li
Q, Sun L and Sun S: Aged oolong tea reduces high-fat diet-induced
fat accumulation and dyslipidemia by regulating the AMPK/ACC
signaling pathway. Nutrients. 10:1872018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu H, Chen X, Hu X, Niu H, Tian R, Wang
H, Pang H, Jiang L, Qiu B, Chen X, et al: Alterations in the gut
microbiome and metabolism with coronary artery disease severity.
Microbiome. 7:682019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bolyen E, Rideout JR, Dillon MR, Bokulich
NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M,
Asnicar F, et al: Reproducible, interactive, scalable and
extensible microbiome data science using QIIME 2. Nat Biotechnol.
37:852–857. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Segata N, Izard J, Waldron L, Gevers D,
Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker
discovery and explanation. Genome Biol. 12:R602011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pannaraj PS, Li F, Cerini C, Bender JM,
Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, et
al: Association between breast milk bacterial communities and
establishment and development of the infant gut microbiome. JAMA
Pediatr. 171:647–654. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Q, Li Y, Li P, Wang M, Wang J, Tang Z,
Wang T, Luo L, Wang C, Wang T and Zhao B: Research progress in the
relationship between type 2 diabetes mellitus and intestinal flora.
Biomed Pharmacother. 117:1091382019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding X, Zhou J, Chai Y, Yan Z, Liu X, Dong
Y, Mei X, Jiang Y and Lei H: A metagenomic study of the gut
microbiome in PTB's disease. Microbes Infect. 24:1048932022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iacobini C, Pugliese G, Fantauzzi CB,
Federici M and Menini S: Metabolically healthy versus metabolically
unhealthy obesity. Metabolism. 92:51–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saad MJ, Santos A and Prada PO: Linking
gut microbiota and inflammation to obesity and insulin resistance.
Physiology (Bethesda). 31:283–293. 2016.PubMed/NCBI
|
|
35
|
Grandl G and Wolfrum C: Hemostasis,
endothelial stress, inflammation, and the metabolic syndrome. Semin
Immunopathol. 40:215–224. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Torres S, Fabersani E, Marquez A and
Gauffin-Cano P: Adipose tissue inflammation and metabolic syndrome.
The proactive role of probiotics. Eur J Nutr. 58:27–43. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Orecchioni M, Ghosheh Y, Pramod AB and Ley
K: Macrophage polarization: Different gene signatures in M1(LPS+)
vs. classically and M2(LPS-) vs. alternatively activated
macrophages. Front Immunol. 10:10842019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mohammad S and Thiemermann C: Role of
metabolic endotoxemia in systemic inflammation and potential
interventions. Front Immunol. 11:5941502021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peterson LW and Artis D: Intestinal
epithelial cells: Regulators of barrier function and immune
homeostasis. Nat Rev Immunol. 14:141–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Suzuki T: Regulation of the intestinal
barrier by nutrients: The role of tight junctions. Anim Sci J.
91:e133572020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Di Tommaso N, Gasbarrini A and Ponziani
FR: Intestinal barrier in human health and disease. Int J Environ
Res Public Health. 18:128362021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang L, Zeng B, Liu Z, Liao Z, Zhong Q, Gu
L, Wei H and Fang X: Green tea polyphenols modulate colonic
microbiota diversity and lipid metabolism in high-fat diet treated
HFA mice. J Food Sci. 83:864–873. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ye X, Liu Y, Hu J, Gao Y, Ma Y and Wen D:
Chlorogenic acid-induced gut microbiota improves metabolic
endotoxemia. Front Endocrinol (Lausanne). 12:7626912021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheng Z, Zhang L, Yang L and Chu H: The
critical role of gut microbiota in obesity. Front Endocrinol
(Lausanne). 13:10257062022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou L, Ni Z, Yu J, Cheng W, Cai Z and Yu
C: Correlation between fecal metabolomics and gut microbiota in
obesity and polycystic ovary syndrome. Front Endocrinol (Lausanne).
11:6282020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Murros KE, Huynh VA, Takala TM and Saris
PEJ: Desulfovibrio bacteria are associated with parkinson's
disease. Front Cell Infect Microbiol. 11:6526172021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu M, Li P, An Y, Ren J, Yan D, Cui J, Li
D, Li M, Wang M and Zhong G: Phloretin ameliorates dextran sulfate
sodium-induced ulcerative colitis in mice by regulating the gut
microbiota. Pharmacol Res. 150:1044892019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu S, Li E, Sun Z, Fu D, Duan G, Jiang M,
Yu Y, Mei L, Yang P, Tang Y and Zheng P: Altered gut microbiota and
short chain fatty acids in Chinese children with autism spectrum
disorder. Sci Rep. 9:2872019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shkoporov AN, Chaplin AV, Khokhlova EV,
Shcherbakova VA, Motuzova OV, Bozhenko VK, Kafarskaia LI and Efimov
BA: Alistipes inops sp. nov. and Coprobacter secundus sp. nov.,
isolated from human faeces. Int J Syst Evol Microbiol.
65:4580–4588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rau M, Rehman A, Dittrich M, Groen AK,
Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P and
Geier A: Fecal SCFAs and SCFA-producing bacteria in gut microbiome
of human NAFLD as a putative link to systemic T-cell activation and
advanced disease. United European Gastroenterol J. 6:1496–1507.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Polansky O, Sekelova Z, Faldynova M,
Sebkova A, Sisak F and Rychlik I: Important metabolic pathways and
biological processes expressed by chicken cecal microbiota. Appl
Environ Microbiol. 82:1569–1576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Venegas DP, De la Fuente MK, Landskron G,
González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN and Hermoso
MA: Short chain fatty acids (SCFAs)-mediated gut epithelial and
immune regulation and its relevance for inflammatory bowel
diseases. Front Immunol. 10:2772019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hosseinkhani F, Heinken A, Thiele I,
Lindenburg PW, Harms AC and Hankemeier T: The contribution of gut
bacterial metabolites in the human immune signaling pathway of
non-communicable diseases. Gut Microbes. 13:1–22. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu W, Luo X, Tang J, Mo Q, Zhong H, Zhang
H and Feng F: A bridge for short-chain fatty acids to affect
inflammatory bowel disease, type 1 diabetes, and non-alcoholic
fatty liver disease positively: By changing gut barrier. Eur J
Nutr. 60:2317–2330. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Peng K, Xia S, Xiao S and Yu Q:
Short-chain fatty acids affect the development of inflammatory
bowel disease through intestinal barrier, immunology, and
microbiota: A promising therapy? J Gastroenterol Hepatol.
37:1710–1718. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kell DB, Heyden EL and Pretorius E: The
biology of Lactoferrin, an iron-binding protein that can help
defend against viruses and bacteria. Front Immunol. 11:12212020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sanders ME, Merenstein DJ, Reid G, Gibson
GR and Rastall RA: Probiotics and prebiotics in intestinal health
and disease: From biology to the clinic. Nat Rev Gastroenterol
Hepatol. 16:605–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Carr LE, Virmani MD, Rosa F, Munblit D,
Matazel KS, Elolimy AA and Yeruva L: Role of human milk bioactives
on Infants' gut and immune health. Front Immunol. 12:6040802021.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Galdeano CM, Cazorla SI, Dumit JM, Velez E
and Perdigon G: Beneficial effects of probiotic consumption on the
immune system. Ann Nutr Metab. 74:115–124. 2019. View Article : Google Scholar : PubMed/NCBI
|