Introduction
Avascular necrosis of the femoral head (ANFH) is a
common orthopedic disease that is caused by disruption of blood
supply near the proximal femur (1). It can bring severe pain, swelling,
decreased leg strength and anesthesia (1). The majority of patients experience
hip joint dysfunction in the development of disease for 1–4 years
(2). There are several
conservative and surgical methods to treat ANFH, including oral
non-steroidal anti-inflammatory drugs, traditional Chinese
medicine, physical therapy, lifestyle modification and even
artificial hip replacement (3).
However, there is still no consensus on the treatment of ANFH and
the optimal treatment strategy should be further explored.
Bone marrow mesenchymal stem cells (BMSCs), a
population of multipotent cells (4), play a crucial role in osteonecrosis
repair by their osteogenic differentiation ability (5). The hypoxia environment in ANFH could
significantly affect the physical function of BMSCs (6,7).
Enhanced adipogenic differentiation ability and decreased
osteogenic differentiation potential of BMSCs have been revealed in
ANFH and the occurrence of ANFH is also closely associated with the
changed differentiation direction of BMSCs (8). Thus, it is a promising strategy to
treat ANFH by targeting BMSCs.
Salvia miltiorrhiza Bunge (also known as
Danshen) is a popular traditional Chinese herbal medicine that
exerts therapeutic effects on dilating blood vessels and
ameliorating blood rheological properties (9). Therefore, it has been used to treat
ANFH (10,11), although the in-depth mechanism
behind how the active components function remains to be further
investigated. Tanshinone IIA (Tan IIA) is one of the most abundant
active compounds in S. miltiorrhiza and it is widely used to
treat cerebrovascular and cardiovascular disease due to its
antioxidant and anti-inflammatory ability (12). Furthermore, emerging studies
indicate potential positive biological effects of Tan IIA for MSCs
(13,14). It has been reported that Tan IIA
can promote the in vitro migration of MSCs by increasing the
expression of C-X-C chemokine receptor type 4 (CXCR-4) (13). Tan IIA also enhances expansion
ability of BMSCs by regulating the progression of S phase of the
cell cycle (14). In addition, Tan
IIA can contribute to osteogenic differentiation of periodontal
ligament stem cells (15) and
mouse myoblast cell line C2C12 (16). Thus, Tan IIA might exert treatment
potential for ANFH by targeting BMSCs. However, the role of Tan IIA
in adipogenesis and osteogenesis of BMSCs remains to be
elucidated.
Therefore, in the current study, the effect and
underlying mechanism of Tan IIA on the adipogenesis and
osteogenesis ability of BMSCs under hypoxic environment were
explored. The findings will help illustrate the in-depth mechanism
of how Tan IIA could treat ANFH by targeting BMSCs and thus
providing solid experimental basis for ANFH treatment by using Tan
IIA.
Materials and methods
Cell isolation, culture,
differentiation and treatment
A total of 12 male C57BL/6 mice (age, 6 weeks;
weight, ~16 gram) were obtained from the Animal Model Research
Center of Nanjing University for BMSC isolation. The mice were
housed under a 12-h light/dark cycle with ad libitum access
to food and water, at a temperature of 20–26°C and humidity of
40–60%. The BMSCs were isolated according to previous literature
(17). Briefly, the mice were
sacrificed and the muscles were dissected from the femurs. The
femurs were cleaned by 1X phosphate buffer solution (PBS) and then
fresh BMSCs were obtained by flushing the femoral cavity.
To mimic the hypoxic microenvironment in ANFH, the
BMSCs were differentiated under hypoxic environment with 5% O2c.
The growth medium contains α-MEM (Gibco; Thermo Fisher Scientific,
Inc.; cat. no. 12571071), 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.; cat. no. 10-013-CV) and 1% Penicillin-Streptomycin (Gibco;
Thermo Fisher Scientific, Inc.; cat. no. 15140-122). The growth
medium was replaced every three days. The BMSCs within three
passages were used for osteogenic or adipogenic
differentiation.
The growth medium was replaced by differentiated
medium until BMSCs reached 90% confluence. Osteogenic medium
(Procell Life Science & Technology Co., Ltd.; cat. no.
PD-003-200) was used in osteogenic induction assays of BMSCs for 21
days. Adipogenic induction medium (AIM), containing high-glucose
DMEM (Gibco; Thermo Fisher Scientific, Inc.; cat. no. 12100-046),
10% FBS, 1 µg/ml insulin (MilliporeSigma; cat. no. I2643), 0.25 µM
dexamethasone (MilliporeSigma; cat. no. D4902), 0.5 mM
3-isobutyl-1-methylxanthine (MilliporeSigma; cat. no. I5879) and 1%
penicillin-streptomycin, was applied in adipogenic differentiation
assays for 14 days.
To explore the effect of Tan IIA, the BMSCs were
induced by adipogenic and osteogenic differentiation medium with or
without 5×10-5 mg/l Tan IIA.
In addition, 10 nM TGFβ signaling inhibitor SB431542
(Cell Signaling Technology, Inc.; cat. no. 14775S) (18) and 10 nM AKT signaling inhibitor
Akti-1/2 (an Akt inhibitor; Tocris Bioscience; cat. no. 5773)
(19) were applied to investigate
the role of related signaling in BMSCs differentiation by being
mixed with the differentiation medium. The differentiated assays
were divided into three groups: DMSO, Tan IIA, Tan IIA + Akti 1/2
or SB431542.
Cell Counting Kit-8 assay
The Cell Counting Kit-8 (Beyotime Institute of
Biotechnology; cat. no. C0038) kit was used according to the
manufacturer's introductions. Briefly, the BMSCs were cultured with
or without 5×10-5 mg/l Tan IIA for 24 h. The CCK8 solutions were
added 20 µl/well. After 1 h, the absorbance at 450 nm was detected
by a microplate reader (Thermo Fisher Scientific, Inc.).
Alkaline phosphatase assay
Alkaline Phosphatase Assay kit (Beyotime Institute
of Biotechnology; cat. no. P0321S) was used to assess the
osteogenic differentiation ability of BMSCs according to
manufacturer's introductions. Briefly, the differentiated BMSCs
were firstly lysed by western and IP lysis (Beyotime Institute of
Biotechnology; cat. no. P0013) for 20 min. Then, the lysed product
was incubated with chromogenic substrate at 37°C for 15 min.
Finally, the absorbance at 405 nm was measured by a microplate
reader (Thermo Fisher Scientific, Inc.).
Alizarin red staining
At three weeks after osteogenic differentiation of
BMSCs, BMSCs were fixed in 4% paraformaldehyde at room temperature
for 20 min and then treated with Alizarin red (Beijing Solarbio
Science & Technology Co., Ltd.; cat. no. G1452) at room
temperature for 15 min. Then, the sample was washed three times by
PBS and images captured under microscope. Cetylpyridinium chloride
(10%) was used to dissolve the dye on the mineralized nodules. The
absorbance values in each well at 490 nm were measured with a
microplate reader and the images were captured by a light BX53
microscope (Olympus Corporation).
Oil red staining
Following adipogenic induction, BMSCs were fixed in
4% paraformaldehyde at room temperature for 20 min and treated with
oil red (Beijing Solarbio Science & Technology Co., Ltd.; cat.
no. G1260) at room temperature for 15 min. Then the sample was
washed three times by 60% ethanol and images were captured under a
light microscope. For lipid quantification analysis, the oil red
was isolated with isopropanol for 5 min and the absorbance at 496
nm was measured by a microplate reader.
Gene expression analysis
BMSCs (90% confluence) were lysed for gene
expression analysis. RNA extraction, cDNA synthesis and
quantitative (q)PCR were performed according to the manufacturers'
protocols. Briefly, total RNA was obtained with EZ-press RNA
Purification Kit (EZ Bioscience; cat. no. B0004DP). M-MuLV Reverse
Transcriptase (New England BioLabs, Inc.; cat. no. M0253L) was used
for reverse transcription (RT). The cDNA was synthesized at 42°C
for 1 h. To exclude contamination with genomic DNA, negative
controls were set up during the reverse transcription step of the
RNA to cDNA conversion. The cDNA was used for subsequent RT-qPCR
with SYBR Green Master (Roche Diagnostics; cat. no. 4913914001) in
a CFX96 System. The qPCR cycling conditions were as follows: 95°C
for 3 min; followed by 45 cycles at 95°C for 5 sec and 60°C for 30
sec; followed by an increase from 65 to 95°C in increments of 0.5°C
for 5 sec, and maintenance at 4°C. 2-ΔΔCq was used to
calculate the relative gene expressions. ΔΔCq indicates the
differences between the difference of targets and control genes
between control and treatment groups (20). The primers of RT-qPCR are listed in
Table I.
 | Table I.Primer sequences of reverse
transcription-quantitative PCR. |
Table I.
Primer sequences of reverse
transcription-quantitative PCR.
| Gene | Sequence |
|---|
| Mouse Lpl
Forward |
5′-TTGCCCTAAGGACCCCTGAA-3′ |
| Mouse Lpl
Reverse |
5′-TTGAAGTGGCAGTTAGACACAG-3′ |
| Mouse Cebpa
Forward |
5′-GCGGGAACGCAACAACATC-3′ |
| Mouse Cebpa
Reverse |
5′-GTCACTGGTCAACTCCAGCAC-3′ |
| Mouse Perilipin 1
Forward |
5′-CTGTGTGCAATGCCTATGAGA-3′ |
| Mouse Perilipin 1
Reverse |
5′-CTGGAGGGTATTGAAGAGCCG-3′ |
| Mouse Gapdh
Forward |
5′-AGGTCGGTGTGAACGGATTTG-3′ |
| Mouse Gapdh
Reverse |
5′-GGGGTCGTTGATGGCAACA-3′ |
| Mouse Adipoq
Forward |
5′-GAAGCCGCTTATGTGTATCGC-3′ |
| Mouse Adipoq
Reverse |
5′-GAATGGGTACATTGGGAACAGT-3′ |
| Mouse Pparg
Forward |
5′-GGAAGACCACTCGCATTCCTT-3′ |
| Mouse Pparg
Reverse |
5′-GTAATCAGCAACCATTGGGTCA-3′ |
| Mouse Fabp4
Forward |
5′-AAGGTGAAGAGCATCATAACCCT-3′ |
| Mouse Fabp4
Reverse |
5′-TCACGCCTTTCATAACACATTCC-3′ |
| Mouse Bsp
Forward |
5′-TCCATCGAAGAATCAAAGCA-3′ |
| Mouse Bsp
Reverse |
5′-AGTAGCGTGGCCGGTACTTA-3′ |
| Mouse Ocn
Forward |
5′-AGACTCCGGCGCTACCTT-3′ |
| Mouse Ocn
Reverse |
5′-CTCGTCACAAGCAGGGTTAAG-3′ |
| Mouse Opn
Forward |
5′-GGAAACCAGCCAAGGTAAGC-3′ |
| Mouse Opn
Reverse |
5′-TGCCAATCTCATGGTCGTAG-3′ |
Western blotting
Total proteins were obtained by western and IP lysis
(Beyotime Institute of Biotechnology; cat. no. P0013) mixed with
phenylmethylsulfonyl fluoride and Dithiothreitol. Protein
concentration was measured using the BCA assay (Beyotime Institute
of Biotechnology; cat. no. P0010S) according to the manufacturer's
instructions. Total proteins (20 µg) in loading buffer were
separated by SDS-PAGE on 10% gels and transferred to nitrocellulose
membranes according to the standard methods (21). Full membranes were blocked with 5%
BSA (Beyotime Institute of Biotechnology; cat. no. ST023) in TBST
for 1 h at room temperature. Primary antibodies were incubated
overnight at 4°C: Anti-rabbit-GAPDH (Cell Signaling Technology,
Inc.; cat. no. 2118L; 1:1,000), anti-rabbit-AKT (Cell Signaling
Technology, Inc.; cat. no. 4865; 1:1,000),
anti-rabbit-phosphorylated (p)-AKT (Cell Signaling Technology,
Inc.; cat. no. 4060; 1:1,000), anti-rabbit-cAMP response
element-binding protein (CREB; Proteintech Group, Inc.; cat. no.
12208-1AP; 1:1,000), anti-rabbit-phospho-CREB (Cell Signaling
Technology, Inc.; cat. no. 9198S; 1:1,000),
anti-rabbit-phospho-Smad3 (Absin Bioscience Inc.; cat. no.
abs140144; 1:1,000), anti-mouse-Smad3 (Cell Signaling Technology,
Inc.; cat. no. 9520; 1:1,000). Then the sample was incubated by
HRP-conjugated secondary anti-rabbit or mouse antibodies (Santa
Cruz Biotechnology, Inc.; cat. no. sc-2357; 1:3,000) for 1 h at
room temperature. The target strips were obtained with ECL Reagent
(ShareBio; cat. no. SBWB012).
Bulk RNA sequencing (RNA-Seq)
Purified mRNA was first obtained (EZ Bioscience,
cat. no. B0004DP) according to the instructions of manufacturer and
RNA sequencing libraries were established by NEB Next Ultra RNA
Library Prep Kit for Illumina (New England BioLabs, Inc.; cat. no.
E7530L). Then, the cDNA library was established with non-stranded
library preparation. Paired-end sequencing library was sequenced on
the Illumina HiSeq xten sequencer (Illumina, Inc.) by 2×150 bp read
length run. Gene abundances were quantified by RSEM (http://deweylab.biostat.wisc.edu/rsem/)
(22). Differential expression
analysis was performed using the DEGseq (23) and genes with P-value ≤0.001 were
considered to indicate a statistically significant difference.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology
(GO) analysis were carried out by KOBAS (http://kobas.cbi.pku.edu.cn/home.do) and Goatools
(https://github.com/tanghaibao/Goatools) (24). RNA sequencing data has been
uploaded to NCBI (https://www.ncbi.nlm.nih.gov/; accession no.:
PRJNA1131477).
Animals
Animal experiments were approved by the animal
ethics committee of Suzhou Wujiang District Second People's
Hospital (approval no. WZY2022056; Suzhou, China). A total of 10
wild-type female mice (age, 8 weeks; weight, ~18 g) were procured
from Shanghai Jihui Laboratory Animal Care Co., Ltd. for subsequent
experiments. The mice were housed as aforementioned. Their health
status was monitored weekly. Tri-bromoethanol (250 mg/kg) was used
as an anesthetic to minimize discomfort. Following the
establishment of an ovariectomy model in mice, tan IIA was
administered to explore its osteogenic potential in vivo.
Over a period of four weeks, intraperitoneal injections were
administered every other day. At the end of an eight-week period
post-surgery, mice were sacrificed with CO2, confirmed by the
cessation of breathing and heartbeat for at least five minutes.
Micro-computed tomography (CT)
analysis
Femurs were collected and fixed in 4%
paraformaldehyde at room temperature for 24 h. The samples were
then washed with PBS before being subjected to micro-CT scanning
(Bruker micro-CT; Bruker Corporation). The micro-CT data were
analyzed to assess bone morphology and density.
Statistical analysis
Statistical analysis was conducted by SPSS version
19.0 for Windows (IBM Corp.). Unpaired Student's t-test was used to
compare two groups of experimental data, whereas one-way analysis
of variance and Tukey test were used to compare quantitative
parameters between more than two groups. All the experiment results
were presented as mean ± standard deviation. Each experiment was
repeated three times. All assays were blindly assessed and analyzed
by two independent investigators. P<0.05 was considered to
indicate a statistically significant difference.
Results
Tan IIA promotes osteogenic
differentiation of BMSCs
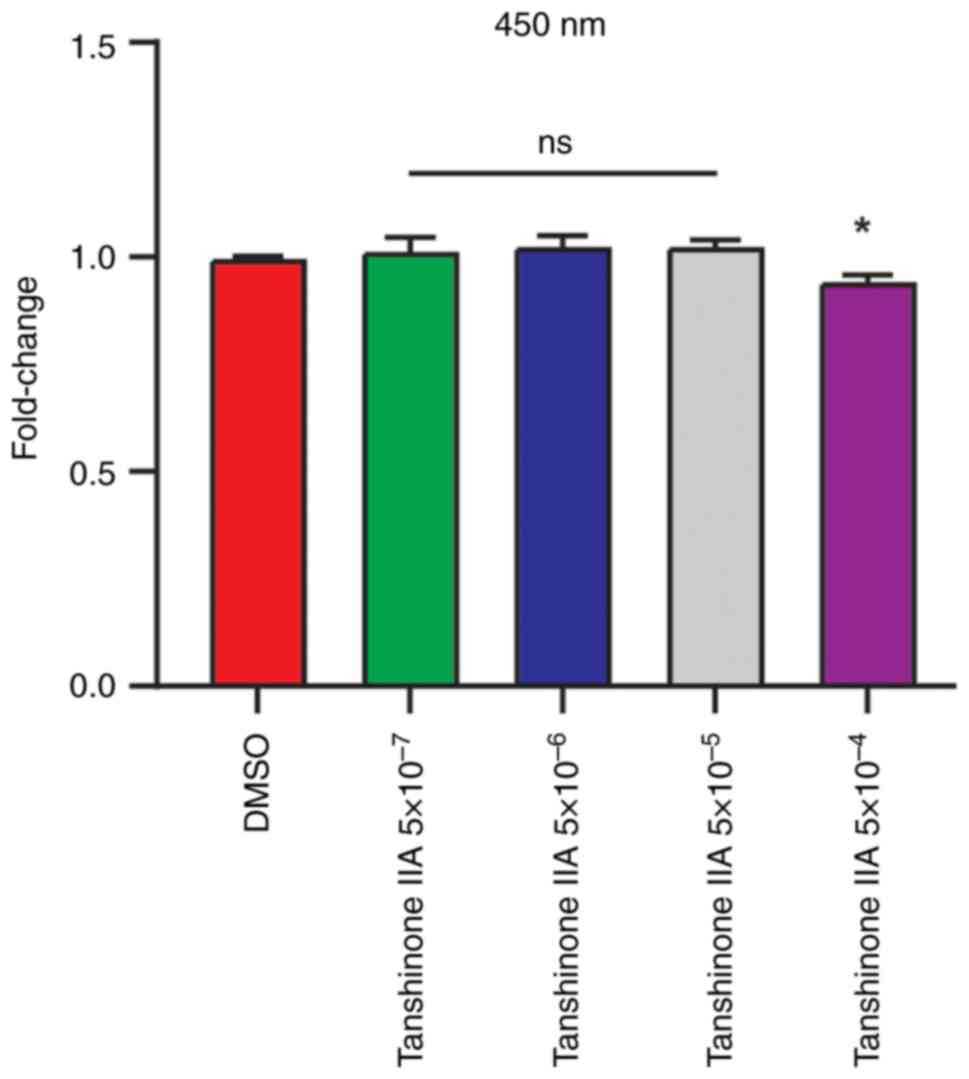
The CCK-8 assay was used to measure the toxicity of
Tan IIA and determine the concentration of Tan IIA in the current
study (Fig. 1). Finally, the
5×10-5 mg/l Tan IIA was used in subsequent experiments. To evaluate
the effects of Tan IIA on osteogenic differentiation of BMSCs, the
BMSCs were cultured in osteogenic induction medium with or without
Tan IIA. At seven days following osteogenic induction of BMSCs, Tan
IIA significantly increased alkaline phosphatase (ALP) level when
compared with control (Fig. 2A and
B). The Alizarin red staining revealed that Tan IIA
significantly enhanced mineralized nodules after 21 days of
osteogenic differentiation (Fig.
2C). RT-qPCR results also found increased expression of
osteogenesis marker genes, including osteocalcin (Ocn), bone
sialoprotein (Bsp) and osteopontin (Opn) and following treatment of
Tan IIA (Fig. 2D). Taken together,
these results demonstrated that Tan IIA promoted osteogenic
differentiation of BMSCs.
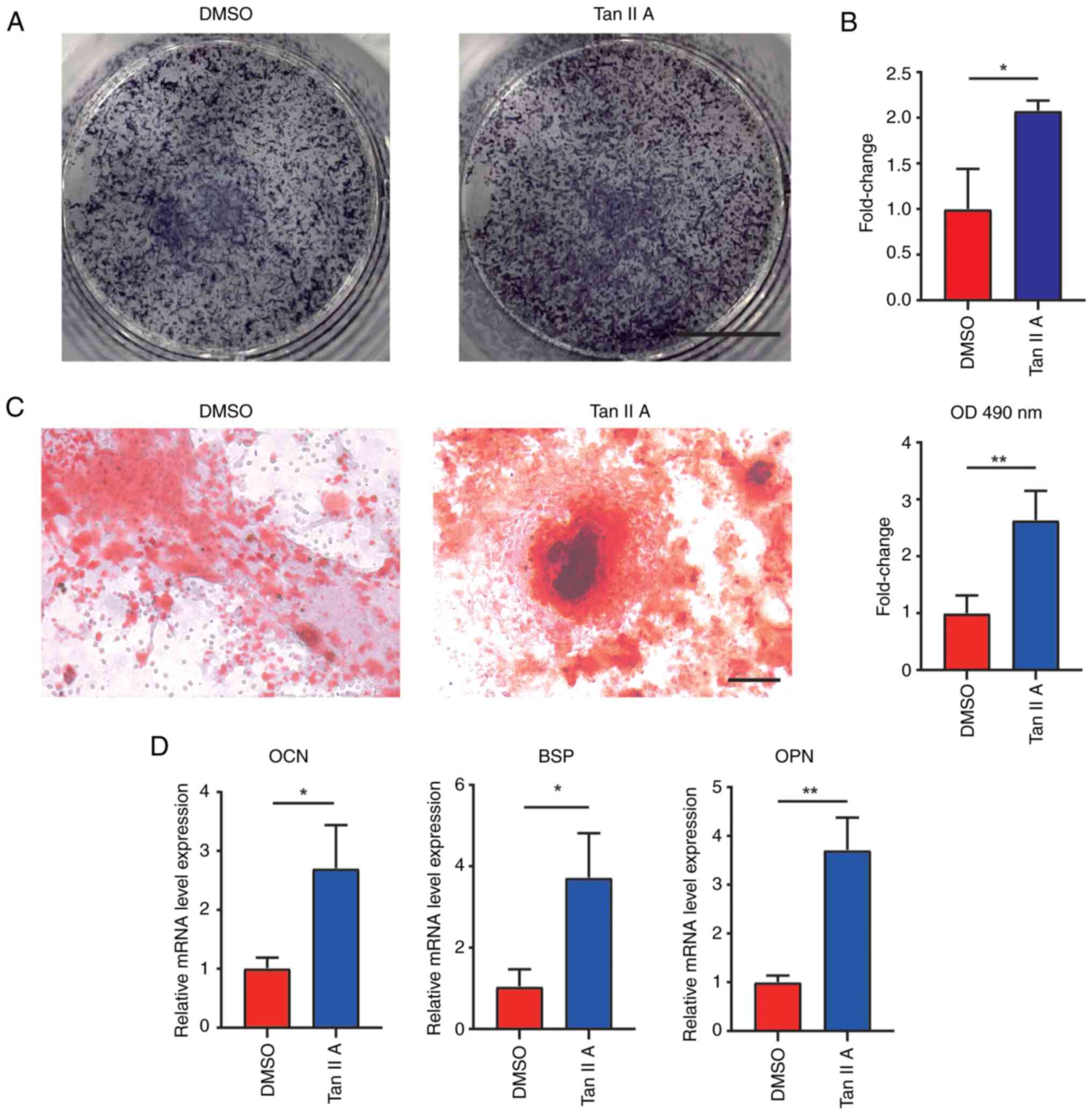 | Figure 2.Tan IIA promotes osteogenic
differentiation of BMSCs. (A) ALP staining for BMSCs and (B)
measurement of ALP in BMSCs following osteogenic differentiation
for 7 days with or without Tan IIA treatments. Scale bar, 5 mm. (C)
Alizarin red staining of BMSCs following osteogenic differentiation
for 21 days in DMSO and Tan IIA groups. Scale bar, 100 µm. (D) Gene
expression of Ocn, Bsp and Opn in BMSCs following osteogenic
differentiation for 21 days in DMSO and Tan IIA groups. *P<0.05,
**P<0.01. Tan IIA, Tanshinone IIA; BMSCs, bone marrow
mesenchymal stem cells; ALP, alkaline phosphatase; Ocn,
osteocalcin; Bsp, bonesialoprotein; Opn, osteopontin. |
Tan IIA suppresses adipogenic
differentiation of BMSCs
To evaluate the effects of Tan IIA on adipogenic
differentiation of BMSCs, the BMSCs were cultured in adipogenic
induction medium with or without Tan IIA for subsequent assays. At
10 days after adipogenic induction of BMSCs, oil red staining
revealed that Tan IIA significantly suppressed formation of lipid
droplets when compared with control (Fig. 3A and B). Furthermore, RT-qPCR
results also found decreased expression of adipogenic marker genes,
including perilipin1 (Plin1), fatty acid-binding protein (Fabp4),
adiponectin (Adipoq), CCAAT/enhancer binding protein alpha (Cebpa),
peroxisome proliferators-activated receptors gamma (Pparg) and
lipoprteinlipase (Lpl) following treatment of Tan IIA (Fig. 3C). Taken together, these results
demonstrated that Tan IIA suppressed the adipogenic differentiation
of BMSCs.
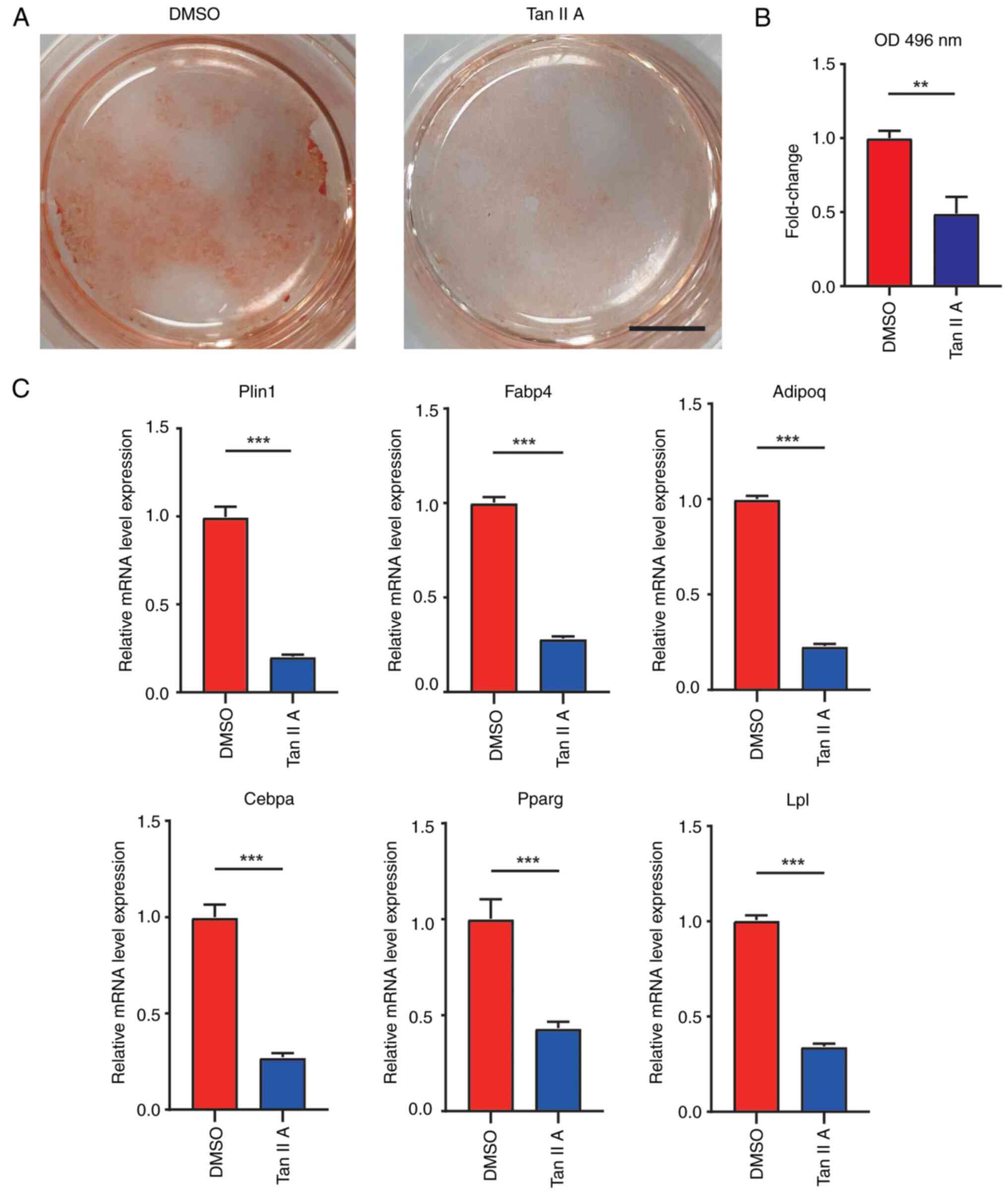 | Figure 3.Tan IIA suppresses adipogenic
differentiation of BMSCs. (A) Oil red staining of BMSCs following
adipogenic differentiation for 14 days in DMSO and Tan IIA groups.
Scale bar, 5 mm. (B) Quantification analysis of oil red staining in
DMSO and Tan IIA groups. (C) Gene expression of Plin1, Fabp4,
Adipoq, Cebpa, Pparg and Lpl of BMSCs following adipogenic
differentiation for 14 days in control and Tan IIA groups.
**P<0.01, ***P<0.001. Tan IIA, Tanshinone IIA; BMSCs, bone
marrow mesenchymal stem cells; Plin1, perilipin1; Fabp4, fatty
acid-binding protein; Adipoq, adiponectin; Cebpa, CCAAT/enhancer
binding protein alpha; Pparg, peroxisome proliferators-activated
receptors gamma; Lpl, lipoprteinlipase. |
RNA-seq reveals activated TGFβ and AKT
signaling of BMSCs following Tan IIA treatment
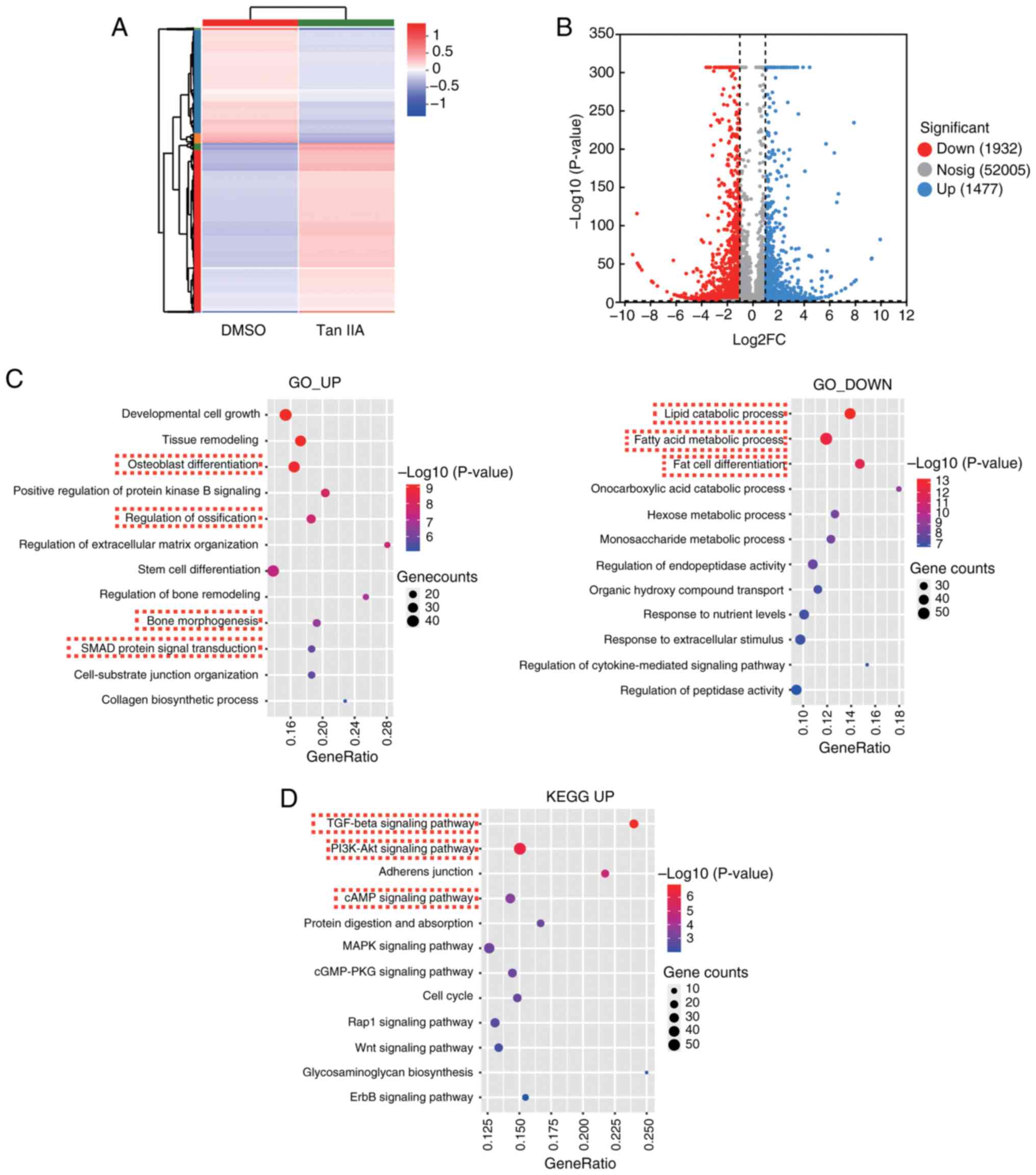
The present study next performed RNA-sequence to
compare the expression profile between BMSCs with or without Tan
IIA treatment. The differentially expressed gene analysis revealed
there were 1,477 upregulated genes and 1932 down-regulated genes
following treatment of Tan IIA (Fig.
4A and B). The GO analysis of upregulated genes enriched key
terms, including osteoblast differentiation, regulation of
ossification and bone morphogenesis (Fig. 4C), which was consistent with
aforementioned phenotype that Tan IIA could promote osteogenic
differentiation of BMSCs. In addition, the GO analysis of
downregulated genes enriched key terms, including lipid catabolic
process, fatty acid metabolic process and fat cell differentiation
(Fig. 4C), which was also
consistent with the results that Tan IIA could inhibit adipogenic
differentiation of BMSCs.
To further investigate the potential mechanism of
how Tan IIA affected differentiation potential of BMSCs, KEGG
analysis of upregulated genes was performed and TGFβ signaling
pathway, AKT signaling pathway and cAMP signaling pathway were
enriched (Fig. 4D). SMAD protein
signaling transduction was also enriched in GO analysis of
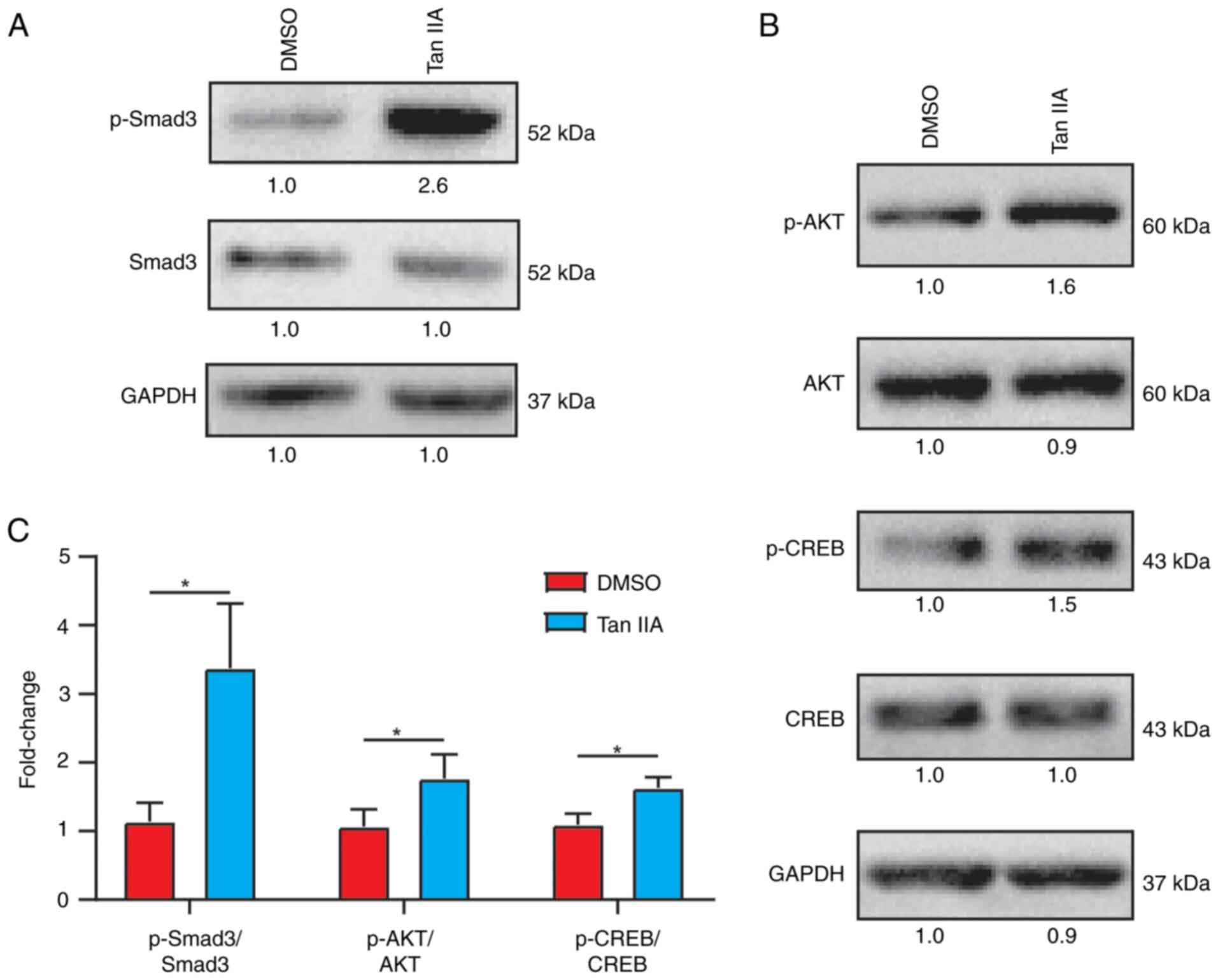
upregulated genes (Fig. 4C). Smad3
is a downstream effector of TGFβ signaling and plays a crucial role
in inhibiting adipogenesis (25–27).
Western blot assay also demonstrated that TGFβ/
Smad3 signaling was activated following Tan IIA treatment (Fig. 5A and C). It is known that CREB is
the classic effector in cAMP and AKT/CREB signaling significantly
contributes to osteogenesis (28–31).
Western blotting also demonstrated that AKT/CREB signaling was
significantly activated following Tan IIA treatment (Fig. 5B and C). Taken together, these data
revealed activated TGFβ and AKT signaling pathways following Tan
IIA treatment.
Tan IIA promoted osteogenic
differentiation of BMSCs through AKT signaling
Next, the present study explored whether Tan IIA
promoted osteogenic differentiation of BMSCs through activating AKT
signaling. Akti 1/2 is a known AKT signaling inhibitor and it was
added in the presence of Tan IIA during the osteogenic
differentiation of BMSCs. At seven days following osteogenic
induction, the group containing both Akti 1/2 and Tan IIA
contributed to decreased ALP levels when compared with Tan IIA
group (Fig. 6A and B). Alizarin
red staining revealed that Akti 1/2 significantly reduced
mineralized nodules after 21 days of osteogenic differentiation
(Fig. 6C). Furthermore, RT-qPCR
results also found decreased expression of osteogenic marker genes,
including Ocn, Opn and Bsp following addition of Akti 1/2 (Fig. 6D). Taken together, these results
demonstrated that Tan IIA promoted osteogenic differentiation of
BMSCs by activating AKT signaling pathway.
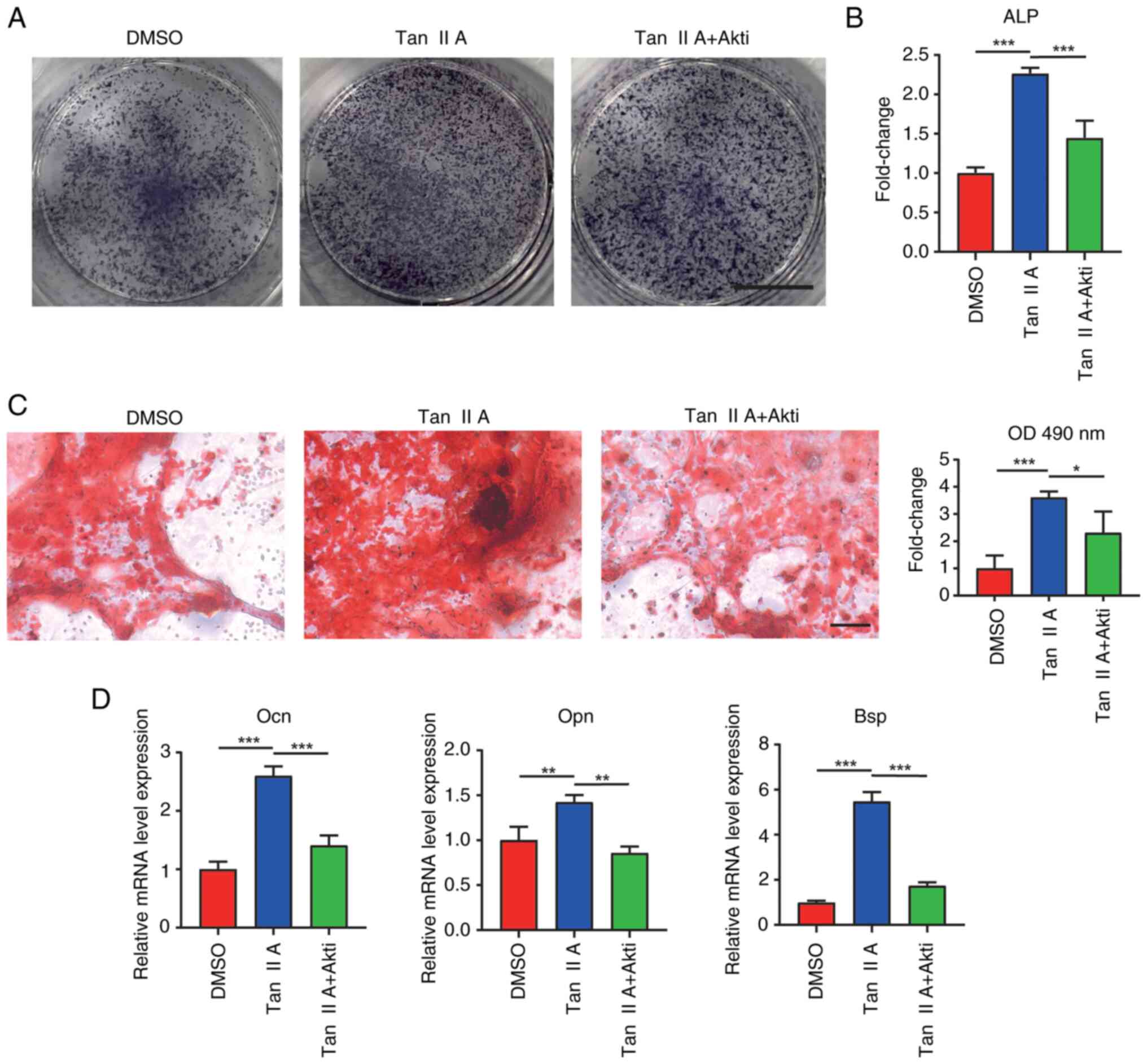 | Figure 6.Tan IIA promotes osteogenic
differentiation of BMSCs through Akt signaling. ALP staining for
BMSCs following osteogenic differentiation for 7 days. The freshly
isolated BMSCs were differentiated in an osteogenic induction
medium (DMSO group) or with Tan IIA treatment (Tan IIA group) or
Tan IIA supplemented with Akt signaling inhibitor Akti 1/2
treatment (Tan IIA + Akti 1/2 group). (A and B) Measurement of ALP
in BMSCs following osteogenic differentiation for 7 days. Scale
bar, 5 mm. (C) Alizarin red staining of BMSCs following osteogenic
differentiation for 21 days with different treatments, Scale bar,
100 µm, and quantification analysis of mineralized nodules of BMSCs
following osteogenic differentiation for 21 days with different
treatments. (D) Gene expression of Ocn, Opn and Bsp in BMSC
following osteogenic differentiation for 21 days with different
treatments. *P<0.05, **P<0.01, ***P<0.001. BMSCs, bone
marrow mesenchymal stem cells; Tan IIA, Tanshinone IIA; Ocn,
osteocalcin; Bsp, bonesialoprotein; Opn, osteopontin. |
Tan IIA suppresses the adipogenic
differentiation of BMSCs through TGFβ signaling
The present study also explored whether Tan IIA
suppressed adipogenic differentiation of BMSCs through activating
TGFβ signaling. SB431542 is a known TGFβ signaling inhibitor and it
was added in the presence of Tan IIA during adipogenic
differentiation of BMSCs. At 10 days after adipogenic induction of
BMSCs, oil red staining revealed that the group containing both Tan
IIA and SB431542 significantly increased formation of lipid
droplets when compared with Tan IIA group (Fig. 7A and B). Furthermore, RT-qPCR
results also found increased expression of adipogenic marker genes
including Adipoq, Cebpa, Pparg, Fabp4, Lpl and Plin1 following
inhibition of TGFβ signaling (Fig.
7C). Although the Fabp4 and Lpl levels did not reach
statistical significance (Fig.
7C). Taken together, these results demonstrated Tan IIA
suppressed the adipogenic differentiation of BMSCs by activating
TGFβ signaling pathway.
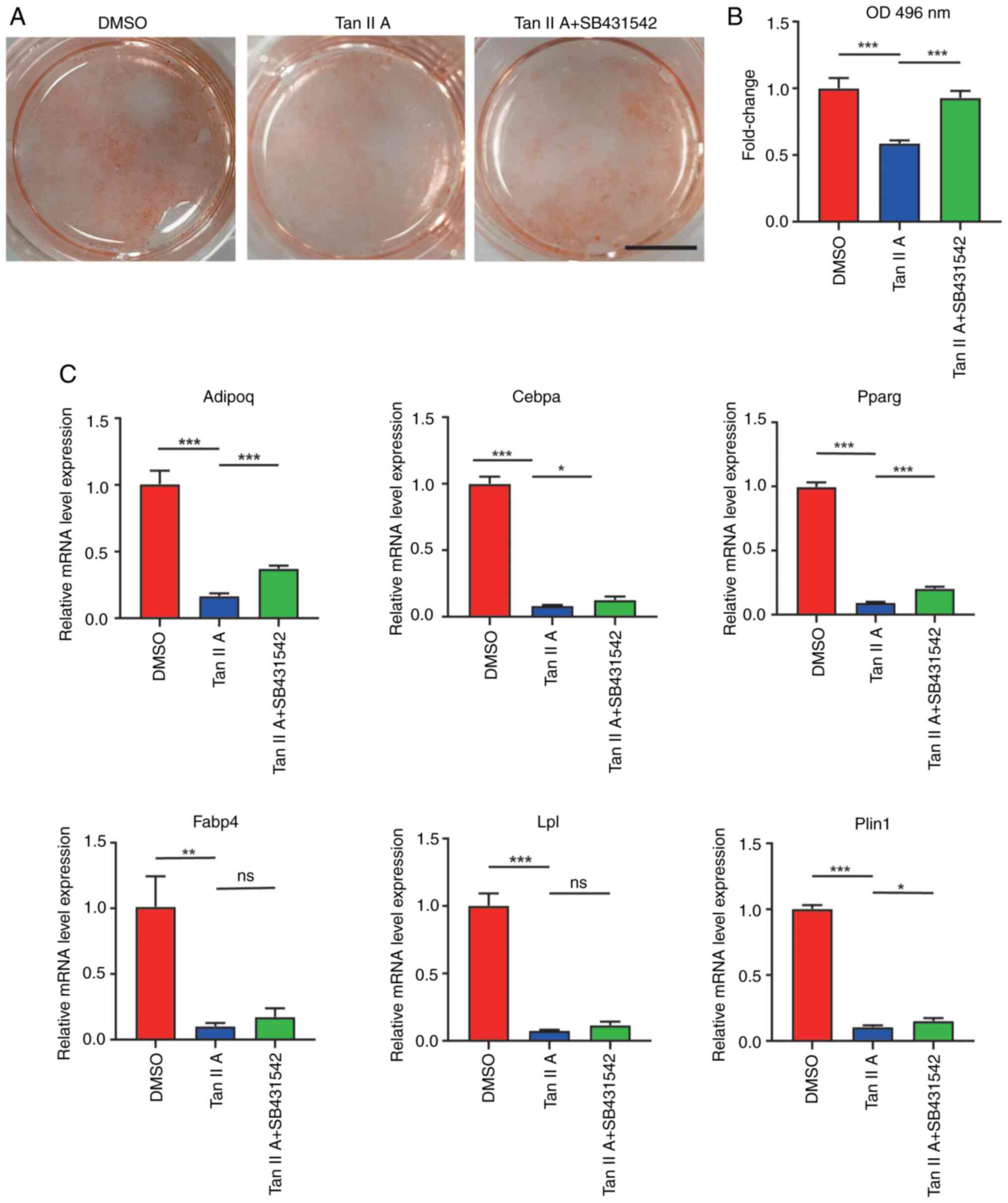 | Figure 7.Tan IIA suppresses the adipogenic
differentiation of BMSCs through TGFβ signaling. (A) Oil red
staining of BMSCs following adipogenic differentiation for 14 days.
The freshly isolated BMSCs was differentiated in an adipogenic
induction medium (DMSO group) or with Tan IIA treatment (Tan IIA
group) or Tan IIA supplemented with TGFβ signaling inhibitor
SB431542 treatment (Tan IIA + SB431542 group). Scale bar, 5 mm. (B)
Quantification analysis of oil red staining of BMSCs following
adipogenic differentiation for 14 days with different treatments.
(C) Gene expression of Plin1, Pparg, Cebpa, Fabp4, Adipoq and Lpl
in BMSC following adipogenic differentiation for 14 days with
different treatments. *P<0.05, **P<0.01, ***P<0.001, ns
P>0.05. Tan IIA, Tanshinone IIA; BMSCs, bone marrow mesenchymal
stem cells; Plin1, perilipin1; Fabp4, fatty acid-binding protein;
Adipoq, adiponectin; Cebpa, CCAAT/enhancer binding protein alpha;
Pparg, peroxisome proliferators-activated receptors gamma; Lpl,
lipoprteinlipase. |
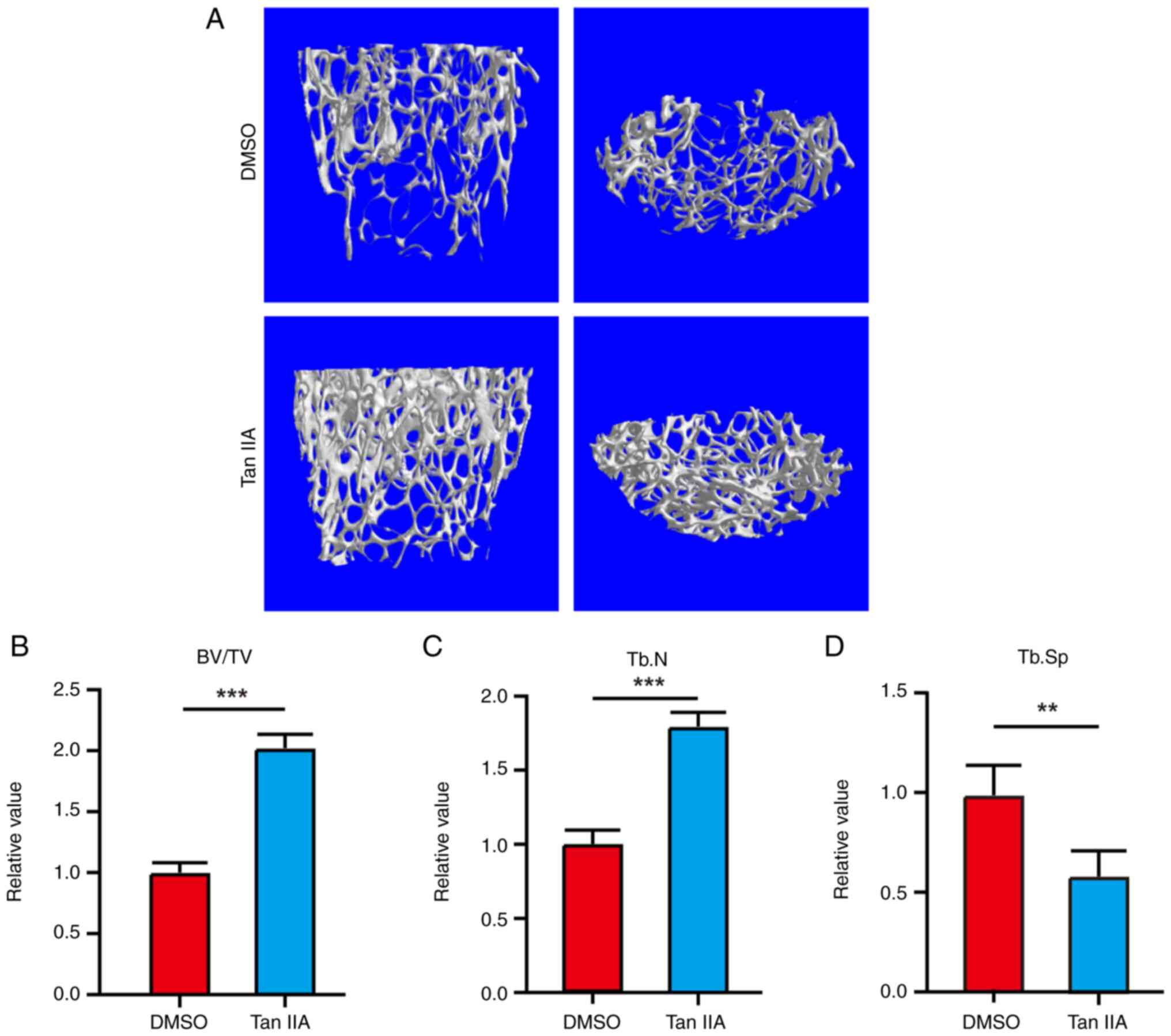
Tan IIA Promotes BMSCs osteogenesis in
vivo
To further investigate the osteogenic potential of
Tan IIA in vivo, an ovariectomy model was established in
mice and treated with Tan IIA. Micro-CT analysis revealed that Tan
IIA inhibited bone loss in the ovariectomy models (Fig. 8A-D), indicating its pro-osteogenic
potential for BMSCs in vivo.
Discussion
ANFH is caused by the interruption of blood supply
to the femoral head, leading to necrosis of localized tissue, which
further affects the femoral head located in the hip joint (1). BMSCs are multipotent stem cells with
the potential for self-renewal and multidirectional differentiation
(adipogenesis, osteogenesis and chondrogenesis) (4).
In the case of osteonecrosis, BMSCs can migrate and
undergo directional differentiation into osteoblasts, actively
participating in the bone repair process (8). However, BMSCs show unsatisfactory
functions in ANFH. There are two main reasons: One is that limited
blood supply of femoral head makes it difficult for MSCs to reach
the necrotic area. Another is that insufficient blood supply leads
to a decreased oxygen level, which can downregulate the expression
of key osteogenic genes [such as BMP-2 and runt-related
transcription factor 2 (RUNX2)], thereby inhibiting the ability of
MSCs to differentiate into osteoblasts (4). Thus, there is decreased osteogenesis
potential and increased adipogenesis ability for BMSCs from ANFH
(8). Effective strategy is
required to treat ANFH by correcting the differentiation direction
of BMSCs.
It has been reported that Salvia miltiorrhiza
Bunge could promote the osteogenesis and inhibit bone resorption
and thus it exerts favorable preventive effects on bone loss
(32). Emerging studies have shown
that some Chinese herbal formulas containing S.
miltiorrhiza, including Gufang Xian, Ling Gu Bao and Huo-gu are
effective in treating ANFH (33,34).
In addition, S. miltiorrhiza could also promote the
migration of MSCs and contribute to the re-ossification and
revascularization of ANFH in rabbit model of ANFH (11). However, the underlying
pharmacological mechanisms of which active component and how it
functions were unknown before the current study, to the best of the
authors' knowledge.
As one of the most abundant active lipophilic
compounds in S. miltiorrhiza, previous studies have
indicated potential positive biological effects of Tan IIA for
MSCs. MSCs treated with Tan IIA show increased CXCR4 and thus
obtain enhanced migration ability (13). In addition, Tan IIA also increases
the ex vivo expansion of human BMSCs by fibroblast growth
factor 2-mediated PI3K/AKT signaling pathways (14). Furthermore, there are studies
indicating the function of Tan IIA in osteogenesis (15,16,35).
Li et al (35) demonstrate
that Tan IIA can block the apoptosis of osteoblasts induced by
dexamethasone via inhibiting Nox4-derived ROS production. Korean
researchers found that Tan IIA can promote the osteogenic
differentiation of mouse muscle cell line C2C12 cells with BMP2
(16). This effect was achieved by
enhancing the activity of P38, which further promoted the activity
of Runx2 (16). Liu et al
(15) demonstrate that Tan IIA can
also contribute to osteogenesis of human periodontal ligament stem
cells via ERK1/2-dependent Runx2 induction. The current study
revealed enhanced osteogenic differentiation ability and decreased
adipogenic differentiation ability of BMSCs following treatment of
Tan IIA. To the best of the authors' knowledge, it is the first
time that effects of Tan IIA for osteogenesis and adipogenesis of
BMSCs under hypoxia environment were investigated. Thus, the
current study provided a promising application prospect for Tan IIA
to treat ANFH by targeting BMSCs.
The transcription factor CREB is a regulatory target
for AKT (36) and the role of
AKT/CREB signaling in osteogenesis has been solidly demonstrated
(28–31). Cao et al (28) found that the osteogenic potency of
MSCs can be maintained by activating AKT/CREB pathway in the
presence of steroids. Kang et al (29) illustrate that phosphorylated AKT
can react with the CREB to promote osteogenic differentiation of
MSCs by upregulating cyclin D1 and cyclin E1. In the current study,
activation of AKT/CREB signaling following Tan IIA treatment was
indicated by RNA-seq and it was further verified that Tan IIA
promoted osteogenesis of BMSCs through the AKT/CREB signaling
pathway. The present study also revealed the importance of
activated TGFβ/Smad3 signaling by Tan IIA in inhibiting adipogenic
differentiation of BMSCs. TGFβ plays crucial roles in a variety of
biological processes through its downstream signaling molecules
Smads (37). Smad3 is one of the
most representative Smads and a number of researchers emphasize the
central role of TGFβ/Smad3 signaling in suppressing adipogenesis
(25–27). Downregulation of Smad3 by RNAi
significantly increases the adipogenic differentiation of MSCs
(25,27), while Smad3 knock out mice show
smaller-size adipocytes when compared with wild-type mice (26). Thus, the present study provided a
detailed mechanism of how Tan IIA promoted osteogenic
differentiation and inhibited adipogenic differentiation ability of
BMSCs.
The current study was not without limitations.
First, although Tan IIA could promote osteogenic differentiation
and inhibit adipogenic differentiation of BMSCs in vitro,
these biological effects should be further verified in animal
models with ANFH due to the more complex microenvironment in
vivo. Furthermore, there was a lack of positive control drug in
current study, so it was hard to determine to what extent Tan IIA
has beneficial effects. Thus, the therapeutic potential of Tan IIA
should be compared with a positive control drug in animal models
with ANFH in further study.
In conclusion, Tan IIA could promote osteogenic
differentiation potential of BMSCs by activating AKT signaling and
suppress adipogenic differentiation potential of BMSCs by
activating TGFβ signaling.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Suzhou Science and
Technology Development Plan (Medical and Health Science and
Technology Innovation) Project (grant no. SKYD2022065).
Availability of data and materials
The RNA-seq data generated in the present study may
be found in the NCBI under accession number PRJNA1131477 or at the
following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1131477.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
WW, HW, SFa and SFe designed the present study,
collected the experimental data and wrote the original draft of the
manuscript. WW, YF and XH conducted the image analysis, statistical
analysis and wrote the original draft of the manuscript. HX and XS
designed the present study and reviewed and edited the manuscript.
YF and SFa administrated the project and edited the manuscript. WW
and SFa confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by the current
the animal ethics committee of Suzhou Wujiang District Second
People's Hospital (approval no. WZY2022056). The present study was
conducted in accordance with ARRIVE guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Konarski W, Poboży T, Śliwczyński A,
Kotela I, Krakowiak J, Hordowicz M and Kotela A: Avascular necrosis
of femoral head-overview and current state of the art. Int J
Environ Res Public Health. 19:73482022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rajpura A, Wright AC and Board TN: Medical
management of osteonecrosis of the hip: A review. Hip Int.
21:385–392. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sai Krishna MLV, Kar S, Kumar R, Singh H,
Mittal R and Digge VK: The role of conservative management in the
avascular necrosis of the femoral head: A review of systematic
reviews. Indian J Orthop. 57:410–420. 2023.PubMed/NCBI
|
|
4
|
Li Z, Wang W, Xu H, Ning Y, Fang W, Liao
W, Zou J, Yang Y and Shao N: Effects of altered CXCL12/CXCR4 axis
on BMP2/Smad/Runx2/Osterix axis and osteogenic gene expressions
during osteogenic differentiation of MSCs. Am J Transl Res.
9:1680–1693. 2017.PubMed/NCBI
|
|
5
|
Shapiro F, Connolly S, Zurakowski D,
Menezes N, Olear E, Jimenez M, Flynn E and Jaramillo D: Femoral
head deformation and repair following induction of ischemic
necrosis: A histologic and magnetic resonance imaging study in the
piglet. J Bone Joint Surg Am. 91:2903–2914. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu N, Liu H, Qu F, Fan J, Mao K, Yin Y,
Liu J, Geng Z and Wang Y: Hypoxia inhibits the differentiation of
mesenchymal stem cells into osteoblasts by activation of Notch
signaling. Exp Mol Pathol. 94:33–39. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benjamin S, Sheyn D, Ben-David S, Oh A,
Kallai I, Li N, Gazit D and Gazit Z: Oxygenated environment
enhances both stem cell survival and osteogenic differentiation.
Tissue Eng Part A. 19:748–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu K, Ge H, Liu C, Jiang Y, Yu Y and Zhou
Z: Notch-RBPJ pathway for the differentiation of bone marrow
mesenchymal stem cells in femoral head necrosis. Int J Mol Sci.
24:62952023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei B, Sun C, Wan H, Shou Q, Han B, Sheng
M, Li L and Kai G: Bioactive components and molecular mechanisms of
Salvia miltiorrhiza Bunge in promoting blood circulation to
remove blood stasis. J Ethnopharmacol. 317:1166972023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun K, Xue Y, Zhang X, Li X, Zhao J, Xu X,
Zhang X and Yang F: Tanshinone I alleviates steroid-induced
osteonecrosis of femoral heads and promotes angiogenesis: In vivo
and in vitro studies. J Orthop Surg Res. 18:4742023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y, Zhang C, Wu J, Han Y and Wu C:
Angiogenesis and bone regeneration by mesenchymal stem cell
transplantation with danshen in a rabbit model of avascular
necrotic femoral head. Exp Ther Med. 18:163–171. 2019.PubMed/NCBI
|
|
12
|
Gao S, Liu Z, Li H, Little PJ, Liu P and
Xu S: Cardiovascular actions and therapeutic potential of
tanshinone IIA. Atherosclerosis. 220:3–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie J, Wang H, Song T, Wang Z, Li F, Ma J,
Chen J, Nan Y, Yi H and Wang W: Tanshinone IIA and astragaloside IV
promote the migration of mesenchymal stem cells by up-regulation of
CXCR4. Protoplasma. 250:521–530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuan P, Qin HY, Wei JY, Chen G and Li X:
Proteomics reveals the potential mechanism of Tanshinone IIA in
promoting the ex vivo expansion of human bone marrow mesenchymal
stem cells. Regen Ther. 21:560–573. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Niu Y, Xie W, Wei D and Du Q:
Tanshinone IIA promotes osteogenic differentiation of human
periodontal ligament stem cells via ERK1/2-dependent Runx2
induction. Am J Transl Res. 11:340–350. 2019.PubMed/NCBI
|
|
16
|
Kim HJ and Kim SH: Tanshinone IIA enhances
BMP-2-stimulated commitment of C2C12 cells into osteoblasts via p38
activation. Amino Acids. 39:1217–1226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abbuehl JP, Tatarova Z, Held W and
Huelsken J: Long-term engraftment of primary bone marrow stromal
cells repairs niche damage and improves hematopoietic stem cell
transplantation. Cell Stem Cell. 21:241–255.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Davaapil H, McNamara M, Granata A, Macrae
RGC, Hirano M, Fitzek M, Aragon-Martin JA, Child A, Smith DM and
Sinha S: A phenotypic screen of Marfan syndrome iPSC-derived
vascular smooth muscle cells uncovers GSK3β as a new target. Stem
Cell Rep. 18:555–569. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiang J and Martinez-Agosto JA: Effects
of mTOR inhibitors on components of the salvador-warts-hippo
pathway. Cells. 1:886–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reggio A, Rosina M, Palma A, Cerquone
Perpetuini A, Petrilli LL, Gargioli C, Fuoco C, Micarelli E,
Giuliani G, Cerretani M, et al: Adipogenesis of skeletal muscle
fibro/adipogenic progenitors is affected by the
WNT5a/GSK3/β-catenin axis. Cell Death Differ. 27:2921–2941. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang L, Feng Z, Wang X, Wang X and Zhang
X: DEGseq: An R package for identifying differentially expressed
genes from RNA-seq data. Bioinformatics. 26:136–138. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39((Web Server Issue)): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo W, Flanagan J, Jasuja R, Kirkland J,
Jiang L and Bhasin S: The effects of myostatin on adipogenic
differentiation of human bone marrow-derived mesenchymal stem cells
are mediated through cross-communication between Smad3 and
Wnt/beta-catenin signaling pathways. J Biol Chem. 283:9136–9145.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsurutani Y, Fujimoto M, Takemoto M,
Irisuna H, Koshizaka M, Onishi S, Ishikawa T, Mezawa M, He P, Honjo
S, et al: The roles of transforming growth factor-β and Smad3
signaling in adipocyte differentiation and obesity. Biochem Biophys
Res Commun. 407:68–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim YJ, Hwang SJ, Bae YC and Jung JS:
MiR-21 regulates adipogenic differentiation through the modulation
of TGF-beta signaling in mesenchymal stem cells derived from human
adipose tissue. Stem Cells. 27:3093–3102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao H, Shi K, Long J, Liu Y, Li L, Ye T,
Huang C, Lai Y, Bai X, Qin L and Wang X: PDGF-BB prevents
destructive repair and promotes reparative osteogenesis of
steroid-associated osteonecrosis of the femoral head in rabbits.
Bone. 167:1166452023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang H, Yang S and Lee J:
Tauroursodeoxycholic acid enhances osteogenic differentiation
through EGFR/p-Akt/CREB1 pathway in mesenchymal stem cells. Cells.
12:14632023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao B, Deng R, Chai Y, Chen H, Hu B, Wang
X, Zhu S, Cao Y, Ni S, Wan M, et al: Macrophage-lineage TRAP+ cells
recruit periosteum-derived cells for periosteal osteogenesis and
regeneration. J Clin Invest. 129:2578–2594. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baker N, Sohn J and Tuan RS: Promotion of
human mesenchymal stem cell osteogenesis by PI3-kinase/Akt
signaling, and the influence of caveolin-1/cholesterol homeostasis.
Stem Cell Res Ther. 6:2382015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han J, Chai Y, Zhang XY, Chen F, Xu ZW,
Feng Z, Yan Q, Wen SB and Wu YK: Gujiansan ameliorates avascular
necrosis of the femoral head by regulating autophagy via the
HIF-1α/BNIP3 pathway. Evid Based Complement Alternat Med.
2021:66830072021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang Z, Fu F, Ye H, Gao H, Tan B, Wang R,
Lin N, Qin L and Chen W: Chinese herbal Huo-Gu formula for the
treatment of steroid-associated osteonecrosis of femoral head: A
14-year follow-up of convalescent SARS patients. J Orthop Translat.
23:122–131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li ZR, Cheng LM, Wang KZ, Yang NP, Yang
SH, He W, Wang YS, Wang ZM, Yang P, Liu XZ, et al: Herbal Fufang
Xian Ling Gu Bao prevents corticosteroid-induced osteonecrosis of
the femoral head-A first multicentre, randomised, double-blind,
placebo-controlled clinical trial. J Orthop Translat. 12:36–44.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, He C, Tong W, Zou Y, Li D, Zhang C
and Xu W: Tanshinone IIA blocks dexamethasone-induced apoptosis in
osteoblasts through inhibiting Nox4-derived ROS production. Int J
Clin Exp Pathol. 8:13695–13706. 2015.PubMed/NCBI
|
|
36
|
Du K and Montminy M: CREB is a regulatory
target for the protein kinase Akt/PKB. J Biol Chem.
273:32377–32379. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tzavlaki K and Moustakas A: TGF-β
signaling. Biomolecules. 10:4872020. View Article : Google Scholar : PubMed/NCBI
|