Introduction
Cervical cancer is one of the leading causes of
cancer deaths in women (1). The
age-standardized incidence rates worldwide is 13.1/100,000 women
(2). Epidemiological
investigations report 445,000 new cases of cervical cancer and
236,000 deaths annually (3), with
~90% of the deaths occurring in developing countries and low and
middle income countries (2). The
burden of cervical cancer remains high in numerous parts of the
world, with incidence rates in most countries above the thresholds
agreed by the WHO Cervical Cancer Elimination Initiative (4,5).
Currently, the incidence and mortality rates of cervical cancer are
progressively decreasing in developed countries (6). In underdeveloped countries, however,
cervical cancer is still one of the most common malignant tumors in
women and is the leading cause of death from malignant tumors
(7). Based on histopathology,
cervical cancer can be divided into squamous cell carcinoma of the
cervix, adenocarcinoma of the cervix and rare types such as
adenosquamous carcinoma, neuroendocrine carcinoma and smooth muscle
sarcoma (8). Among them, the most
common type of cervical cancer is cervical squamous cell carcinoma
(CESC), which accounts for ~80% of the total number of cases
(9,10). It is now generally accepted that
human papillomavirus (HPV) is the leading contributor to cervical
cancer development and has been categorized into low-risk and
high-risk strains on the basis of their oncogenic ability (11). Of the >40 established HPV
species that can infect human genitalia, 18 HPV strains have been
categorized as high-risk genotypes (12). Almost all cervical cancers are due
to high-risk HPV, with serotypes 16 and 18 accounting for 70% of
all cases (13). Patients with
early-stage cervical cancer have favorable prognosis (14). However, for advanced cervical
cancer, cisplatin-based chemotherapy is preferred, but its efficacy
is unsatisfactory, with only 1/5 of patients responding to
cisplatin-based chemotherapy modalities or
radiotherapy-chemotherapy combinations (15). Therefore, searching for new
prognostic molecules and effective target molecules for early
diagnosis, and establishing new and effective therapeutic measures
for CESC are urgent clinical problems.
Collagen, as the most abundant protein, provides a
scaffold for the assembly of the extracellular matrix (ECM) and is
considered a ‘highway’ for cancer cell migration and invasion
(16). The extent of collagen
lysine hydroxylation influences the stability of intermolecular
collagen cross-links (17).
Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2), also
termed lysyl hydroxylase 2, is a key enzyme that mediates the
formation of stable collagen cross-links by catalyzing the
hydroxylation of lysine (18).
Studies have shown that PLOD2 is significantly overexpressed in
head and neck squamous cell carcinoma and promotes cancer cell
proliferation, migration and invasion (19,20).
RNA sequencing identified PLOD2 as a key gene marker for
HPV-associated oropharyngeal squamous cell carcinoma (21). In addition, database analysis
revealed that PLOD2 was significantly upregulated in cervical
esophageal carcinoma (CESC), which is a potent prognostic marker
and associated with immune infiltration in HPV-associated CESC
(22). It is worth noting that all
of the aforementioned cancers can be caused by HPV infection
(23,24). Therefore, it was hypothesized that
there is an association between PLOD2 and HPV infection. Several
studies have revealed that PLOD2 is overexpressed in bone
carcinoma, hepatocellular carcinoma, pancreatic carcinoma and
squamous cell carcinoma of the head and neck, accompanied by
promoted proliferation, migration and invasion of cancer cells
(19,25). Nevertheless, studies on PLOD2 in
CESC have not yet been published. Hence, the objective of the
present study was to characterize the role of PLOD2 in CESC cells
and the mechanism by which PLOD2 impedes the progression of
CESC.
Materials and methods
Data mining using public
databases
The CESC-related dataset GSE64217 was screened and
downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and used to
mine PLOD2 gene expression in patients with CESC samples (26). The differentially expressed genes
of P<0.05 were screened out. UALCAN database (https://ualcan.path.uab.edu) analyzed PLOD2 expression
in CESC tissues from the TCGA database (https://portal.gdc.cancer.gov/) and the prognostic
impact of PLOD2 in patients with CESC (27,28).
In addition, the BioGrid v4.4 website (https://thebiogrid.org) (29) predicted the molecules that PLOD2
may bind to and HDock (http://hdock.phys.hust.edu.cn/) was utilized to verify
the binding of PLOD2 to Yes-associated protein 1 (YAP1).
Cell culture
Human cervical endometrial cell line End1/E6E7 and
CESC cell lines C33A, SiHa, HT-3 and MS751 were obtained from
Cellverse Bioscience Technology Co., Ltd. Cells were incubated in
Dulbecco's modified Eagle's medium containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin at 37°C under 5% CO2.
Cell transfection
The specific siRNA targeting PLOD2 (siRNA-PLOD2#1/2)
and the corresponding control siRNA (siRNA-NC), the pc-DNA3.1
vector containing the whole length of YAP1 (Ov-YAP1) and the empty
vector (Ov-NC) were synthesized by GenePharma. The sequence
information of siRNAs used for cell transfection are shown in
Table I. Using
Lipofectamine® 2000 reagent, 100 nM vectors was
transfected into SiHa cells following a typical protocol at 37°C
for 48 h (30). Cells were
harvested 48 h after transfection for subsequent
experimentation.
 | Table I.Sequence information of siRNAs used
for cell transfection. |
Table I.
Sequence information of siRNAs used
for cell transfection.
| si-RNA | Direction | Sequence
(5′-3′) |
|---|
| siRNA-PLOD2#1 | Sense |
ACUAUACGGUUGACAUAUGGA |
|
| Antisense |
CAUAUGUCAACCGUAUAGUUC |
| siRNA-PLOD2#2 | Sense |
AUCGAAUUCACAAAGAGUGCA |
|
| Antisense |
CACUCUUUGUGAAUUCGAUAC |
| siRNA-NC | Sense |
UUCUCCGAACGUGUCACGUTT |
|
| Antisense |
ACGUGACACGUUCGGAGAATT |
Cell counting kit-8 (CCK-8) assay
SiHa cells were inoculated into 96-well plates,
followed by transfection with siRNA-PLOD2 with or without Ov-YAP1
for 48 h. After which, 10 µl WST-8 (Beyotime Institute of
Biotechnology) was added to each well to grow the cells for 2 h,
and the absorbance was calculated at 450 nm with a microplate
reader (Bio-Rad Laboratories, Inc.).
Wound healing assay
Transfected SiHa cells were initially inoculated
into a six-well plate. After the cells reached 90% confluence, the
cell monolayers were scratched with a white pipette tip (31). Following 24 h of incubation
(without serum), the migratory rate was determined by a light
microscope. Image J v1.50 (National Institutes of Health) was used
to quantify the wound healing percentage. The formula was as
follows: Initial wound size-healing wound size/initial wound size
×100%.
Transwell assay
Cell suspension was prepared by serum-free medium
and the transfected SiHa cell suspension (2×105
cells/ml) was loaded into the upper chamber. Then the medium
containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was
added to the lower chamber. After incubating at 37°C for 24 h, the
bottom of the cell chamber was fixed with 100% methanol and stained
with 0.1% crystal violet at room temperature for 10 min (31). The number of migrating cells was
counted by a fluorescent microscope.
Cell apoptosis analysis
Transfected SiHa cells were washed with precooled
PBS. The cells were then stained with Annexin V-FITC for 15 min,
followed by the addition of propidium iodide (10 mg/ml) for 5 min
at room temperature in darkness. Apoptosis was analyzed by CytoFLEX
flow cytometer (Beckman Coulter, Inc.) recognized using FlowJo
software v10.8 (Tree Star, Inc.).
SA-β staining
After treatment, SiHa cells were washed three times
with HBSS and treated with 1 ml β-galactosidase fixative for 15 min
at room temperature. After which, the cell fixative was separated
and the cells were allowed to incubate with the staining solution
overnight at 37°C (32). The
plates were viewed under an inverted microscope.
Co-immunoprecipitation (co-IP)
Total protein was extracted from SiHa cells by IP
lysate (NCM Biotech; cat. no. P70100) containing protease inhibitor
and was incubated with rabbit IgG (1 µg; cat. no. Sc-2027, Santa
Cruz Biotechnology, Inc.) or IP-indicating antibody (1 µg) at 4°C
overnight, while an appropriate amount of the extracted protein was
used as an input control. The following antibodies were used: PLOD2
(Abcam; cat. no. ab313765) and YAP1 (Abcam; cat. no. ab52771).
Protein A/G PLUS-Agarose (20 µl) was applied and incubated at 4°C
for 2 h to form the immune mixture, and then centrifuged at 1,000 ×
g at 4°C for 3 min to isolate the complexes. After washing 4 times
with 1 ml cold lysis buffer and boiling for 5 min in the
appropriate protein sample buffer, the supernatant was collected in
a new tube to carry out western blotting as mentioned below to
analyze the immuno-complexes.
Immunofluorescence staining
SiHa cells were fixed with 4% polyoxymethylene at
room temperature for 10 min and permeabilized with 0.5%
Trition-X100. Following blocking with 10% BSA (Biofroxx; neoFroxx)
in PBS for 1 h at room temperature, the cells were incubated with
primary antibodies (1:100) overnight at 4°C and secondary
antibodies (1:500) for 2 h at room temperature, and stained with
DAPI at room temperature for 10 min. The following antibodies were
used: p21 (Proteintech Group, Inc.; cat. no. 67362-1-Ig), p53
(Proteintech Group, Inc.; cat. no. 60283-2-Ig) and
CoraLite488-conjugated Donkey Anti-IgG (H+L; Proteintech Group,
Inc.; cat. no. SA00013-5). The samples were visualized under a
confocal microscope.
RT-qPCR
The total RNA used in the present study was isolated
from SiHa cells using Trizol® reagent. The cDNA was
synthesized using a cDNA reverse transcription kit (Applied
Materials, Inc.) at 37°C for 15 min. qPCR was performed using the
SYBR Green PCR Master Mix (Applied Biosystems, cat. no. 4367659)
and MiniOpticon qPCR detection System (Bio-Rad Laboratories, Inc.).
Thermocycling conditions used for qPCR comprised a preincubation
step at 95°C for 60 sec, followed by 40 cycles: 95°C for 15 sec for
denaturation; 63°C for 25 sec for annealing and extension. The
results were estimated based on the 2−ΔΔCq method
(33) with GAPDH used as the
internal reference gene. The primers used are shown in Table II.
 | Table II.Primer sequences used in the
RT-qPCR. |
Table II.
Primer sequences used in the
RT-qPCR.
| Gene name | Direction | Sequence
(5′-3′) |
|---|
| PLOD2 | Forward |
GACAGCGTTCTCTTCGTCCTCATC |
|
| Reverse |
ACCACCTCCCTGAAAGTCTTCTCC |
| YAP1 | Forward |
CGTCATGGGTGGCAGCAACTC |
|
| Reverse |
TCAGCCGCAGCCTCTCCTTC |
| IL-6 | Forward |
GGTGTTGCCTGCTGCCTTCC |
|
| Reverse |
GTTCTGAAGAGGTGAGTGGCTGTC |
| IL-1β | Forward |
GGACAGGATATGGAGCAACAAGTGG |
|
| Reverse |
CAACACGCAGGACAGGTACAGATTC |
| IL-8 | Forward |
GGACCACACTGCGCCAACAC |
|
| Reverse |
CCCTCTGCACCCAGTTTTCCTTG |
| CCL20 | Forward |
TGCTGTACCAAGAGTTTGCTCCTG |
|
| Reverse |
CTTCTGATTCGCCGCAGAGGTG |
| GAPDH | Forward |
GTGGACCTGACCTGCCGTCTAG |
|
| Reverse |
GAGTGGGTGTCGCTGTTGAAGTC |
Western blotting
Total proteins from SiHa cells were extracted using
RIPA lysis buffer (NCM Biotech; cat. no. WB3100). The protein
samples of each group were quantified by BCA kit (NCM Biotech) and
20 µg/lane protein samples were loaded on 10% SDS-PAGE gel. After
10% SDS-PAGE separation, equal amounts of proteins were transferred
to a PVDF membrane. After blocking with 5% BSA (Biofroxx; neoFroxx)
for 1 h at room temperature, the membranes were cultured with
primary antibodies PLOD2 (1:1,000; Abcam; cat. no. ab313765), MMP2
(1:3,000; Proteintech Group, Inc.; cat. no. 66366-1-Ig), MMP9
(1:500; Proteintech Group, Inc.; cat. no. 27306-1-AP), Bcl-2
(1:1,000; Affinity Biosciences; cat. no. AF6139), Bax (1:1,000;
Affinity Biosciences; cat. no. AF0120), caspase 3 (1:1,000; CST
Biological Reagents Co., Ltd.; cat. no. 9662), cleaved caspase 3
(1:1,000; CST Biological Reagents Co., Ltd.; cat. no. 9661), YAP1
(1:5,000; Abcam; cat. no. ab52771) and GAPDH (Proteintech Group,
Inc.; cat. no. 60004-1-Ig; 1:5,000) overnight at 4°C and with
secondary antibodies HRP-conjugated Goat Anti-Mouse IgG (H+L;
1:2,000; Proteintech Group, Inc.; cat. no. SA00001-1) or
HRP-conjugated Goat Anti-Rabbit IgG (H+L; 1:2,000; Proteintech
Group, Inc.; cat. no. SA00001-2) at 37°C for 2 h. The ELC A
solution was mixed with the B solution in equal proportions and
visualization of the protein bands was achieved using the ECL
detection system (Thermo Fisher Scientific, Inc.) in accordance
with standard protocols (34), and
analysis of protein density was assessed by Image J software v1.50
(National Institutes of Health).
Statistical analysis
Statistical analyses were performed using SPSS 22.0
(IBM Corp.) and GraphPad Prism 6 software (Dotmatics). Data are
presented as the mean ± standard deviation of three independent
experiments. Results were obtained using one-way ANOVA followed by
Bonferroni post hoc tests for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
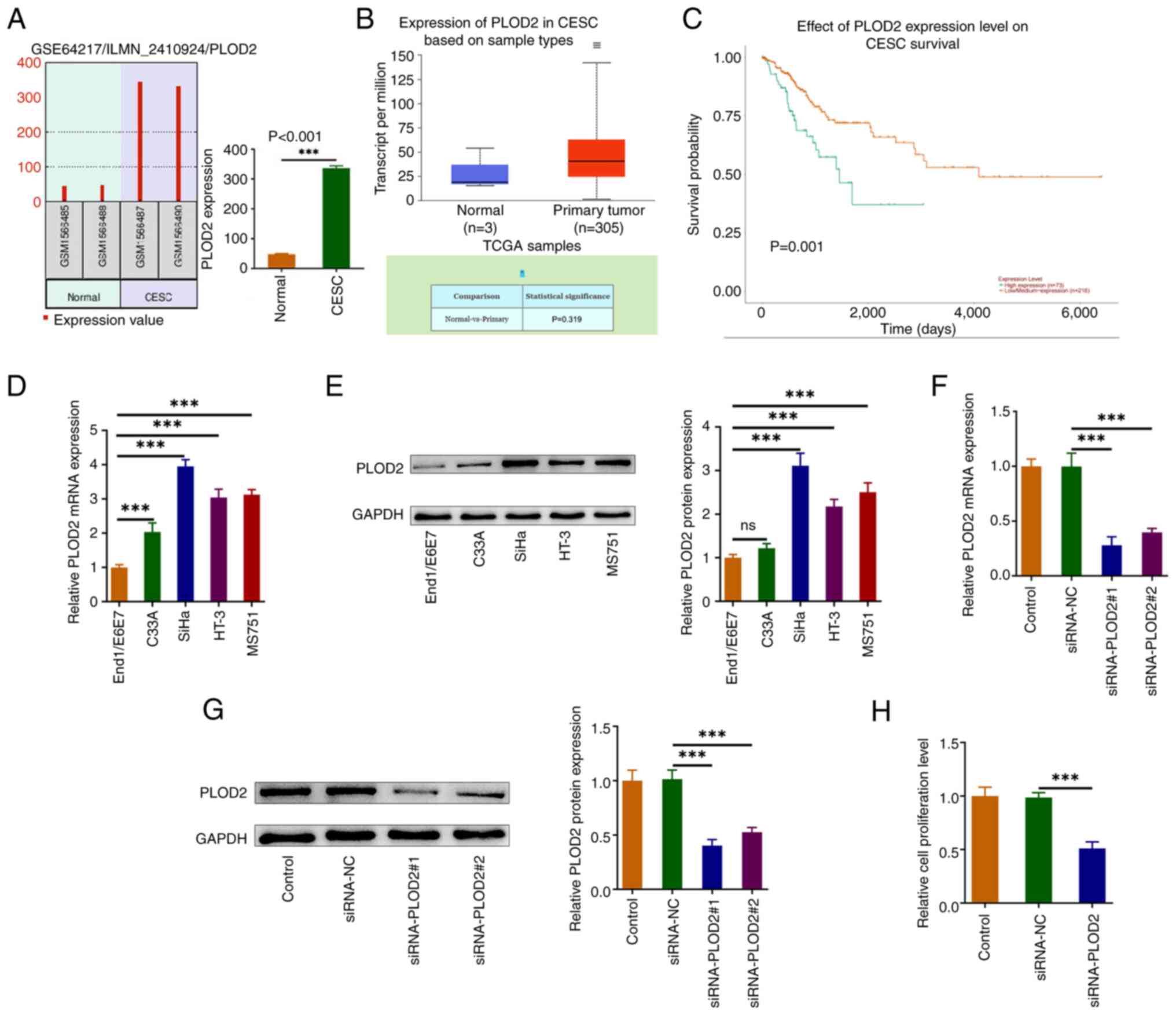
PLOD2 expression is upregulated in
CESC tissues and cell lines, and inhibition of PLOD2 decreases CESC
cell proliferation
To determine whether PLOD2 plays a role in CESC
tumorigenesis, bioinformatic analyses were performed. PLOD2
expression was markedly enhanced in CESC tissues in public
databases, as shown in Fig. 1A and
B. High expression of PLOD2 was associated with poor prognosis
in CESC tissues (Fig. 1C). In
addition, by comparative analysis it was found that PLOD2 levels
were specifically increased in CESC cell lines compared with the
End1/E6E7 cell line. Among them, the expression of PLOD2 was
strongest in SiHa cells, so it was selected for subsequent
experiments (Fig. 1D and E). Next,
PLOD2 expression was silenced and the transfection efficiency was
shown in Fig. 1F and G. The
findings exhibited that the transfection efficiency of
siRNA-PLOD2-1 was higher, hence siRNA-PLOD2-1 (termed siRNA-PLOD2)
was selected for the subsequent experiments. CCK-8 assay indicated
that silencing PLOD2 significantly decreased the proliferation of
SiHa cells when compared with the control group (Fig. 1H).
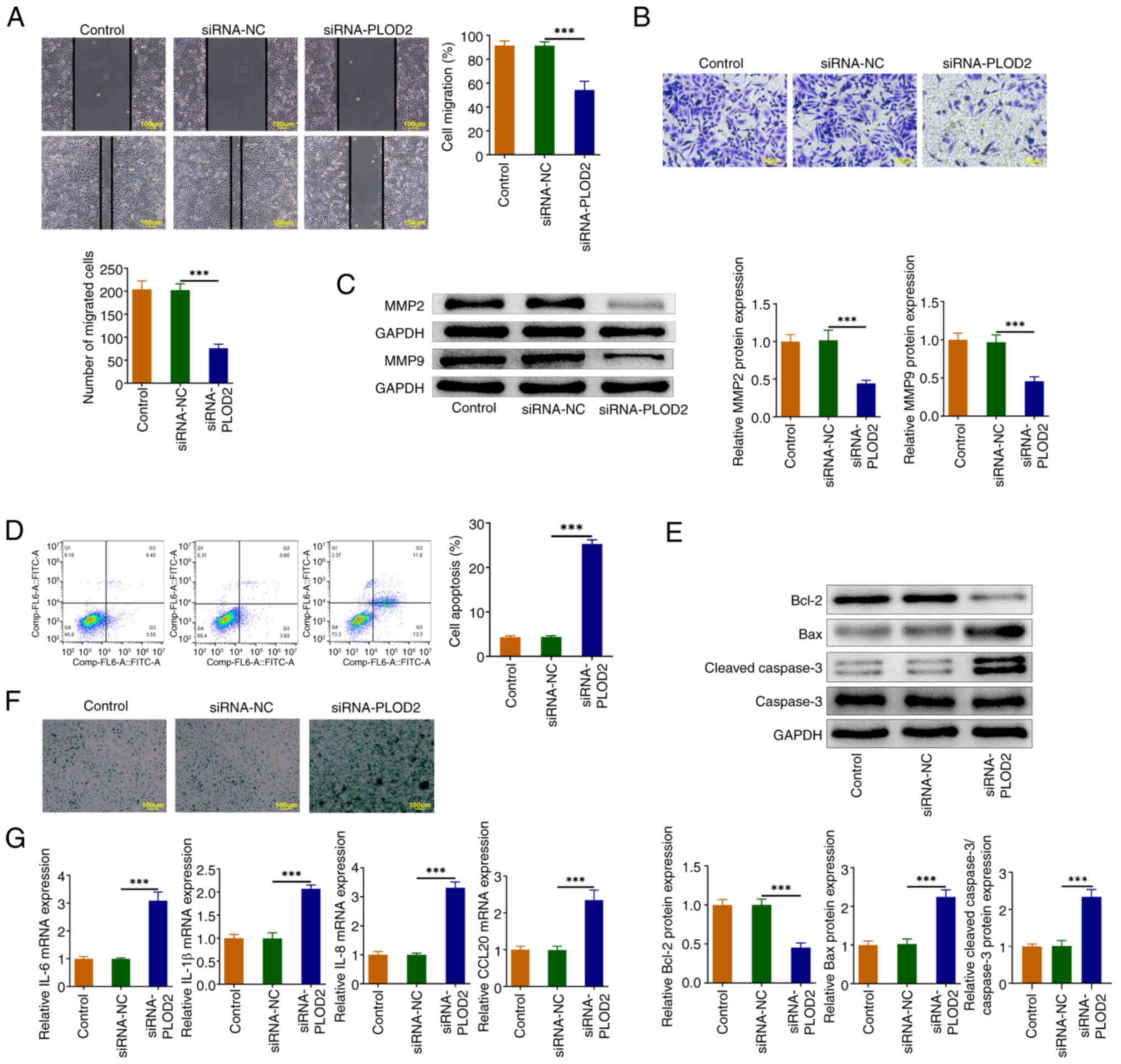
PLOD2 silencing suppresses the
migration, and promotes apoptosis and senescence in SiHa cells
As illustrated in Fig.
2A and B, PLOD2 knockdown diminished cell migration. Silencing
PLOD2 resulted in suppression of the protein levels of MMP2 and
MMP9 (Fig. 2C). In addition, flow
cytometry analysis demonstrated a notable increase in apoptosis
following transfection with siRNA-PLOD2 (Fig. 2D), which was consistent with the
Western blotting results whereby the level of Bcl-2 was declined in
cells silenced with PLOD2, but the levels of Bax and cleaved
caspase 3 were augmented (Fig.
2E). Moreover, the knockdown of PLOD2 facilitated the levels of
senescence-associated β-galactosidase (SA-β-Gal) and mRNA
expressions of senescence-associated secretory phenotype (SASP)
genes IL-6, IL-1β, IL-8 and CCL20 (Fig. 2F and G).
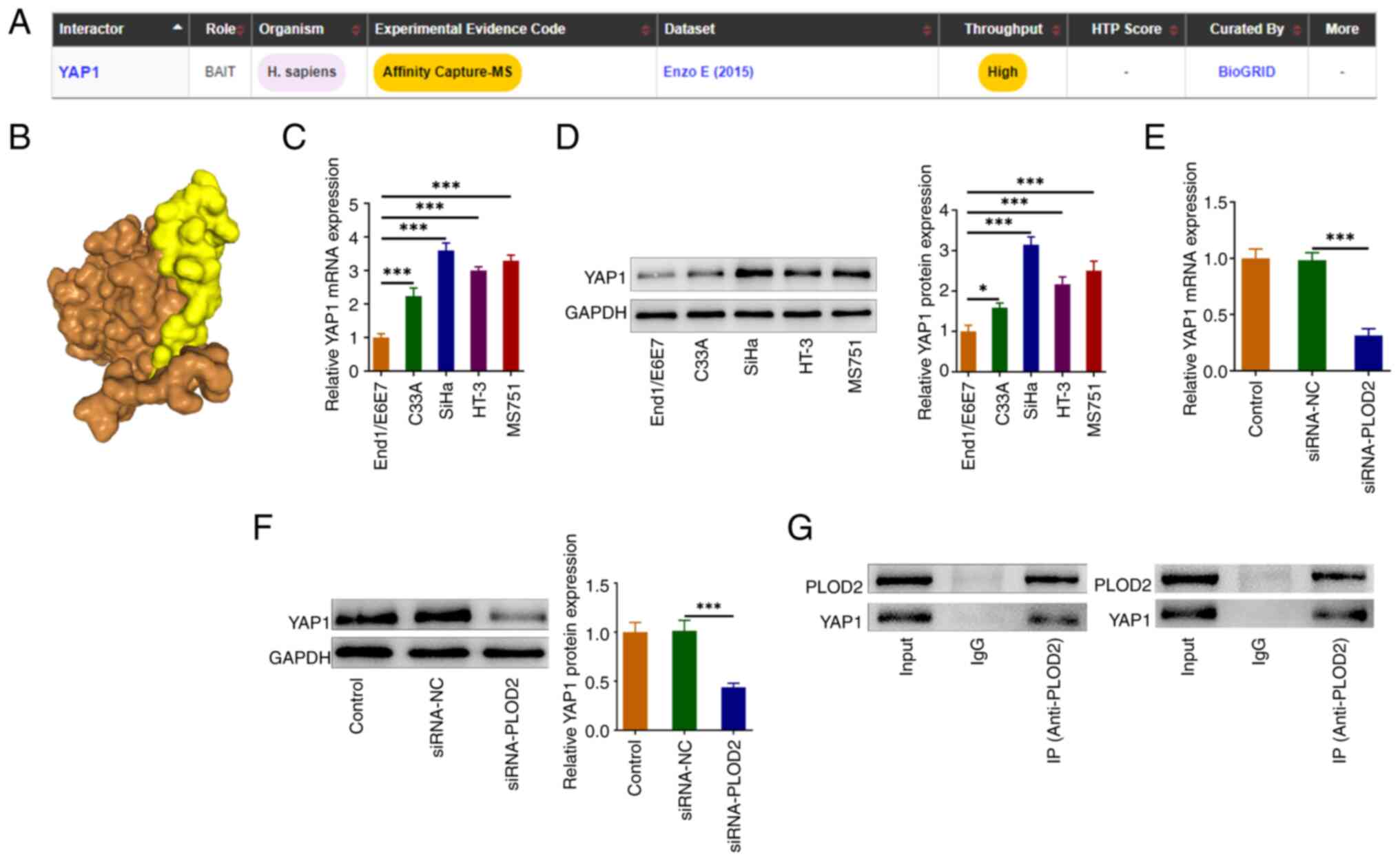
PLOD2 binds to YAP1
The possible mechanism of PLOD2 in CESC was then
explored. Using the BioGrid website, PLOD2 was predicted to bind to
YAP1 (Fig. 3A). The combination of
PLOD2 and YAP1 was further verified by HDock software (Fig. 3B). RT-qPCR and western blotting
results revealed that YAP1 expression was evidently increased in
CESC cell lines when compared with the control End1/E6E7 cells
(Fig. 3C and D). The data also
revealed that PLOD2 silencing significantly reduced the protein
level of YAP1 compared with the negative control group (Fig. 3E and F). The co-IP experiment
verified that PLOD2 could combine with YAP1 (Fig. 3G).
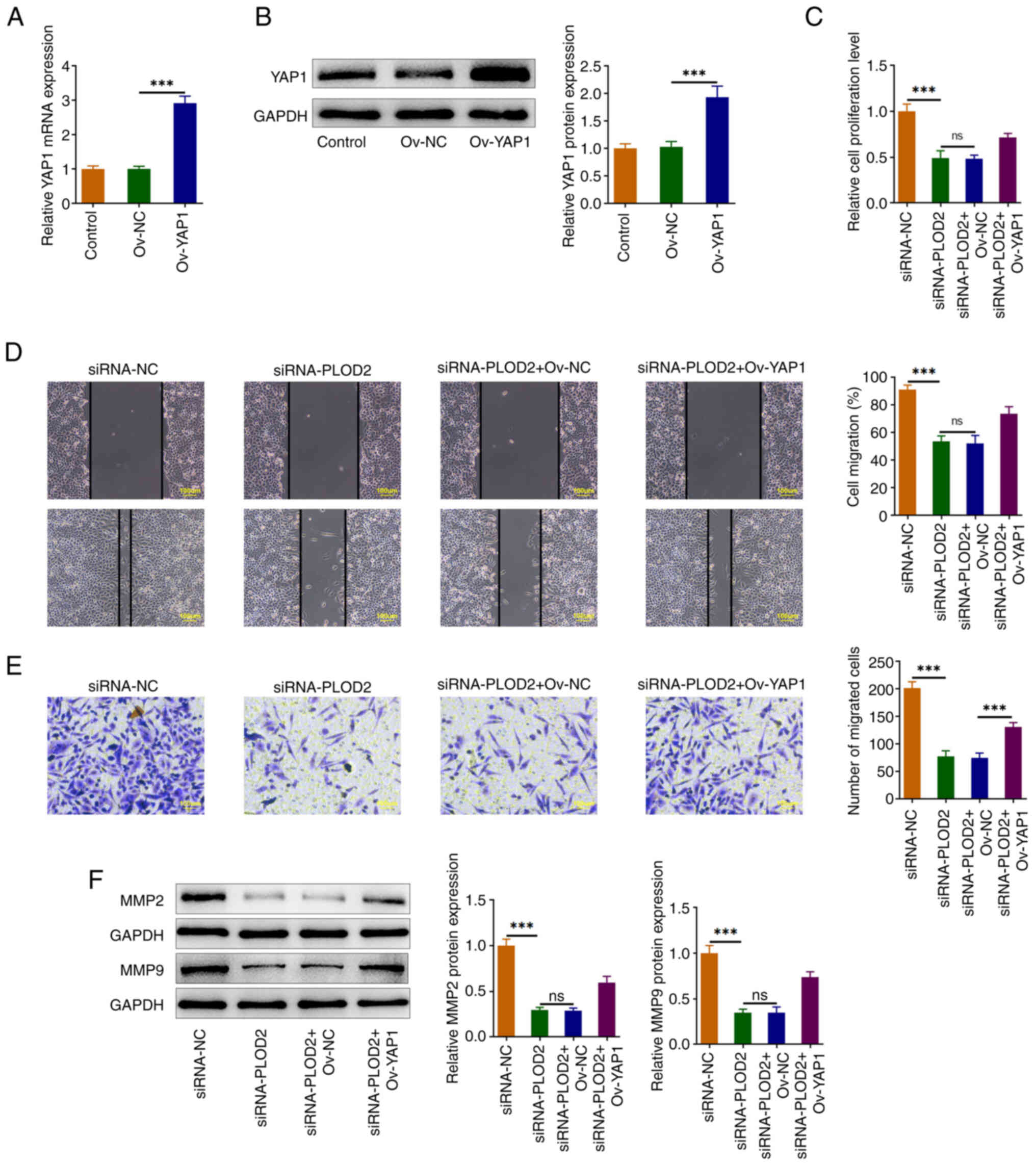
PLOD2 regulates YAP1 to promote the
proliferation and migration of SiHa cells
To explore the biological roles of YAP1 in SiHa
cells, YAP1 was overexpressed and transfection efficiency was shown
in Fig. 4A and B. CCK-8 assay
indicated that YAP1 elevated the cell proliferation of
PLOD2-modulated SiHa cells (Fig.
4C). The migration of SiHa cells was enhanced after YAP1
overexpression (Fig. 4D and E).
Moreover, the upregulation of YAP1 reversed the decreased levels of
MMP2 and MMP9 in PLOD2-silenced SiHa cells (Fig. 4F).
PLOD2 silencing promotes apoptosis and
senescence in SiHa cells by binding to YAP1 and regulates the p53
pathway
As shown in Fig.
5A, the rate of cell apoptosis in YAP1-overexpressed SiHa cells
was significantly reduced compared with the PLOD2-silenced SiHa
cells alone, which was consistent with the western blotting results
where Bcl-2 levels were increased and the protein levels of cleaved
caspase 3 and Bax were reduced (Fig.
5B). YAP1 overexpression in turn alleviates the level of
SA-β-Gal and SASP genes (Fig. 5C and
D). Furthermore, PLOD2 silencing increased the levels of p21
and p53 in SiHa cells, which was reversed by YAP1 overexpression
(Fig. 5E and F).
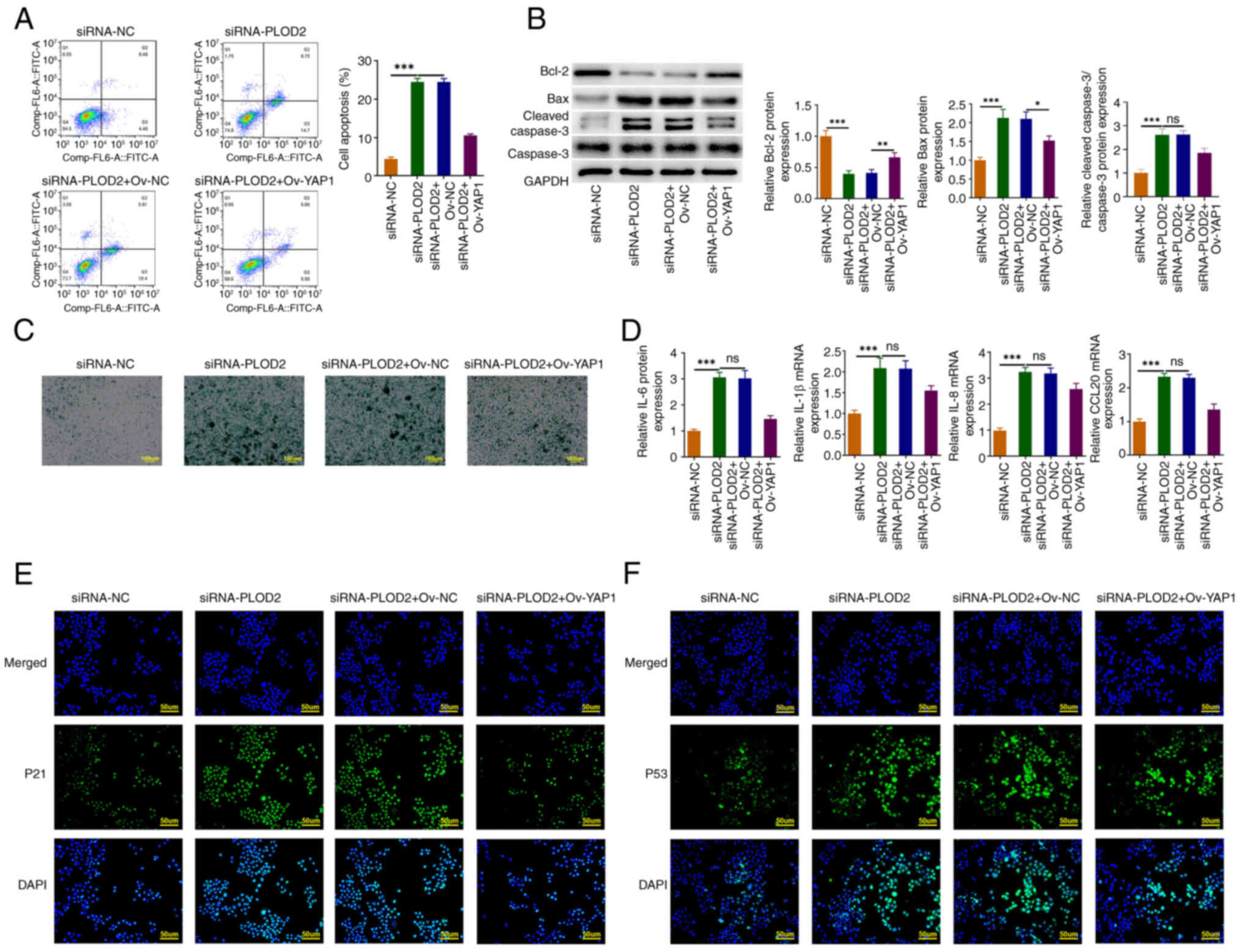 | Figure 5.PLOD2 silencing promoted apoptosis
and senescence in SiHa cells by binding to YAP1 and regulated the
p53 pathway. (A) Cell apoptosis was examined by flow cytometric
analysis. (B) Western blotting was used to assess apoptotic-related
proteins (Bcl-2, Bax, cleaved caspase 3 and caspase 3) levels in
SiHa cells. (C) The expression level of SA-β-Gal was detected by
SA-β-Gal staining. (D) The levels of senescence-associated
secretory phenotype genes (IL-6, IL-1β, IL-8 and CCL20) were
identified by RT-qPCR. Immunofluorescence staining was carried out
to test the levels of (E) p21 and (F) p53 in SiHa cells. Results
are the mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
SA-β-Gal, β-galactosidase; PLOD2, procollagen-lysine 2-oxoglutarate
5-dioxygenase 2; YAP1, Yes-associated protein 1; siRNA-NC, small
interfering RNA-negative control; siRNA-PLOD2, small interfering
RNA-PLOD2. |
Discussion
Present studies have illustrated that apoptosis and
cellular senescence are considered to be the two primary mechanisms
for preventing the development of cancer (35). Cellular senescence is currently
defined as the cell cycle arrest in the G1 phase (36), and senescent cells are
characterized by an increase in SA-β-gal, a flattened cellular
morphology, and large and vacuolated cell size (37). Goodwin et al (38) demonstrated that E6 and E7 proteins,
which are strongly expressed in cervical cancer cells, actively
prevent cervical cancer cell senescence. Consequently, activation
of the endogenous senescence pathway in cancer cells has been
proposed as a therapy for cervical cancer, including CESC. In the
present study, it was confirmed that suppression of PLOD2 resulted
in repression of CESC cell proliferation, migration and
exacerbation of apoptosis and senescence. It was discovered that
PLOD2 binds to YAP1 and plays a modulatory role in SiHa cells,
which may be related to p53 signaling.
PLOD2, a member of the PLOD family, is a critical
enzyme in the process of forming collagen cross-links (39). PLOD2 fosters the aggressive
progression of a number of tumors, including breast, hepatocellular
and non-small-cell lung cancers (40). PLOD2 is positively associated with
poor prognosis in cancers by acting on the morphologic changes in
collagen fibers and facilitating the development of tumor
metastatic ‘highways’ (41,42).
The KEGG pathway and GO biological processes of PLOD2 as well as
interacting genes revealed that PLOD2 is involved in protein
digestion/absorption pathways and collagen fibre organization
processes, which are closely associated with the ECM (22). This implies that PLOD2 expression
may promote the migration and adhesion ability of cervical cancer
cells by affecting the ECM. At present, the function and pathways
involved in PLOD2 have only been assessed by bioinformatics methods
and still need to be further validated by in vitro and in
vivo experiments in future research (22). In the present study, PLOD2
expression was significantly higher in samples from patients with
CESC compared with healthy samples (logFC≥1 or logFC≤-1; P<0.05)
as indicated by differential analysis of the CESC-related dataset
GSE64217. The UALCAN database further confirmed that PLOD2 was
significantly higher expressed in samples from patients with CESC
compared with healthy samples, and the lower the expression, the
better the prognosis. Additionally, when PLOD2 was knocked down,
the migration of SiHa cells was diminished while the apoptosis and
senescence were accelerated.
To explore the mechanism of PLOD2 in CESC, the
molecules that PLOD2 may bind to were predicted using the BioGrid
website. YAP1 was obtained and further simulations were performed
to validate the binding of PLOD2 to YAP1 through H Dock. YAP1, also
termed YAP, is a member of the FOX family of transcription factors
(43). The hyperactivation of YAP1
can drive the onset and progression of cervical cancer, including
CESC (44). Activated YAP
upregulates TGF-α, amphiregulin and EGFR, thus forming a positive
signaling loop to drive cervical cancer cell proliferation
(45). Liu et al (44) showed that YAP expression in the
cytoplasm of samples from patients with CESC was significantly
higher than that in normal cervical tissues. He et al
(46) demonstrated that YAP1
hyperactivation in cervical epithelial cells increased HPV
receptors, disrupted innate immunity of the host cells, and
promoted HPV infection, which promotes cervical cancer development
and progression. Deng et al (47) demonstrated that large tumor
suppressor kinase 1 (LATS1) inhibited cervical cancer cell
proliferation and invasion by regulating YAP1, and LATS1
overexpression decreased the protein level of YAP1 and increased
YAP1 phosphorylation. In the present study, the binding of PLOD2 to
YAP1 was confirmed by co-IP. After YAP1 was overexpressed, the
migration, apoptosis and senescence of SiHa cells regulated by
PLOD2 silencing were all reversed, indicating the role of YAP1 in
PLOD2-silenced SiHa cells.
It was observed that cell senescence is partly
induced by the activation of p53 (48). For example, DeFilippis et al
(49) demonstrated that activation
of p53 by inhibiting E6 protein expression triggered cervical
cancer cell senescence and apoptosis, which in turn inhibited the
pathological development of cervical cancer. In addition,
activation of p53 can inhibit the pathological development of CESC,
for example, up-regulation of p53 by LncRNA WT1-AS inhibits the
proliferation of CESC cells (50).
Thus, activation of p53 can mediate CESC cell senescence and
inhibit the pathological development of CESC. In addition, CLP36
can promote the pathological development of p53-deficient tumors
through upregulation of YAP1 (51). Xu et al (52) demonstrated that knockdown of YAP1
in glial cells significantly promotes premature senescence of glial
cells, including reduced cell proliferation, morphological
hypertrophy, increased SA-β-Gal activity and upregulation of
several senescence-associated genes such as p16, p53 and NF-κB. It
was also found that the levels of p53 and p21 were affected by
PLOD2 silencing and YAP1 overexpression, which was in agreement
with the aforementioned findings.
To conclude, the findings of the present study
indicated that the expression of PLOD2 was elevated in CESC tissues
and cell lines, and PLOD2 silencing caused the inhibition of CESC
cell proliferation and migration, and promotion of apoptosis and
senescence of CESC cells. PLOD2 was predicted to be bound to YAP1
and YAP1 overexpression reversed the effects of PLOD2 silencing on
CESC cell proliferation, cell migration, apoptosis and senescence.
In addition, PLOD2 facilitated CESC progression by regulating the
p53 pathway through YAP1. These findings demonstrated the impacts
of PLOD2 silencing on CESC cells and reported the role of the
binding of PLOD2 and YAP1 in CESC cells, which suggests that PLOD2
could be a prospective therapeutic target for CESC.
However, there were some limitations to the present
study. Firstly, generalizability and clinical translational
potential of the present findings were not verified by animal
models or clinical samples. In the current in vitro
experiments, a single cervical cancer cell line was used, and the
results of the aforementioned experiments require validation with
multiple cervical cancer cell lines. Moreover, the overexpression
or silencing of PLOD2 at different concentrations must be
investigated to comprehensively evaluate the effects of PLOD2 on
CESC cells at different expression levels for a more comprehensive
understanding of the mechanism of action of PLOD2. Although the
present study reveals the mechanism by which PLOD2 regulates the
p53 signaling pathway through YAP1 to promote CESC progression, the
specific molecular mechanisms, such as the mechanisms by which
PLOD2 binds to YAP1, what their binding sites are and how YAP1
regulates p53, still need to be further investigated by
bioinformatics and experimentally. Therefore, future endeavors will
further investigate the aforementioned research elements.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MY conceptualized the study and wrote the original
draft of the manuscript; YW performed the investigation and the
formal analysis; TQ was responsible for conceptualization,
methodology, writing, review, editing and revising the manuscript
critically for important intellectual content. All authors read and
approved the final version of the manuscript. MY, YW and TQ confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chatterjee K, Mukherjee S, Vanmanen J,
Banerjee P and Fata JE: Dietary polyphenols, resveratrol and
pterostilbene exhibit antitumor activity on an HPV E6-positive
cervical cancer model: An in vitro and in vivo analysis. Front
Oncol. 9:3522019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Romli R, Mohd Hashim S, Abd Rahman R, Chew
KT, Mohamad EMW and Mohammed Nawi A: Understanding cervical cancer
screening motivations from women and health practitioners'
perspectives: A qualitative exploration. Gynecol Oncol Rep.
52:1013492024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alqarni SS, Alshehri SM, Alkhateeb MA and
Alsudias LS: Assessing Saudi women's awareness about human
papillomavirus (HPV) and their susceptibility to receive the
vaccine. Hum Vaccin Immunother. 20:23950862024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh D, Vignat J, Lorenzoni V, Eslahi M,
Ginsburg O, Lauby-Secretan B, Arbyn M, Basu P, Bray F and
Vaccarella S: Global estimates of incidence and mortality of
cervical cancer in 2020: A baseline analysis of the WHO global
cervical cancer elimination initiative. Lancet Glob Health.
11:e197–e206. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jensen JE, Becker GL, Jackson JB and
Rysavy MB: Human papillomavirus and associated cancers: A review.
Viruses. 16:6802024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
US Preventive Services Task Force, . Curry
SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, Doubeni
CA, Epling JW Jr, Kemper AR, et al: Screening for cervical cancer:
US preventive services task force recommendation statement. JAMA.
320:674–686. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meng Y, Chu T, Lin S and Wu P, Zhi W, Peng
T, Ding W, Luo D and Wu P: Clinicopathological characteristics and
prognosis of cervical cancer with different histological types: A
population-based cohort study. Gynecol Oncol. 163:545–551. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mayadev JS, Ke G, Mahantshetty U, Pereira
MD, Tarnawski R and Toita T: Global challenges of radiotherapy for
the treatment of locally advanced cervical cancer. Int J Gynecol
Cancer. 32:436–445. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ran Z, Wu S, Ma Z, Chen X, Liu J and Yang
J: Advances in exosome biomarkers for cervical cancer. Cancer Med.
11:4966–4978. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walboomers JM, Jacobs MV, Manos MM, Bosch
FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ and Muñoz N:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pimple SA and Mishra GA: Global strategies
for cervical cancer prevention and screening. Minerva Ginecol.
71:313–320. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rerucha CM, Caro RJ and Wheeler VL:
Cervical cancer screening. Am Fam Physician. 97:441–448.
2018.PubMed/NCBI
|
|
14
|
Zhou Y, Wang W, Tang J, Hu K and Zhang F:
Comparison of outcomes between early-stage cervical cancer patients
without high-risk factors undergoing adjuvant concurrent
chemoradiotherapy and radiotherapy alone after radical surgery. BMC
Cancer. 24:5482024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nuchpramool P and Hanprasertpong J:
Preoperative neutrophil-lymphocyte ratio and platelet-lymphocyte
ratio are not clinically useful in predicting prognosis in early
stage cervical cancer. Surg Res Pract. 2018:91629212018.PubMed/NCBI
|
|
16
|
Shi R, Zhang Z, Zhu A, Xiong X, Zhang J,
Xu J, Sy MS and Li C: Targeting type I collagen for cancer
treatment. Int J Cancer. 151:665–683. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Terajima M, Taga Y, Chen Y, Cabral WA,
Hou-Fu G, Srisawasdi S, Nagasawa M, Sumida N, Hattori S, Kurie JM,
et al: Cyclophilin-B modulates collagen cross-linking by
differentially affecting lysine hydroxylation in the helical and
telopeptidyl domains of tendon type I collagen. J Biol Chem.
291:9501–9512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piersma B and Bank RA: Collagen
cross-linking mediated by lysyl hydroxylase 2: An enzymatic
battlefield to combat fibrosis. Essays Biochem. 63:377–387. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sato K, Parag-Sharma K, Terajima M,
Musicant AM, Murphy RM, Ramsey MR, Hibi H, Yamauchi M and Amelio
AL: Lysyl hydroxylase 2-induced collagen cross-link switching
promotes metastasis in head and neck squamous cell carcinomas.
Neoplasia. 23:594–606. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gong S, Wu C, Duan Y, Fu J, Wang Y, Wu H,
Zhang B, Tang J and Wu P: PLODs: Novel prognostic biomarkers and
potential immunotherapy targets for head and neck squamous cell
carcinoma. Heliyon. 9:e134792023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sekaran K, Varghese RP, Krishnan S, Zayed
H, El Allali A and Doss GPC: Dissecting crucial gene markers
involved in HPV-associated oropharyngeal squamous cell carcinoma
from RNA-sequencing data through explainable artificial
intelligence. Front Biosci (Landmark Ed). 29:2202024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li G, Wang X and Liu G: PLOD2 is a potent
prognostic marker and associates with immune infiltration in
cervical cancer. Biomed Res Int. 2021:55123402021.PubMed/NCBI
|
|
23
|
Liu R, He X, Bao W and Li Z: Enhancement
of HPV therapeutic peptide-based vaccine efficacy through
combination therapies and improved delivery strategies: A review.
Hum Vaccin Immunother. 20:23967102024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suhaila K, Mukherjee A, Maharjan B, Dhakal
A, Lama M, Junkins A, Khakurel U, Jha AN, Jolly PE, Lhaki P and
Shrestha S: Human papillomavirus, related diseases, and
vaccination: Knowledge and awareness among health care students and
professionals in Nepal. J Cancer Educ. 37:1727–1735. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sada M, Ohuchida K, Horioka K, Okumura T,
Moriyama T, Miyasaka Y, Ohtsuka T, Mizumoto K, Oda Y and Nakamura
M: Hypoxic stellate cells of pancreatic cancer stroma regulate
extracellular matrix fiber organization and cancer cell motility.
Cancer Lett. 372:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu B and Xi S: Bioinformatics analysis of
differentially expressed genes and pathways in the development of
cervical cancer. BMC Cancer. 21:7332021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
S S Shukla V, Khan GN, Eswaran S, Adiga D
and Kabekkodu SP: Integrated bioinformatic analysis of miR-15a/16-1
cluster network in cervical cancer. Reprod Biol. 21:1004822021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li CJ, Chiu YH, Chang C, Chang YCI, Sheu
JJ and Chiang AJ: Acetyl coenzyme A synthase 2 acts as a prognostic
biomarker associated with immune infiltration in cervical squamous
cell carcinoma. Cancers (Basel). 13:31252021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein sci.
30:187–200. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dalby B, Cates S, Harris A, Ohki EC,
Tilkins ML, Price PJ and Ciccarone VC: Advanced transfection with
Lipofectamine 2000 reagent: Primary neurons, siRNA, and
high-throughput applications. Methods. 33:95–103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, He H, Ren X, Chen Y, Liu W, Pu R,
Fang L, Shi Y, Liu D, Zhao J, et al: APOBEC3A suppresses cervical
cancer via apoptosis. J Cancer. 14:3429–3443. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Q, Sun X, Luo X, Wang J, Hu J and
Feng Y: PIK3R3 inhibits cell senescence through p53/p21 signaling.
Cell Death Dis. 11:7982020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lv S, Wang Y, Xu W and Dong X: Serum
exosomal miR-17-5p as a promising biomarker diagnostic biomarker
for breast cancer. Clin Lab. 66:2020. View Article : Google Scholar
|
|
34
|
Zhong C, Ju G, Yang S, Zhao X, Chen J and
Li N: Total flavonoids of polygala fallax hemsl induce apoptosis of
human ectopic endometrial stromal cells through PI3K/AKT/Bcl-2
signaling pathway. Gynecol Obstet Invest. 88:197–213. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Childs BG, Baker DJ, Kirkland JL, Campisi
J and van Deursen JM: Senescence and apoptosis: Dueling or
complementary cell fates? EMBO Rep. 15:1139–1153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ou HL, Hoffmann R, González-López C,
Doherty GJ, Korkola JE and Muñoz-Espín D: Cellular senescence in
cancer: From mechanisms to detection. Mol Oncol. 15:2634–2671.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tzoneva R: Special issue ‘role of
apoptosis and cellular senescence in cancer and aging’. Int J Mol
Sci. 25:21032024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio
D and Hwang ES: Rapid induction of senescence in human cervical
carcinoma cells. Proc Natl Acad Sci U S A. 97:10978–10983. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lan J, Zhang S, Zheng L, Long X, Chen J,
Liu X, Zhou M and Zhou J: PLOD2 promotes colorectal cancer
progression by stabilizing USP15 to activate the AKT/mTOR signaling
pathway. Cancer Sci. 114:3190–3202. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu Q, Kong N, Zhao Y, Wu Q, Wang X, Xun X
and Gao P: Pan-cancer analyses reveal oncogenic and immunological
role of PLOD2. Front Genet. 13:8646552022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li K, Niu Y, Li K, Zhong C, Qiu Z, Yuan Y,
Shi Y, Lin Z, Huang Z, Zuo D, et al: Dysregulation of PLOD2
promotes tumor metastasis and invasion in hepatocellular carcinoma.
J Clin Transl Hepatol. 11:1094–1105. 2023.PubMed/NCBI
|
|
42
|
Yang YS, Jin X, Li Q, Chen YY, Chen F,
Zhang H, Su Y, Xiao Y, Di GH, Jiang YZ, et al: Superenhancer drives
a tumor-specific splicing variant of MARCO to promote
triple-negative breast cancer progression. Proc Natl Acad Sci USA.
119:e22072011192022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zheng L, Qian B, Tian D, Tang T, Wan S,
Wang L, Zhu L and Geng X: FOXA1 positively regulates gene
expression by changing gene methylation status in human breast
cancer MCF-7 cells. Int J Clin Exp Pathol. 8:96–106.
2015.PubMed/NCBI
|
|
44
|
Liu T, Liu Y, Gao H, Meng F, Yang S and
Lou G: Clinical significance of yes-associated protein
overexpression in cervical carcinoma: The differential effects
based on histotypes. Int J Gynecol Cancer. 23:735–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
He C, Mao D, Hua G, Lv X, Chen X,
Angeletti PC, Dong J, Remmenga SW, Rodabaugh KJ, Zhou J, et al: The
Hippo/YAP pathway interacts with EGFR signaling and HPV
oncoproteins to regulate cervical cancer progression. EMBO Mol Med.
7:1426–1449. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He C, Lv X, Huang C, Angeletti PC, Hua G,
Dong J, Zhou J, Wang Z, Ma B, Chen X, et al: A Human
Papillomavirus-Independent Cervical Cancer Animal Model Reveals
Unconventional Mechanisms of Cervical Carcinogenesis. Cell Rep.
26:2636–2650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Deng J, Zhang W, Liu S, An H, Tan L and Ma
L: LATS1 suppresses proliferation and invasion of cervical cancer.
Mol Med Rep. 15:1654–1660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sheekey E and Narita M: p53 in
senescence-it's a marathon, not a sprint. FEBS J. 290:1212–1220.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
DeFilippis RA, Goodwin EC, Wu L and DiMaio
D: Endogenous human papillomavirus E6 and E7 proteins
differentially regulate proliferation, senescence, and apoptosis in
HeLa cervical carcinoma cells. J Virol. 77:1551–1563. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Y, Na R and Wang X: LncRNA WT1-AS
up-regulates p53 to inhibit the proliferation of cervical squamous
carcinoma cells. BMC Cancer. 19:10522019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lu Y, Mu Y, Chen J, Guan X, Guo L and Wu
C: CLP36 promotes p53 deficient sarcoma progression through
suppression of atrophin-1 interacting protein-4 (AIP-4)-dependent
degradation of YAP1. Theranostics. 12:5051–5068. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu X, Shen X, Wang J, Feng W, Wang M, Miao
X, Wu Q, Wu L, Wang X Ma Y, et al: YAP prevents premature
senescence of astrocytes and cognitive decline of Alzheimer's
disease through regulating CDK6 signaling. Aging Cell.
20:e134652021. View Article : Google Scholar : PubMed/NCBI
|