Introduction
Hepatocellular carcinoma (HCC) is a highly
aggressive malignancy associated with high rates of mortality and
morbidity. It is predicted that Asia accounts for 72% of the cases
(with China alone contributing >50%), while Europe has accounts
for 10%, Africa for 7.8%, North America for 5.1%, Latin America for
4.6% and Oceania for 0.5% (1). Due
to the lack of symptoms, the majority of patients with HCC are
diagnosed at advanced stages, resulting in a poor prognosis
(2). At present, the methods used
for diagnosing HCC in the clinic include ultrasound, computed
tomography, detection of α-fetoprotein (AFP) levels and
pathological biopsy. Pathological biopsy has been recognized as the
criterion for accurate diagnosis of HCC (3,4).
Since pathological biopsy is an invasive examination that can cause
bleeding and tissue damage, and imaging examinations lack
specificity and sensitivity, serum AFP is currently the most
frequently used screening biomarker for HCC in clinical practice
(5). However, its diagnostic
performance is relatively limited, particularly for patients with
early stage HCC (6). Therefore, it
is important to identify noninvasive biomarkers to improve the
specificity and sensitivity of HCC diagnosis and prognosis.
Long noncoding RNAs (lncRNAs) are defined as
noncoding RNA molecules >200 nucleotides in length, which were
once regarded as transcriptional noise (7). Increasing evidence has demonstrated
that lncRNAs serve crucial role in diverse biological processes,
such as cell migration, metastasis and angiogenesis (8–10).
Moreover, lncRNAs have been verified to participate in the
occurrence and development of various tumors, including HCC
(11). For example, the lncRNAs
HOTTIP, PVT1 and HOTAIR have been confirmed to be closely
associated with hepatocarcinogenesis (12–14).
Additionally, it has been shown that lncRNAs are stably present in
body fluids (15). Serum lncRNA
LINC01535 has been suggested to be a novel biomarker of diagnosis,
prognosis and disease progression in breast cancer (16). Additional lncRNAs have been
verified as promising biomarkers for HCC diagnosis and prognosis,
including MALAT1, UBE2CP3 and NETA-1; however, the diagnostic
performance of previously reported lncRNAs for HCC varies
considerably among different studies (17–19),
and the majority of serum lncRNAs associated with HCC need further
investigation.
The lncRNA PTTG3P is a processed pseudogene located
at chromosome 8q13.1, which is involved in the development of
different types of cancer, such as colorectal cancer, pancreatic
cancer, osteosarcoma and non-small cell lung cancer (20–23).
Moreover, our previous study confirmed that lncRNA PTTG3P acts as
an oncogene in HCC; through elevating mRNA PTTG1 and activating
PI3K/AKT signaling, lncRNA PTTG3P was shown to promote tumor growth
and metastasis in HCC (24).
Currently, there are few studies concerning the application of
serum lncRNA PTTG3P and mRNA PTTG1 as diagnostic markers for HCC
(25,26). Thus, to investigate the diagnostic
efficacy of serum lncRNA PTTG3P, mRNA PTTG1 and their combinations
for the diagnosis of HCC, the present study aimed to evaluate the
serum expression of lncRNA PTTG3P and mRNA PTTG1 in patients with
HCC, chronic hepatitis B (CHB) and liver cirrhosis (LC), and in
healthy controls (HCs).
Materials and methods
Patients
In the present study, 373 participants were
enrolled, including 73 patients with HCC, 100 patients with CHB,
100 patients with LC and 100 HCs. The participants were recruited
from The Second Affiliated Hospital of Guangzhou University of
Chinese Medicine (Guangzhou, China) between July 2022 and March
2023. The patients with HCC were diagnosed for the first time by
histological examination and did not receive any treatment, whereas
those with LC were diagnosed according to the American Association
for the Study of Liver Diseases Practice Guidelines (27), and the patients with CHB were
diagnosed according to the 2017 Clinical Practice Guidelines on the
management of Hepatitis B virus infection of the European
Association for the Study of the Liver (28). The 100 HCs were recruited during
routine medical examinations at the same hospital during the
aforementioned time period. Moreover, patient clinical data were
collected from the hospital medical records for further study,
including sex, age and HBV surface antigen (HBsAg). The present
study was performed according to the principles of The Declaration
of Helsinki. Each subject provided written informed consent and the
research protocol was approved by the Ethics Committee of The
Second Affiliated Hospital of Guangzhou University of Chinese
Medicine (approval no. BE2020-211-01).
Sample collection
Peripheral blood samples were collected in separate
vacuum tubes from patients prior to surgery, chemotherapy or
pharmacological intervention. Paired preoperative and postoperative
plasma samples were obtained from 36 patients with HCC, with
postoperative samples collected 10 days after surgery. All serum
samples were stored at −80°C for further analysis.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Serum RNA was extracted from serum samples using a
HiPure Liquid RNA Kit (cat. no. R416303; Magen Biotechnology Co.,
Ltd.) according to the manufacturer's instructions. DNase On Column
Kit B (cat. no. R4911B; Magen Biotechnology Co., Ltd.) was used to
remove DNA. The quantity and purity of RNA were verified using a
NanoDrop 2000c Spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc.). Subsequently, cDNA was generated using Evo M-MVL
RT Premix for qPCR (cat. no. AG11706; Hunan Accurate Bio-Medical
Technology Co., Ltd.) according to manufacturer's protocol. RNA
expression levels were assessed by qPCR using SYBR®
Green Premix Pro Taq HS qPCR Kit (cat. no. AG11701 Hunan Accurate
Bio-Medical Technology Co., Ltd.), which was performed on an LC480
Real Time PCR system (serial no. 28833; Roche Diagnostics GmbH).
qPCR was performed under the following conditions: 95°C for 30 sec,
followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. U6 and
β-actin were used as internal controls. All results are presented
as the mean ± standard deviation (SD) of ≥3 independent
experiments. Comparative quantification was performed using the
2−ΔΔCq method (29).
The primer sequences used in the present study are shown in
Table I.
 | Table I.Primers used for reverse
transcription-quantitative PCR in the present study. |
Table I.
Primers used for reverse
transcription-quantitative PCR in the present study.
| Gene symbol | Sequences |
|---|
| U6 | Sense
CTCGCTTCGGCAGCACA |
|
| Antisense
AACGCTTCACGAATTTGCGT |
| PTTG3P | Sense
GGGGTCTGGACCTTCAATCAA |
|
| Antisense
GCTTTAGGTAAGGATGTGGGA |
| PTTG1 | Sense
ACCCGTGTGGTTGCTAAGG |
|
| Antisense
ACGTGGTGTTGAAACTTGAGAT |
| β-actin | Sense
TGGCACCCAGCACAATGAA |
|
| Antisense
CTAAGTCATAGTCCGCCTAGAA |
|
| GCA |
Detection of liver function and serum
AFP
Liver function-related indicators were detected on a
Roche Cobas 8000 c702 chemistry analyzer (serial no. 16K9-07; Roche
Diagnostics GmbH) according to the manufacturer's protocol,
including g-glutamyl transpeptidase (GGT; cat. no. 05168775190),
aspartate transaminase (AST; cat. no. 05850819190), alanine
aminotransferase (ALT; cat. no. 05850797190), alkaline phosphatase
(ALP; cat. no. 05166888190), total protein (TP; cat. no.
05171385190), albumin (ALB; cat. no. 05166861190), total bilirubin
(TBIL; cat. no. 05795419190) and direct bilirubin (DBIL; cat. no.
05975921190) (all from Roche Diagnostics GmbH). Serum AFP (cat. no.
04481798190; Roche Diagnostics GmbH) was detected using the Roche
Cobas 8000 e602 electrochemiluminescence immunoanalyzer (serial no.
16U3-11; Roche Diagnostics GmbH) according to the manufacturer's
instructions.
Data analysis based on The Cancer
Genome Atlas (TCGA)-liver hepatocellular carcinoma
RNA-sequencing expression (level 3) profiles and
corresponding clinical information of 370 patients with HCC were
downloaded from TCGA database (https://portal.gdc.cancer.gov/). Heatmap of PTTG1 was
plotted using ‘pheatmap’ R package with zero-mean normalization
(30). The time ROC (v 0.4)
analysis (https://cran.r-project.org/web/packages/timeROC/index.html)
(31) was used to compare the
predictive accuracy of PTTG1 mRNA. For Kaplan-Meier curves,
P-values and hazard ratio (HR) with 95% confidence interval (CI)
were generated by log-rank tests and univariate Cox proportional
hazards regression. All of the analysis methods and R packages were
implemented by R (Foundation for Statistical Computing 2020)
version 4.0.3 (32).
Statistical analysis
SPSS 17.0 software (SPSS, Inc.) was used to perform
statistical analysis. The Kolmogorov-Smirnov test was used to
evaluate the normality of the data distribution. Data are presented
as the mean ± SD, and were analyzed by unpaired two-tailed
Student's t-test if the variables were normally distributed.
Comparisons between more than two groups were made by one-way ANOVA
followed by the Dunnett T3 post hoc test. For nonparametric data,
the Mann-Whitney U test was applied for comparisons between groups.
For non-normally distributed variables, the Spearman's correlation
coefficient test was performed. The χ2 test or Fisher's
exact test was used to evaluate the association between lncRNA
PTTG3P expression levels and clinical characteristics. Wilcoxon
signed-rank test was used to compare the preoperative and
postoperative levels of lncRNA PTTG3P and mRNA PTTG1. Receiver
operating characteristic (ROC) curve and area under the curve (AUC)
analyses were used to assess the diagnostic performance of lncRNA
PTTG3P, mRNA PTTG1, AFP, ALT, AST, GGT, ALP and their combinations.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristics
A total of 373 participants were enrolled in the
present study, including 73 patients with HCC, 100 patients with
CHB, 100 patients with LC and 100 HCs. Detailed demographic
information of all participants is presented in Table II. The mean age of patients with
HCC, CHB and LC, and of HCs was 55 years (range, 29–83 years), 44
years (range, 21–72 years), 53 years (range, 23–80 years) and 42
years (23–84 years), respectively. There were 62 men and 11 women
in the HCC group, 70 men and 30 women in the LC group, 64 men and
36 women in the CHB group, and 48 men and 52 women in the HC group.
Additionally, the clinical features of the HCC, LC and CHB groups
were compared with those of HC group. Compared with in the HC
group, the HCC group exhibited significant differences in all
parameters, whereas the LC group showed significant differences
compared with the HC group in all parameters with the exception of
AFP and TP. By contrast, only ALT, AST, ALP and ALB were
significantly different between the CHB and HC groups.
 | Table II.Demographics of HCs, and patients
with LC, CHB and HCC. |
Table II.
Demographics of HCs, and patients
with LC, CHB and HCC.
| Feature | HCC |
P-valuea | LC |
P-valuea | CHB |
P-valuea | HCs |
|---|
| Number | 73 | / | 100 | / | 100 | / | 100 |
| Mean age, years
(range) | 55 (29–83) | / | 53 (23–80) | / | 44 (21–72) | / | 42 (23–84) |
| Male/Female | 62/11 | / | 70/30 | / | 64/36 | / | 48/52 |
| Mean AFP, ng/ml
(range) | 8,129.42
(1.02–60,501.00) |
<0.001b | 4.16
(0.61–48.49) | 0.810 | 3.61
(0.63–49.82) | 0.794 | 2.84
(0.61–9.45) |
| Mean GGT, U/l
(range) | 129.89
(9.00–623.00) |
<0.001b | 33.64
(7.00–123.00) |
<0.001b | 25.46
(7.00–154.00) | 0.779 | 28.77
(9.00–178.00) |
| Mean ALT, U/l
(range) | 49.26
(8.00–266.00) |
<0.001b | 26.93
(6.00–75.00) |
<0.001b | 29.87
(8.00–483.00) | 0.046b | 23.39
(5.00–182.00) |
| Mean AST, U/l
(range) | 64.83
(12.00–452.00) |
<0.001b | 27.03
(3.00–78.00) |
<0.001b | 26.16
(14.00–194.00) | 0.014b | 21.85
(10.00–72.00) |
| Mean ALP, U/l
(range) | 119.81
(50.00–606.00) |
<0.001b | 83.74
(39.00–260.00) |
<0.001b | 72.93
(31.00–128.00) | 0.018b | 68.61
(27.00–160.00) |
| Mean TP, g/l
(range) | 70.20
(31.30–95.00) |
<0.001b | 74.15
(56.80–86.10) | 0.127 | 75.42
(42.00–87.70) | 0.288 | 75.07
(63.20–85.60) |
| Mean ALB, g/l
(range) | 41.00
(26.60–191.00) |
<0.001b | 45.12
(24.70–52.20) |
<0.001b | 45.94
(28.00–52.90) | 0.032b | 46.92
(25.80–53.10) |
| Mean TBIL, µmol/l
(range) | 22.11
(4.10–588.00) | 0.009b | 14.69
(6.00–54.70) |
<0.001b | 10.89
(4.80–21.30) | 0.934 | 11.15
(3.50–27.10) |
| Mean DBIL, µmol/l
(range) | 12.31
(1.50–403.90) | 0.001b | 6.41
(1.60–45.00) |
<0.001b | 4.18
(0.60–7.50) | 0.886 | 4.31
(1.50–10.60) |
Serum expression levels of lncRNA
PTTG3P and mRNA PTTG1 in patients with HCC, LC and CHB, and in
HCs
In the present study, blood samples obtained from
patients with HCC, LC and CHB, and HCs were evaluated by RT-qPCR to
screen for the presence of serum lncRNA PTTG3P and mRNA PTTG1. The
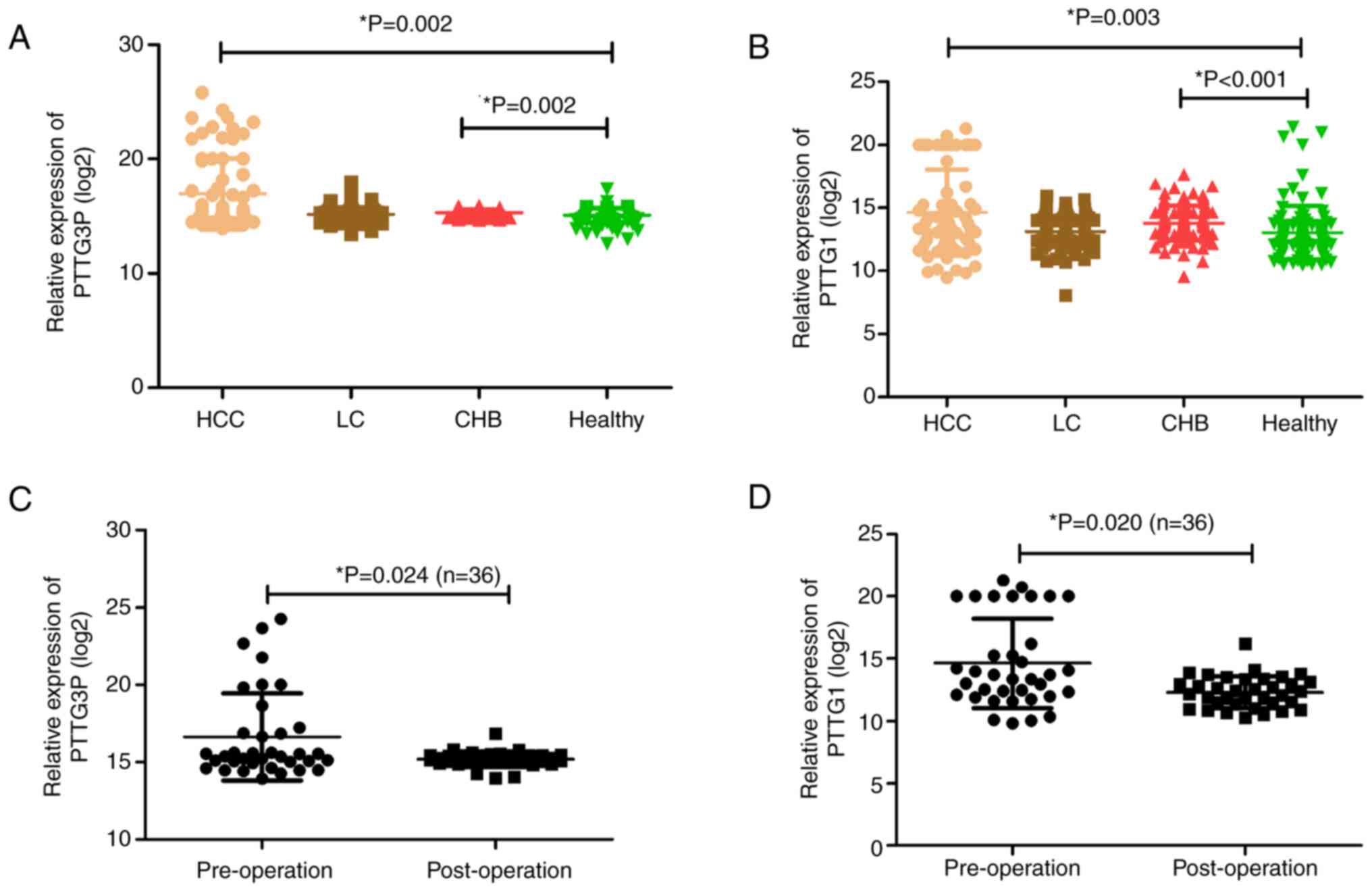
results showed that lncRNA PTTG3P levels were markedly increased in
patients with HCC and CHB compared with those in HCs (Fig. 1A). Since mRNA PTTG1 is a target of
lncRNA PTTG3P (24), its
expression was detected in the same cohort. The results indicated
consistency with lncRNA PTTG3P; namely, mRNA PTTG1 was highly
expressed in patients with HCC and CHB compared with in HCs
(Fig. 1B).
Serum expression levels of lncRNA
PTTG3P and mRNA PTTG1 in patients with HCC before and after
surgery
To verify whether the combination of lncRNA PTTG3P
and mRNA PTTG1 may be a potential biomarker for monitoring the
prognosis of patients with HCC, the preoperative and postoperative
expression levels of lncRNA PTTG3P and mRNA PTTG1 were compared in
36 patients with HCC. Among them, the group consisted of 34 men and
2 women, aged between 29 and 83 years (mean age, 53 years). The
postoperative levels of lncRNA PTTG3P and mRNA PTTG1 were both
significantly lower than the corresponding preoperative levels
(Fig. 1C and D).
Survival analysis of PTTG1 in patients
with HCC from TCGA dataset
In order to investigate the prognostic impact of
PTTG1 on HCC, the present study used TCGA database. The median
expression level of PTTG1 was used as the cutoff value; 185
patients were included in the low expression PTTG1 group and 185
patients were included in the high expression PTTG1 group. The
results showed that in the high expression group a higher
proportion of patients were dead than in the low expression group
(Fig. S1A). In addition, the HR
of the low-expression group relative to the high-expression group
was 2.048 (95% CI: 1.435–2.923), and the median survival time of
the high expression group was shorter than that of the low
expression group (Fig. S1B). The
heatmap results indicated that PTTG1 expression was elevated in the
majority of patients with HCC (Fig.
S1C). Time ROC analysis results showed that the 1-year AUC,
3-year AUC and 5-year AUC were 0.716 (95% CI: 0.657–0.775), 0.670
(95% CI: 0.605–0.735) and 0.634 (95% CI: 0.550–0.718), respectively
(Fig. S1D), suggesting that PTTG1
had predictive ability regarding 1-, 3- and 5-year survival. These
findings indicated that higher expression of PTTG1 may be a risk
factor for patients with HCC.
Association between serum lncRNA
PTTG3P and clinical characteristics
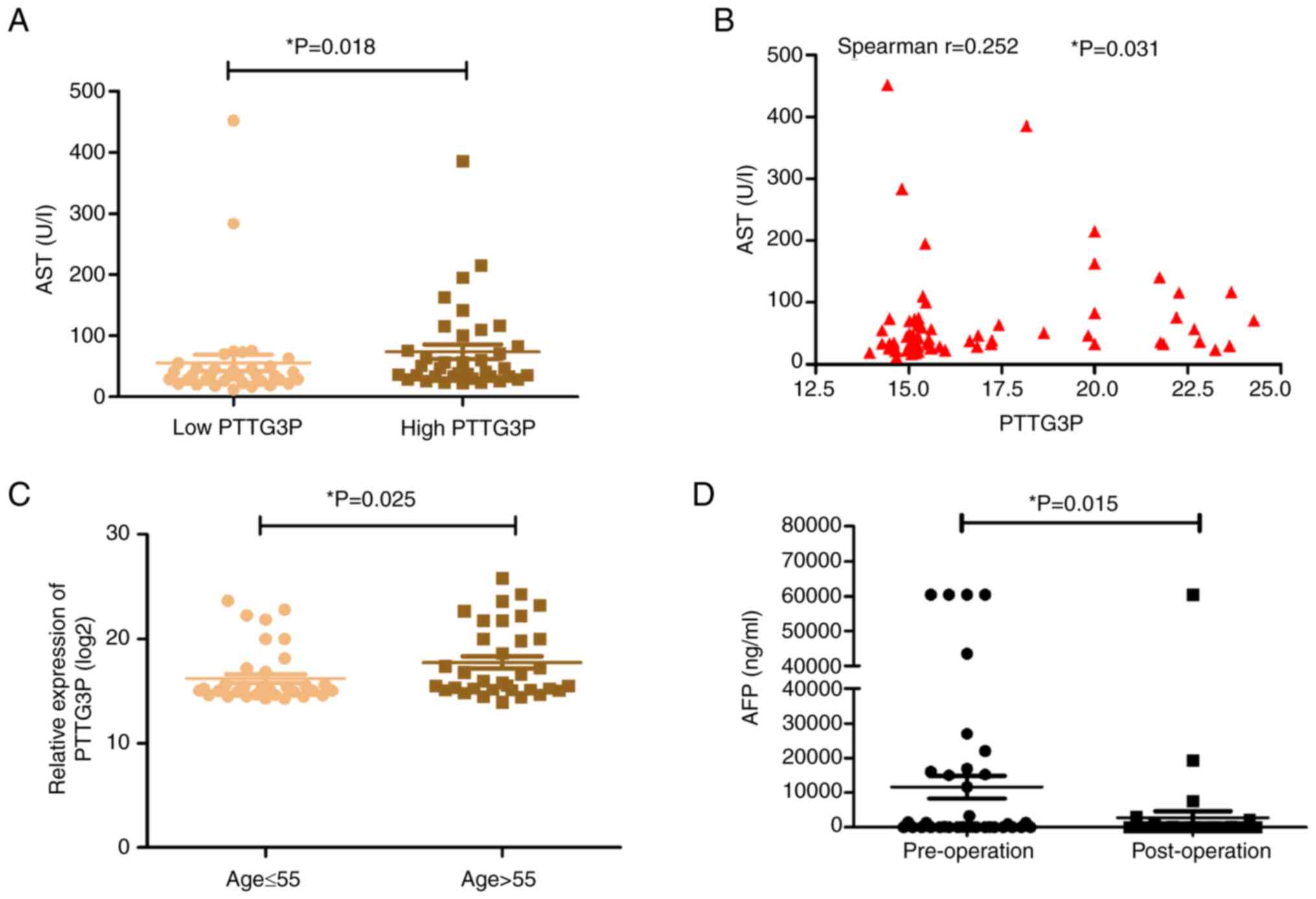
In the current study, the associations between the
serum expression levels of lncRNA PTTG3P and clinical parameters in
patients with HCC were explored. The median expression level of
lncRNA PTTG3P was used as the cutoff value; 36 patients were
included in the low lncRNA PTTG3P expression group and 37 patients
were included in the high lncRNA PTTG3P expression group. The
results indicated that serum lncRNA PTTG3P expression was weakly
associated with AST (Spearman's correlation test r=0.252, P=0.031;
Table III; Fig. 2A and B). Furthermore, patients with
HCC aged >55 years were confirmed to have an elevated level of
lncRNA PTTG3P (Table III;
Fig. 2C). However, there were no
associations between serum lncRNA PTTG3P expression and sex, HBsAg,
AFP level, cirrhosis, tumor size, tumor number, Edmondson grade
(33), lymph node metastasis,
portal vein invasion, ALT, GGT, ALB, ALP, TP, TBIL or DBIL
(Table III). In addition, the
clinical features of 36 patients with HCC before and after surgery
were explored. As shown in Table
IV and Fig. 2D, the AFP level
post-surgery was significantly lower than that in pre-surgery,
while other parameters showed no significant differences, including
GGT, ALT, AST, ALP, TP, ALB, TBIL and DBIL.
 | Table III.Association between serum expression
levels of PTTG3P and clinicopathological characteristics in 73
patients with HCC. |
Table III.
Association between serum expression
levels of PTTG3P and clinicopathological characteristics in 73
patients with HCC.
|
| PTTG3P
expression |
|
|---|
|
|
|
|
|---|
| Feature | Low (n=36) | High (n=37) | P-value |
|---|
| Sex |
|
| 0.515a |
|
Male | 32 | 30 |
|
|
Female | 4 | 7 |
|
| Age |
|
| 0.025b |
| ≤55
years | 24 | 15 |
|
| >55
years | 12 | 22 |
|
| HBsAg |
|
| 0.955 |
|
Positive | 29 | 30 |
|
|
Negative | 7 | 7 |
|
| AFP level |
|
| 0.279 |
| ≤20
ng/ml | 13 | 18 |
|
| >20
ng/ml | 23 | 19 |
|
| Cirrhosis |
|
| 0.955 |
|
With | 29 | 30 |
|
|
Without | 7 | 7 |
|
| Tumor size |
|
| 0.741 |
| ≤5
cm | 13 | 12 |
|
| >5
cm | 23 | 25 |
|
| Tumor number |
|
|
>0.999a |
|
Single | 5 | 6 |
|
|
Multiple | 31 | 31 |
|
| Edmondson
grade |
|
| 0.345 |
|
I–II | 9 | 13 |
|
|
III–IV | 27 | 24 |
|
| Lymph node
metastasis |
|
| 0.281 |
|
With | 22 | 27 |
|
|
Without | 14 | 10 |
|
| Portal vein
invasion |
|
| 0.384 |
|
With | 24 | 21 |
|
|
Without | 12 | 16 |
|
| Mean GGT, U/l
(range) | 129.19
(9.00–623.00) | 130.57
(15.00–340.00) | 0.244 |
| Mean ALT, U/l
(range) | 40.63
(8.00–179.00) | 57.65
(12.00–266.00) | 0.141 |
| Mean AST, U/l
(range) | 55.75
(12.00–452.00) | 73.68
(23.00–386.00) | 0.018b |
| Mean ALP, U/l
(range) | 123.17
(50.00–606.00) | 116.54
(56.00–227.00) | 0.529 |
| Mean TP, g/l
(range) | 72.24
(50.80–95.00) | 68.21
(31.30–85.30) | 0.055 |
| Mean ALB, g/l
(range) | 43.98
(26.60–191.00) | 38.10
(27.80–45.50) | 0.153 |
| Mean TBIL, µmol/l
(range) | 14.02
(4.10–40.90) | 29.98
(4.90–588.00) | 0.570 |
| Mean DBIL, µmol/l
(range) | 6.44
(2.10–26.00) | 18.02
(1.50–403.90) | 0.069 |
 | Table IV.Demographics of patients with HCC
pre-operation and post-operation (n=36). |
Table IV.
Demographics of patients with HCC
pre-operation and post-operation (n=36).
| Feature | Pre-operation | Post-operation | P-value |
|---|
| Mean AFP, ng/ml
(range) | 11,624.14
(1.33–60,501.00) | 2,781.92
(1.16–60,501.00) | 0.015a |
| Mean GGT, U/l
(range) | 130.42
(15.00–340.00) | 286.94
(11.00–4,412.00) | 0.706 |
| Mean ALT, U/l
(range) | 52.47
(15.00–256.00) | 35.85
(9.00–141.00) | 0.086 |
| Mean AST, U/l
(range) | 63.69
(17.00–452.00) | 46.47
(18.00–129.00) | 0.289 |
| Mean ALP, U/l
(range) | 113.69
(63.00–210.00) | 151.55
(55.00–881.00) | 0.285 |
| Mean TP, g/l
(range) | 70.22
(50.80–85.30) | 68.70
(56.40–82.90) | 0.292 |
| Mean ALB, g/l
(range) | 38.93
(26.60–53.70) | 36.51
(26.80–44.50) | 0.065 |
| Mean TBIL, µmol/l
(range) | 12.70
(4.10–25.70) | 21.60
(4.60–185.70) | 0.338 |
| Mean DBIL, µmol/l
(range) | 5.70
(1.50–16.10) | 12.42
(1.40–135.80) | 0.235 |
Diagnostic efficiency of serum lncRNA
PTTG3P and mRNA PTTG1 expression in patients with HCC
ROC curves were applied to evaluate the diagnostic
efficacy of lncRNA PTTG3P and mRNA PTTG1 expression. When comparing
patients with HCC with HCs, the AUC was 0.636 (95% CI: 0.547–0.724)
for PTTG3P, 0.634 (95% CI: 0.547–0.722) for mRNA PTTG1, 0.891 (95%
CI: 0.836–0.946) for AFP, 0.762 (95% CI: 0.692–0.833) for ALT,
0.879 (95% CI: 0.827–0.931) for AST, 0.886 (95% CI: 0.837–0.936)
for GGT and 0.840 (95% CI: 0.782–0.899) for ALP (Table V; Fig.
3A-G). The diagnostic value of PTTG3P exhibited 37.0%
sensitivity and 97.0% specificity, whereas that of mRNA PTTG1
showed 77% specificity and 50.7% sensitivity (Table V; Fig.
3A). These results suggested that PTTG3P and PTTG1 have certain
diagnostic efficacy in assisting the diagnosis of HCC.
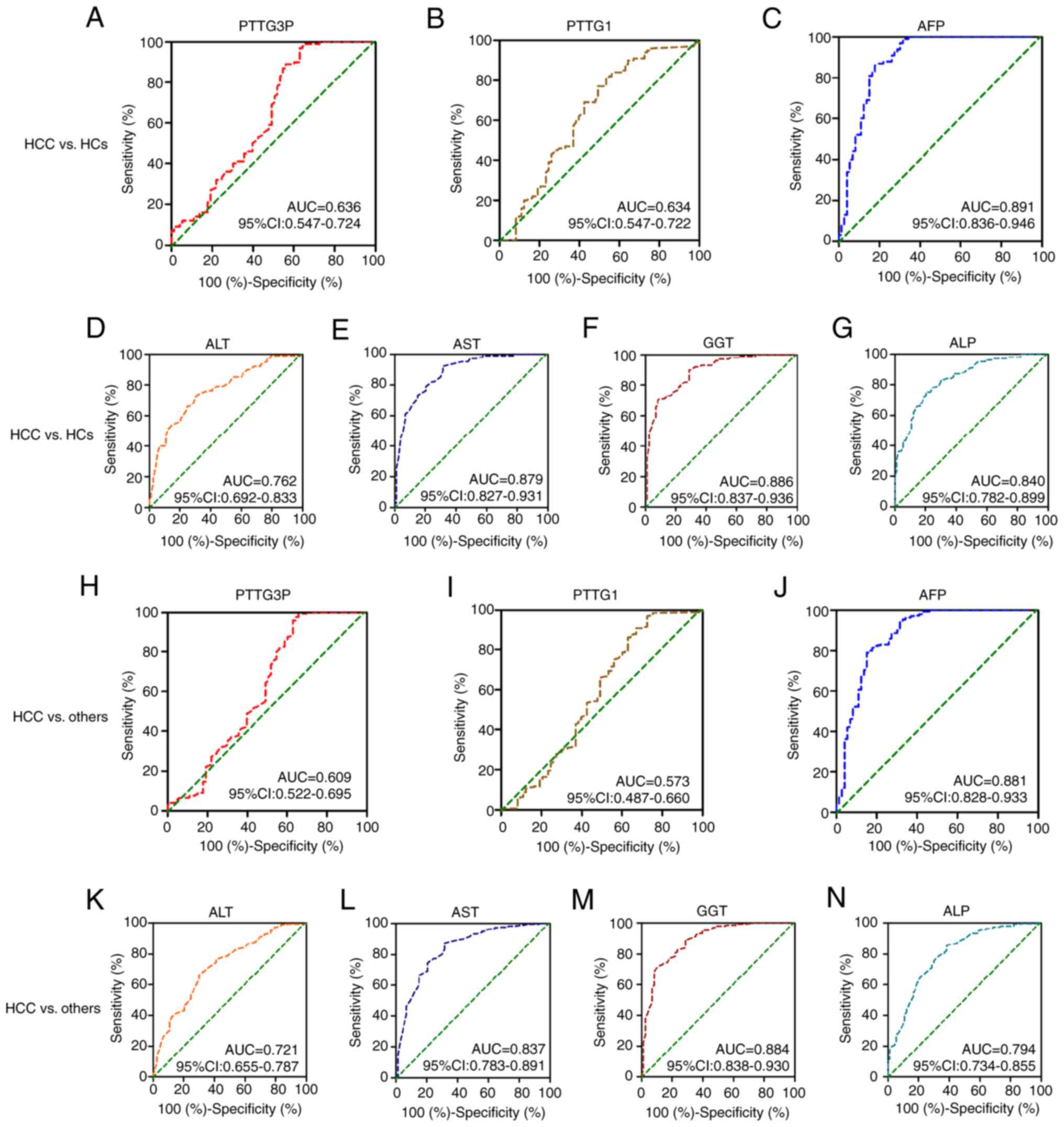 | Figure 3.Diagnostic value of serum lncRNA
PTTG3P and mRNA PTTG1 in patients with HCC. The diagnostic value of
(A) PTTG3P, (B) PTTG1, (C) AFP, (D) ALT, (E) AST, (F) GGT and (G)
ALP was estimated by receiver operating characteristic curve
analysis when comparing patients with HCC with HCs. Diagnostic
efficacy of (H) PTTG3P, (I) PTTG1, (J) AFP, (K) ALT, (L) AST, (M)
GGT and (N) ALP to discriminate patients with HCC from those with
liver cirrhosis and chronic hepatitis B, and HCs. lncRNA, long
noncoding RNA; HCC, hepatocellular carcinoma; HC, healthy control;
AFP, α-fetoprotein; GGT, γ-glutamyl transpeptidase; AST, aspartate
transaminase; ALT, alanine aminotransferase; ALP, alkaline
phosphatase; AUC, area under the curve. |
 | Table V.Performance of AFP, ALT, AST, GGT,
ALP, PTTG3P, PTTG1 and the combinations in HCs and patients with
LC, CHB, and HCC. |
Table V.
Performance of AFP, ALT, AST, GGT,
ALP, PTTG3P, PTTG1 and the combinations in HCs and patients with
LC, CHB, and HCC.
|
| HCC vs. HCs | HCC vs.
othersa |
|---|
|
|
|
|
|---|
| Method | AUC (95% CI) | SEN, % | SPE, % | P-value | AUC (95% CI) | SEN, % | SPE, % | P-value |
|---|
| AFP | 0.891
(0.836–0.946) | 0.822 | 0.860 |
<0.001b | 0.881
(0.828–0.933) | 0.849 | 0.790 |
<0.001b |
| ALT | 0.762
(0.692–0.833) | 0.699 | 0.720 |
<0.001b | 0.721
(0.655–0.787) | 0.699 | 0.663 |
<0.001b |
| AST | 0.879
(0.827–0.931) | 0.685 | 0.920 |
<0.001b | 0.837
(0.783–0.891) | 0.685 | 0.873 |
<0.001b |
| GGT | 0.886
(0.837–0.936) | 0.918 | 0.700 |
<0.001b | 0.884
(0.838–0.930) | 0.904 | 0.707 |
<0.001b |
| ALP | 0.840
(0.782–0.899) | 0.795 | 0.750 |
<0.001b | 0.794
(0.734–0.855) | 0.661 | 0.853 |
<0.001b |
| PTTG3P | 0.636
(0.547–0.724) | 0.370 | 0.970 | 0.003b | 0.609
(0.522–0.695) | 0.356 | 0.973 | 0.004b |
| PTTG1 | 0.634
(0.547–0.722) | 0.507 | 0.770 | 0.030b | 0.573
(0.487–0.660) | 0.274 | 0.963 | 0.052 |
| AFP + PTTG3P | 0.908
(0.856–0.960) | 0.795 | 0.970 |
<0.001b | 0.861
(0.795–0.927) | 0.767 | 0.973 |
<0.001b |
| AFP + PTTG1 | 0.907
(0.856–0.957) | 0.822 | 0.900 |
<0.001b | 0.853
(0.790–0.916) | 0.726 | 0.933 |
<0.001b |
| PTTG1 + PTTG3P | 0.748
(0.671–0.824) | 0.452 | 0.990 |
<0.001b | 0.693
(0.612–0.774) | 0.452 | 0.983 |
<0.001b |
| AFP + PTTG3P +
PTTG1 | 0.927
(0.881–0.973) | 0.822 | 0.960 |
<0.001b | 0.884
(0.825–0.943) | 0.781 | 0.980 |
<0.001b |
| AFP + ALT + AST +
GGT +ALP + PTTG3P + PTTG1 | 0.959
(0.925–0.993) | 0.904 | 0.980 |
<0.001b | 0.948
(0.911–0.985) | 0.890 | 0.950 |
<0.001b |
Moreover, whether lncRNA PTTG3P and mRNA PTTG1
expression could distinguish patients with HCC from others (CHB, LC
and HCs) was investigated. The ROC curve analysis results indicated
that AFP, PTTG3P, ALT, AST, GGT and ALP exhibited diagnostic
efficacy for distinguishing patients with HCC from others (CHB, LC,
and HCs) (Table V; Fig. 3H and J-N). However, mRNA PTTG1
[AUC: 0.573; 95% CI: 0.487–0.660] showed no diagnostic efficacy for
discriminating patients with HCC from others (CHB, LC, and HCs)
(P>0.05; Table V; Fig. 3I). The diagnostic efficacy of
lncRNA PTTG3P displayed 97.3% specificity and 35.6% sensitivity
(Table V; Fig. 3H).
AFP is the most commonly used biomarker for HCC
screening and diagnosis in the clinic. Additionally, ALT, AST, GGT
and ALP have been utilized as biomarkers for HCC. Therefore, the
combinations of lncRNA PTTG3P and mRNA PTTG1 with the
aforementioned five biomarkers were estimated for HCC diagnosis.
Table V shows the diagnostic
efficacy parameters for the combinations of lncRNA PTTG3P, mRNA
PTTG1, and AFP, ALT, AST, GGT and ALP to distinguish patients with
HCC from HCs and from others (CHB, LC, and HCs). For the
discrimination of patients with HCC from HCs, compared with AFP
alone, the AUC values of the combinations of lncRNA PTTG3P and mRNA
PTTG1 with AFP to predict HCC were elevated (Table V; Fig.
4A, B and D). Moreover, the combination of lncRNA PTTG3P, mRNA
PTTG1, AFP, ALT, AST, GGT and ALP presented the highest accuracy,
with an AUC of 0.959 (95% CI: 0.925–0.993), 90.4% sensitivity and
98.0% specificity (Table V;
Fig. 4E). Furthermore, combining
lncRNA PTTG3P and mRNA PTTG1 showed a higher AUC than that of
lncRNA PTT3P or mRNA PTTG1 alone (Table V; Fig.
4C and H). However, the AUC values of lncRNA PTTG3P and AFP, or
mRNA PTTG1 and AFP were lower than that of AFP alone when used to
distinguish patients with HCC from others (CHB, LC and HCs)
(Table V; Fig. 4F and G).
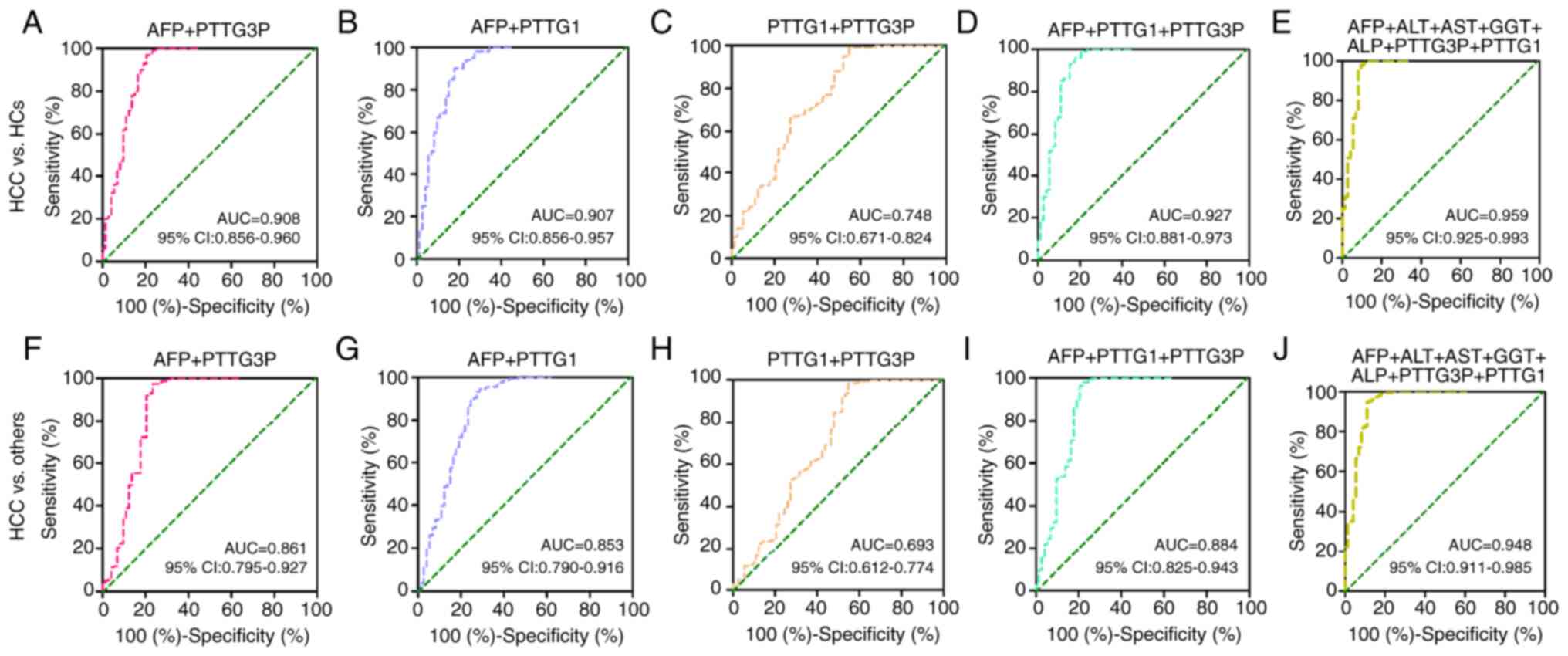 | Figure 4.Diagnostic efficacy of the
combinations of lncRNA PTTG3P and mRNA PTTG1 with other biomarkers
in patients with HCC. The diagnostic value of the combinations of
(A) PTTG3P and AFP, (B) PTTG1 and AFP, (C) PTTG1 and PTTG3P, (D)
PTTG3P, PTTG1 and AFP, and (E) PTTG3P, PTTG1, AFP, ALT, AST, GGT
and ALP were estimated by receiver operating characteristic curve
analysis when comparing patients with HCC and HCs. Diagnostic
efficacy of the combinations of (F) PTTG3P and AFP, (G) PTTG1 and
AFP, (H) PTTG1 and PTTG3P, (I) PTTG3P, PTTG1 and AFP, and (J)
PTTG3P, PTTG1, AFP, ALT, AST, GGT and ALP to discriminate patients
with HCC from patients with liver cirrhosis and chronic hepatitis
B, and HCs. lncRNA, long noncoding RNA; HCC, hepatocellular
carcinoma; HC, healthy control; AFP, α-fetoprotein; GGT, γ-glutamyl
transpeptidase; AST, aspartate transaminase; ALT, alanine
aminotransferase; ALP, alkaline phosphatase; AUC, area under the
curve. |
Discussion
HCC is one of the most aggressive tumors worldwide
and it is associated with a poor prognosis; therefore, the
identification of novel potential serum biomarkers for detecting
early-stage HCC remains a challenge. Research has revealed that
lncRNAs are stably present in serum or plasma, and can be detected
in patients with different types of cancer (15). Notably, several HCC-related lncRNAs
have been detected in the serum of patients with HCC, including
lncRNA HULC, HEIH, SCARNA10 and lnc34a (34–37).
Taken together, these findings suggested that serum lncRNAs may be
potential noninvasive biomarkers for HCC diagnosis and
prognosis.
Our previous study indicated that the interaction
between lncRNA PTTG3P and mRNA PTTG1 served a vital role in the
occurrence and development of HCC (24). A schematic model of the functions
of lncRNA PTTG3P during HCC tumor growth and metastasis is shown in
Fig. S2. PTTG3P may enhance
cellular proliferation and metastatic capabilities by increasing
the expression of PTTG1 and stimulating the PI3K/AKT signaling
pathway. Subsequently, this modulation could affect its downstream
signaling cascades, encompassing various cell-cycle regulators and
epithelial-mesenchymal transition-associated factors in HCC. In the
present study, the diagnostic efficacy of serum lncRNA PTTG3P, mRNA
PTTG1 and their combinations for the diagnosis of HCC was
evaluated. The results indicated that the expression levels of
lncRNA PTTG3P and mRNA PTTG1 were markedly elevated in patients
with HCC and CHB compared with those in HCs. In addition, the
preoperative and postoperative expression levels of lncRNA PTTG3P
and mRNA PTTG1 were compared in 36 patients with HCC. The data
showed that the postoperative expression levels of lncRNA PTTG3P
and mRNA PTTG1 were significantly lower than the preoperative
levels, indicating that they may be applied as promising prognostic
markers for HCC. lncRNA PTTG3P is a processed pseudogene, which has
been confirmed to serve a key role in the development and
progression of cancer. Liu et al (38) reported that the lncRNA PTTG3P
levels were markedly elevated in colorectal cancer (CRC), and were
closely related to the incidence of lymph node metastasis and
distant metastasis in patients with CRC. Furthermore, lncRNA PTTG3P
could downregulate the expression of microRNA-155-5P to promote the
invasion and migration of CRC. Huang et al (23) demonstrated that PTTG3P was
associated with non-small cell lung cancer (NSCLC) cell
proliferation; these previous results suggested that PTTG3P could
serve as a new therapeutic and prognostic target for NSCLC. In
addition, previous studies have regarded mRNA PTTG1 as an oncogenic
factor in various types of cancer, such as pancreatic
adenocarcinoma and lung adenocarcinoma (39,40).
A long follow-up of patients could provide valuable information on
disease progression and recurrence; however, the present study
recruited patients between July 2022 and March 2023. Furthermore,
at the time of writing, 90% of patients recruited were alive, which
is not suitable for survival analysis. We aim to follow up the
patients recruited between July 2022 and March 2023 to provide
valuable information on disease progression and recurrence in a
further study.
To investigate the prognostic impact of PTTG3P and
PTTG1 on HCC, the present study used TCGA database. Data from TCGA
program showed that low expression of PTTG1 indicated improved
prognosis compared with high expression of PTTG1; thus, high
expression of PTTG1 appears to be a risk factor for HCC prognosis.
The interaction between lncRNA PTTG3P and mRNA PTTG1 crucially
participates in the development of cancer. The specific mechanisms
underlying the interaction between lncRNA PTTG3P and mRNA PTTG1 in
cancer have been reported in previous studies. Huang et al
(41) suggested that PTTG3P can
promote the resistance of prostate cancer cells to
androgen-deprivation therapy via upregulating PTTG1. Guo et
al (42) indicated that PTTG3P
may promote the growth and metastasis of cervical cancer through
PTTG1. Zhang and Shi (43) showed
that PTTG3P is distinctly upregulated and may serve an oncogenic
role in a PTTG1 and PTTG2-mediated manner in esophageal squamous
cell carcinoma. Moreover, Huang et al (25) reported that the levels of serum
PTTG3P were significantly higher in patients with HCC than in
patients with benign liver diseases and HCs, which is in accordance
with the present data. Overall, the aforementioned studies further
confirm the present results that serum lncRNA PTTG3P and mRNA PTTG1
may have potential as novel biomarkers for HCC.
Subsequently, the associations between the serum
levels of lncRNA PTTG3P and clinical parameters of patients with
HCC were investigated in the present study. The results showed that
serum lncRNA PTTG3P expression was slightly associated with AST.
AST is an enzyme mainly used to evaluate hepatic outcomes. Previous
research has revealed that high levels of AST may be indicative of
liver damage (44); therefore,
monitoring the levels of AST may be an appropriate way to evaluate
liver function (45). Previous
studies have reported that patients with HCC exhibiting high AST
levels show poorer survival (46,47).
Additionally, the current data indicated that patients with HCC
aged >55 years exhibited elevated expression levels of lncRNA
PTTG3P, whereas younger patients tended to have lower expression
levels of lncRNA PTTG3P, thus suggesting that there may be a
certain association between lncRNA PTTG3P levels and the age of
onset of HCC. However, there were no associations detected between
serum lncRNA PTTG3P expression and sex, HBsAg, AFP level,
cirrhosis, tumor size, tumor number, Edmondson grade, lymph node
metastasis, portal vein invasion, ALT, GGT, ALB, ALP, TP, TBIL or
DBIL, which was consistent with the results of Huang et al
(25). The aforementioned negative
results may be due to the limited sample size of the present study.
Furthermore, the clinical features of 36 patients with HCC before
and after surgery were explored in the present study, and a
significantly decreased AFP level was observed in patients
post-operation. AFP is the most frequently used marker for the
screening and diagnosis of HCC in clinical practice (48,49).
Tian et al (50) reported
that the concentration of AFP in the HCC group was significantly
higher than that in the other groups, while the levels of AFP after
liver cancer surgery were significantly lower than those before
surgery, which is in agreement with the present data. Accordingly,
it may be concluded that increased serum expression levels of
lncRNA PTTG3P could serve as an unfavorable prognostic factor for
HCC.
To explore the diagnostic efficacy of lncRNA PTTG3P
and mRNA PTTG1, ROC curve analysis was applied. The present data
indicated that both serum lncRNA PTTG3P and mRNA PTTG1 exhibited
good diagnostic accuracy in distinguishing patients with HCC from
HCs. The diagnostic value of PTTG3P exhibited 37.0% sensitivity and
97.0% specificity, whereas that of mRNA PTTG1 showed 50.7%
sensitivity and 97.0% specificity. Moreover, ROC curve analysis
showed that AFP, ALT, AST, GGT, ALP and PTTG3P had predictive value
for discriminating patients with HCC from others (CHB, LC, and
HCs). However, mRNA PTTG1 showed no diagnostic efficacy for
distinguishing patients with HCC from others (CHB, LC and HCs).
In recent decades, the early diagnosis of HCC has
relied on surveillance with serological assessments of AFP;
however, the specificity and sensitivity of AFP is not sufficiently
satisfactory to detect early-onset HCC (51). Thus, the combinations of lncRNA
PTTG3P and mRNA PTTG1 with AFP for HCC diagnosis were estimated in
the present study. The data indicated that the AUC values of the
combinations were greater than that of AFP alone for discriminating
HCC from HC. Moreover, ALT, AST, GGT and ALP have also been
recognized as prognostic biomarkers for HCC (52–54).
In the present study, the diagnostic efficacy of the combination of
the aforementioned biomarkers with lncRNA PTTG3P and mRNA PTTG1 was
investigated. The combination of lncRNA PTTG3P and mRNA PTTG1 with
AFP, ALT, AST, GGT and ALP exhibited the best diagnostic efficacy,
yielding an AUC of 0.959 with 90.4% sensitivity and 98.0%
specificity. The present results are consistent with the literature
(25). Thus, the current study
demonstrated that serum lncRNA PTTG3P and mRNA PTTG1 could be
potential biomarkers for the prognosis and diagnosis of HCC.
Notably, there are certain limitations in the
present study. Firstly, the present study was conducted with a
limited sample size, since it is a single-center study. Secondly,
no validation cohort was included in the study. Finally, the
present study lacks follow-up data. Therefore, large-scale
multicenter studies and further research are recommended to
evaluate the potential of serum lncRNA PTTG3P and mRNA PTTG1 as
novel biomarkers for the diagnosis and prognosis of HCC.
In conclusion, the current study demonstrated that
the combinations of serum lncRNA PTTG3P, mRNA PTTG1, AFP, ALT, AST,
GGT and ALP exhibited a good performance for the diagnosis of HCC.
Moreover, the postoperative expression levels of lncRNA PTTG3P and
mRNA PTTG1 were significantly lower than the preoperative levels in
36 paired patients with HCC. Thus, it was concluded that serum
lncRNA PTTG3P and mRNA PTTG1 may be novel and noninvasive
biomarkers for the diagnosis and prognosis of HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Scientific Research Project of
Traditional Chinese Medicine Bureau of Guangdong Province (grant
no. 20202067), the Guangzhou Basic and Applied Basic Research
Project (grant no. 202102020101), the Natural Science Foundation of
Guangdong Province (grant no. 2021A1515220163), the Special Project
of Dampness Syndrome of Traditional Chinese Medicine Bureau of
Guangdong Province (grant no. 20225005), the Guangdong Province
Basic and Applied Basic Research Fund Project (grant no.
2023A1515220084) and the Guangdong Provincial Key Laboratory of
Research on Emergency in TCM (grant no. 2023B1212060062).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SWC conceived and proposed the idea. SWC and PFK
designed the work. FZ, XYC and SKK contributed to acquisition,
analysis and interpretation of data. XRZ, TTL, YHS and CMK
contributed to collection of clinical samples and interpretation of
data. SQ, HMW, YW and SZL contributed to the interpretation of
data, and drafted and revised the manuscript. SWC and FZ confirm
the authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was performed according to the
principles of The Declaration of Helsinki. Each subject provided
written informed consent and the research protocol was approved by
the Ethics Committee of the Second Affiliated Hospital of Guangzhou
University of Chinese Medicine (Guangzhou, China; approval no.
BE2020-211-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singal A, Lampertico P and Nahon P:
Epidemiology and surveillance for hepatocellular carcinoma: New
trends. J Hepatol. 72:250–261. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lazzaro A and Hartshorn KL: A
comprehensive narrative review on the history, current landscape,
and future directions of hepatocellular carcinoma (HCC) systemic
therapy. Cancers (Basel). 15:25062023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schlosser S, Tümen D, Volz B, Neumeyer K,
Egler N, Kunst C, Tews HC, Schmid S, Kandulski A, Müller M and
Gülow K: HCC biomarkers-state of the old and outlook to future
promising biomarkers and their potential in everyday clinical
practice. Front Oncol. 12:10169522022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Omar MA, Omran MM, Farid K, Tabll AA,
Shahein YE, Emran TM, Petrovic A, Lucic NR, Smolic R, Kovac T and
Smolic M: Biomarkers for hepatocellular carcinoma: From origin to
clinical diagnosis. Biomedicines. 11:18522023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bertino G, Ardiri A, Malaguarnera M,
Malaguarnera G, Bertino N and Calvagno GS: Hepatocellualar
carcinoma serum markers. Seminars Oncol. 39:410–433. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanif H, Ali MJ, Susheela AT, Khan IW,
Luna-Cuadros MA, Khan MM and Lau DT: Update on the applications and
limitations of alpha-fetoprotein for hepatocellular carcinoma.
World J Gastroenterol. 28:216–229. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu SJ, Dang HX, Lim DA, Feng FY and Maher
CA: Long noncoding RNAs in cancer metastasis. Nat Rev Cancer.
21:446–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Verma S, Sahu BD and Mugale MN: Role of
lncRNAs in hepatocellular carcinoma. Life Sci. 325:1217512023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ahmad M, Weiswald L, Poulain L, Denoyelle
C and Meryet-Figuiere M: Involvement of lncRNAs in cancer cells
migration, invasion and metastasis: cytoskeleton and ECM crosstalk.
J Exp Clin Cancer Res. 42:1732023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ge WJ, Huang H, Wang T, Zeng WH, Guo M,
Ren CR, Fan TY, Liu F and Zeng X: Long non-coding RNAs in
hepatocellular carcinoma. Pathol Res Pract. 248:1546042023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei H, Xu Z, Chen L, Wei Q, Huang Z, Liu
G, Li W, Wang J, Tang Q and Pu J: Long non-coding RNA PAARH
promotes hepatocellular carcinoma progression and angiogenesis via
upregulating HOTTIP and activating HIF-1α/VEGF signaling. Cell
Death Dis. 13:1022022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang B, Yang B, Wang Q, Zheng X, Guo Y
and Lu W: lncRNA PVT1 promotes hepatitis B virus-positive liver
cancer progression by disturbing histone methylation on the c-Myc
promoter. Oncol Rep. 43:718–726. 2020.PubMed/NCBI
|
|
14
|
Wang BR, Chu DX, Cheng MY, Jin Y, Luo HG
and Li N: Progress of HOTAIR-microRNA in hepatocellular carcinoma.
Hered Cancer Clin Pract. 20:42022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen T: Circulating non-coding RNAs as
potential diagnostic biomarkers in hepatocellular carcinoma. J
Hepatocell Carcinoma. 9:1029–1040. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng Y, Huang X and Wang H: Serum lncRNA
LINC01535 as biomarker of diagnosis, prognosis, and disease
progression in breast cancer. Clin Breast Cancer. 23:620–627. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bao H, Jiang Y, Wang N, Su H and Han X:
Long noncoding RNAs MALAT1 and HOTTIP Act as serum biomarkers for
hepatocellular carcinoma. Cancer Control. 31:107327482412848212024.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao SW, Huang JL, Chen J, Hu YW, Hu XM,
Ren TY, Zheng SH, Lin JD, Tang J, Zheng L and Wang Q: Long
non-coding RNA UBE2CP3 promotes tumor metastasis by inducing
epithelial-mesenchymal transition in hepatocellular carcinoma.
Oncotarget. 8:65370–65385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tripathi SK, Pal A, Ghosh S, Goel A,
Aggarwal R, Banerjee S and Das S: LncRNA NEAT1 regulates
HCV-induced hepatocellular carcinoma by modulating the miR-9-BGH3
axis. J Gen Virol. 103:2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng Y, Wang Y, Liu Y, Xie L, Ge J, Yu G
and Zhao G: N6-methyladenosine modification of PTTG3P contributes
to colorectal cancer proliferation via YAP1. Front Oncol.
11:6697312021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu W, Tang J, Zhang H, Kong F, Zhu H, Li
P, Li Z, Kong X and Wang K: A novel lncRNA
PTTG3P/miR-132/212-3p/FoxM1 feedback loop facilitates tumorigenesis
and metastasis of pancreatic cancer. Cell Death Discov. 6:1362020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Y, Wang D, Chen G, Shan Z and Li D:
Exploring the molecular landscape of osteosarcoma through PTTG
family genes using a detailed multi-level methodology. Front Genet.
15:14316682024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang HT, Xu YM, Ding SG, Yu XQ, Wang F,
Wang HF, Tian X and Zhong CJ: The novel lncRNA PTTG3P is
downregulated and predicts poor prognosis in non-small cell lung
cancer. Arch Med Sci. 16:931–940. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang JL, Cao SW, Ou QS, Yang B, Zheng SH,
Tang J, Chen J, Hu YW, Zheng L and Wang Q: The long non-coding RNA
PTTG3P promotes cell growth and metastasis via up-regulating PTTG1
and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol
Cancer. 17:932018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang J, Zheng Y, Xiao X, Liu C, Lin J,
Zheng S, Yang B and Ou Q: A circulating long noncoding RNA panel
serves as a diagnostic marker for hepatocellular carcinoma. Dis
Markers. 2020:54175982020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tien S, Zhou H, Zhou Q, Liu H, Wu B and
Guo Y: PTTG1 alleviates acute alcoholic liver injury by inhibiting
endoplasmic reticulum stress-induced hepatocyte pyroptosis. Liver
Int. 43:840–854. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heathcote EJ: Management of primary
biliary cirrhosis. The American association for the study of liver
diseases practice guidelines. Hepatology. 31:1005–1013. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
European Association for the Study of the
Liver. Electronic address, . simpleeasloffice@easloffice.eu;
European Association for the Study of the Liver: EASL 2017 clinical
practice guidelines on the management of hepatitis B virus
infection. J Hepatol. 67:370–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kolde RJR: Pheatmap: Pretty heatmaps. R
package version. 61:9152012.
|
|
31
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Z, Lin E, Zhuang H, Xie L, Feng X,
Liu J and Yu Y: Construction of a novel gene-based model for
prognosis prediction of clear cell renal cell carcinoma. Cancer
Cell Int. 20:272020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gaber DA, Shaker O, Younis AT and
El-Kassas M: LncRNA HULC and miR-122 expression pattern in
HCC-related HCV Egyptian patients. Genes (Basel). 13:16692022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ismail M, Fadul MM, Taha R, Siddig O,
Elhafiz M, Yousef BA, Jiang Z, Zhang L and Sun L: Dynamic role of
exosomal long non-coding RNA in liver diseases: Pathogenesis and
diagnostic aspects. Hepatol Int. Sep 21–2024.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han Y, Jiang W, Wang Y, Zhao M, Li Y and
Ren L: Serum long non-coding RNA SCARNA10 serves as a potential
diagnostic biomarker for hepatocellular carcinoma. BMC Cancer.
22:4312022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Niu H, Yang P, Ma J, Yuan BY,
Zeng ZC and Xiang ZL: Serum lnc34a is a potential prediction
biomarker for bone metastasis in hepatocellular carcinoma patients.
BMC Cancer. 21:1612021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu N, Dou L and Zhang X: LncRNA PTTG3P
sponge absorbs microRNA-155-5P to promote metastasis of colorectal
cancer. Onco Targets Ther. 13:5283–5291. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Long L, Gao J and Zhang R: PTTG1 enhances
oncolytic adenovirus 5 entry into pancreatic adenocarcinoma cells
by increasing CXADR expression. Viruses. 15:11532023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Z, Cao K, Hou Y, Lu F, Li L, Wang L,
Xia Y, Zhang L, Chen H, Li R, et al: PTTG1 knockdown enhances
radiation-induced antitumour immunity in lung adenocarcinoma. Life
Sci. 277:1195942021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang S, Liao Q, Li W, Deng G, Jia M, Fang
Q, Ji H and Meng M: The lncRNA PTTG3P promotes the progression of
CRPC via upregulating PTTG1. Bull Cancer. 108:359–368. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo XC, Li L, Gao ZH, Zhou HW, Li J and
Wang QQ: The long non-coding RNA PTTG3P promotes growth and
metastasis of cervical cancer through PTTG1. Aging (Albany NY).
11:1333–1341. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Z and Shi Z: The pseudogene PTTG3P
promotes cell migration and invasion in esophageal squamous cell
carcinoma. Open Med (Wars). 14:516–522. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Du S, Zhang X, Jia Y, Peng P, Kong Q,
Jiang S, Li Y, Li C, Ding Z and Liu L: Hepatocyte HSPA12A inhibits
macrophage chemotaxis and activation to attenuate liver
ischemia/reperfusion injury via suppressing glycolysis-mediated
HMGB1 lactylation and secretion of hepatocytes. Theranostics.
13:3856–3871. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Maqsood Q, Sumrin A, Iqbal M, Younas S,
Hussain N, Mahnoor M and Wajid A: Hepatitis C virus/hepatitis B
virus coinfection: Current prospectives. Antivir Ther.
28:135965352311896432023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Casadei-Gardini A, Rimini M, Kudo M,
Shimose S, Tada T, Suda G, Goh MJ, Jefremow A, Scartozzi M, Cabibbo
G, et al: Real life study of lenvatinib therapy for hepatocellular
carcinoma: RELEVANT study. Liver Cancer. 11:527–539. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kaewdech A, Sripongpun P, Assawasuwannakit
S, Wetwittayakhlang P, Jandee S, Chamroonkul N and Piratvisuth T:
FAIL-T (AFP, AST, tumor sIze, ALT, and tumor number): A model to
predict intermediate-stage HCC patients who are not good candidates
for TACE. Front Med (Lausanne). 10:10778422023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Johnson P, Zhou Q, Dao DY and Lo YMD:
Circulating biomarkers in the diagnosis and management of
hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol.
19:670–681. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hu X, Chen R, Wei Q and Xu X: The
landscape of alpha fetoprotein in hepatocellular carcinoma: Where
are we? Int J Biol Sci. 18:536–551. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tian S, Chen Y, Zhang Y and Xu X: Clinical
value of serum AFP and PIVKA-II for diagnosis, treatment and
prognosis of hepatocellular carcinoma. J Clin Lab Anal.
37:e248232023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang W and Wei C: Advances in the early
diagnosis of hepatocellular carcinoma. Genes Dis. 7:308–319. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lei K, Deng Z, Wang J, Wang H, Hu R, Li Y,
Wang X, Xu J, You K and Liu Z: A novel nomogram based on the
hematological prognosis risk scoring system can predict the overall
survival of patients with hepatocellular carcinoma. J Cancer Res
Clin Oncol. 149:14631–14640. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao Z, Zhu Y, Ni X, Lin J, Li H, Zheng L,
Zhang C, Qi X, Huo H, Lou X, et al: Serum GGT/ALT ratio predicts
vascular invasion in HBV-related HCC. Cancer Cell Int. 21:5172021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang CW, Wu TH, Hsu HY, Pan KT, Lee CW,
Chong SW, Huang SF, Lin SE, Yu MC and Chen SM: Reappraisal of the
role of alkaline phosphatase in hepatocellular carcinoma. J Pers
Med. 12:5182022. View Article : Google Scholar : PubMed/NCBI
|