Introduction
Epidermal growth factor receptor (EGFR) is a 170-kDa
transmembrane glycoprotein receptor that exhibits tyrosine kinase
activity, which regulates cell growth. EGFR expression was
frequently observed in squamous cell carcinomas of the head and
neck (SCCHN) (1). Since EGFR
signaling was found to control not only cell growth, but also
angiogenesis and DNA repair (2,3), it
may be a molecular target in patients with SCCHN (4,5).
Gefitinib (ZD1839, Iressa®), an
anilinoquinazoline-based inhibitor specific for EGFR tyrosine
kinase, was shown to have clinical efficacy in patients with
non-small cell lung carcinoma (NSCLC) (1). Since mutations in the tyrosine kinase
domain of the EGFR gene were reported to increase the
binding affinity of gefitinib, the presence of EGFR mutations in
this domain is predictive of the clinical benefits of gefitinib
treatment for NSCLC patients (6,7).
Gefitinib has also shown clinical efficacy in some patients without
EGFR mutations (4,5). Thus, in such patients, EGFR
gene amplifications are critical for gefitinib efficacy (6,7).
Previous studies have shown that mutations in the kinase domain of
the EGFR gene occur less frequently in SCCHN than in NSCLC
(1,4). The clinical efficacy of gefitinib is
due not only to its direct action on EGFR tyrosine kinase, but on
its indirect action through activation of the immune system
(9). For example, to demonstrate
the direct action of EGFR tyrosine kinase, it was shown that SCCHN
cell lines with heterozygous mutations (G/G→G/A) at nucleotide 2607
of EGFR are more sensitive to gefitinib than wild-type
EGFR (4). To further
investigate these observations, the mechanism by which the G/A
mutation confers sensitivity to gefitinib was examined.
Materials and methods
Cell lines and cell culture
The 16 human SCCHN cell lines and their sites of
origin included YCU-M862, YCU-M911 and KCC-M871, from tumors of the
mesopharynx; YCU-H891, from a tumor of the hypopharynx; KCC-L871
and YCU-L891, from tumors of the larynx; KCC-T871, YCU-T891,
YCU-T892 and KCC-T873, from tumors of the tongue; KCC-TCM902,
KCC-TCM903 and KCC-TCM901, from metastatic tumors of 3 tongue
carcinomas; KCC-OR891, from tumors of the oral floor; and KCC-MS871
and YCU-MS861, from tumors of the maxillary sinus. The cell lines
were maintained in RPMI-1640 supplemented with 10% fetal bovine
serum and cultured at 37°C in a humidified atmosphere of 5%
CO2/95% air.
Fluorescence in situ hybridization
(FISH)
The cells were fixed in carunor solution, placed on
glass slides and hybridized with LSI EGFR Dual Color Probe (Abbott
Molecular, CA, USA) (1,2). The signals were counted for 60 cells
in each cell line and data were expressed as mean ± SD.
mRNA half life
Each cell line was treated with 5 μg/ml actinomycin
D (Wako Pure Chemical Industries, Osaka, Japan) and total RNA was
purified using Isogen RNA purification kit (Nippon Gene, Tokyo,
Japan). Total RNA was then reverse-transcribed to cDNA using avian
myeloblastosis virus (AMV) reverse transcriptase (Takara, Tokyo,
Japan) and amplified by real-time quantitative PCR
(Stratagene® MX3000P; Agilent Technologies, Santa Clara,
CA, USA) with Stratagene Brilliant II Fast SYBR® Green
QPCR Master Mix (Agilent Technologies). The specific primer sets
are shown in Table I. The
amplification protocol consisted of 30 cycles of denaturation at
95°C for 30 sec, annealing at 55°C for 1 min and extension at 72°C
for 30 sec.
 | Table IQPCR primer sets. |
Table I
QPCR primer sets.
| Genes | Sequence (5′→3′) |
|---|
| EGFR | Forward:
TTCGATGATCAACTCACGGAAC |
| Reverse:
GCCACCCATATGTACCATCGAT |
| β-actin | Forward:
CAGCTGAGAGGGAAATCGTG |
| Reverse:
GCTGGTTGCCAATAGTGAATG |
Statistical analysis
The groups were compared using paired Student’s
t-tests and Chi-square tests. Linear regression analysis was used
to compare the EGFR mRNA half life with the EGFR gene
copy number, the steady state levels of EGFR mRNA and the
EGFR protein levels. P<0.05 was considered to be statistically
significant.
Results
EGFR copy number
The 16 SCCHN cell lines were divided into two
groups: those heterozygous for the mutation (G/A) at nucleotide
2607 of the EGFR gene (9 cell lines) and those with the
wild-type (G/G) sequence (7 cell lines). The mean number of
EGFR copies in the cell lines was 3.59±2.14 and 1.58±0.32,
respectively (Fig. 1, Table II) (4), or 2.27-fold higher in cell lines with
the mutant sequence. It was found that 10 of the 16 cell lines had
≥2 copies of the EGFR gene, whereas the other 6 had <2
copies. The Chi-square test showed a statistically significant
correlation between the G/A mutation and EGFR copy number
(Table III). The EGFR copy
number was significantly higher in YCU-OR891 than in the remaining
cell lines.
 | Table IIA summary of EGFR protein, mRNA,
gefitinib IC50 value, SNP, FISH copy number ratio and
mRNA half life. |
Table II
A summary of EGFR protein, mRNA,
gefitinib IC50 value, SNP, FISH copy number ratio and
mRNA half life.
| Cell line | EGFR proteina | EGFR mRNA | IC50
valuea | Genotype at
2607b | FISH X/Y | mRNA half life
(h) |
|---|
| YCU-MS861 | 0.15 | 0.20 | 66.3 | G/G | 1.13 | 4.3 |
| YCU-M862 | 1.85 | 0.59 | 64.8 | G/G | 1.33 | 5.4 |
| KCC-M871 | 0.46 | 0.10 | 52.1 | G/G | 1.47 | 3.9 |
| KCC-L871 | 0.58 | 0.04 | 58.0 | G/G | 1.54 | 5.6 |
| YCU-T892 | 2.25 | 4.43 | 64.8 | G/G | 1.68 | 3.0 |
| YCU-M911 | 0.95 | 0.55 | 61.4 | G/G | 1.82 | 5.2 |
| KCC-T873 | 2.96 | 0.85 | 85.0 | G/G | 2.12 | 5.6 |
| KCC-TCM903 | 0.38 | 0.25 | 51.4 | G/A | 2.02 | 6.0 |
| KCC-T871 | 0.60 | 0.09 | 44.4 | G/A | 2.55 | 6.0 |
| KCC-TCM901 | 0.77 | 0.25 | 43.9 | G/A | 2.80 | 5.8 |
| YCU-H891 | 0.21 | 2.38 | 69.0 | G/A | 2.89 | 13.0 |
| YCU-L891 | 0.91 | 1.14 | 52.2 | G/A | 2.89 | 8.5 |
| YCU-T891 | 0.56 | 2.46 | 56.0 | G/A | 2.89 | 18.0 |
| KCC-MS871 | 0.23 | 1.35 | 41.6 | G/A | 3.53 | 15.0 |
| KCC-TCM902 | 0.25 | 1.16 | 50.2 | G/A | 3.60 | 11.2 |
| YCU-OR891 | 0.47 | 0.16 | 53.6 | G/A | 9.16 | 8.5 |
| Average | 0.85 | 1.00 | 57.2 | | 2.71 | 7.8 |
 | Table IIIHeterozygous EGFR mutants contain a
high copy number of EGFR. |
Table III
Heterozygous EGFR mutants contain a
high copy number of EGFR.
Association between EGFR copy number and
mRNA half life
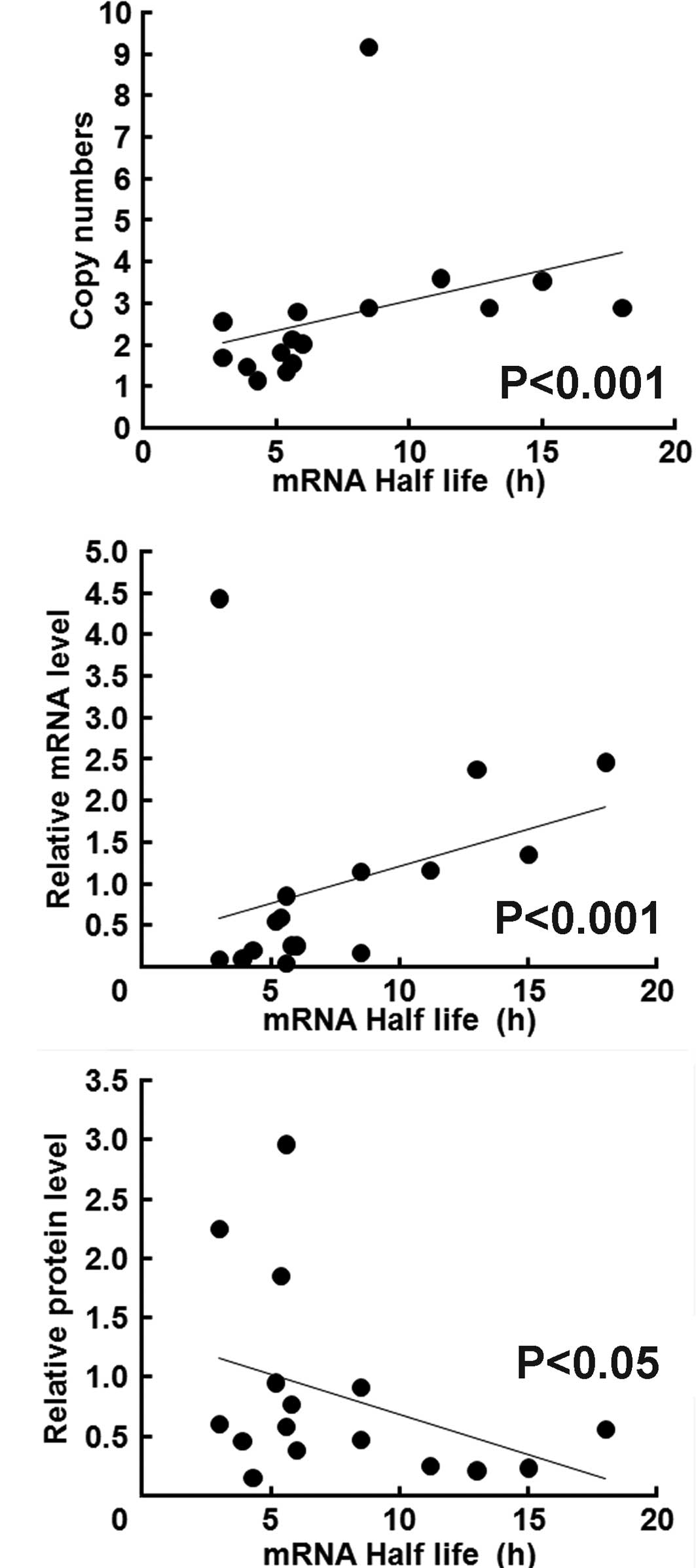
Real-time qPCR analysis revealed that the
EGFR gene copy number was positively correlated with the
steady state levels of EGFR mRNA and the EGFR mRNA
half life (Fig. 2C). Notably, the
mRNA half life was inversely correlated with EGFR protein
concentration (Fig. 2A).
Discussion
The efficacy of gefitinib in patients with NSCLC was
thought to depend on specific EGFR gene mutations as opposed
to the concentration of EGFR protein. In particular, mutations in
the tyrosine kinase domain, consisting of exons 18–21, were
regarded as critical for a response to gefitinib. Dominant
mutations or deletions of 2–15 nucleotides between codons 740 and
753 in exon 19 were found to increase the tyrosine kinase
inhibitory activity of gefitinib due to conformational changes at
the ATP-binding site (4). We found
that a number of SCCHN cell lines have the G/A genotype, consisting
of a heterozygous G→A transition at nucleotide 2607 in exon 20 of
the EGFR gene (4). Cell
lines with the G/A genotype were significantly more sensitive to
gefitinib (lower IC50 values) than cell lines with the
wild-type G/G genotype (P=0.016). We then focused on the
relationship between gene expression and sensitivity to gefitinib
in vitro. The FISH assay showed that cell lines carrying the
G/A mutation have a significantly higher EGFR copy number
than wild-type cell lines. Moreover, the steady state levels of
EGFR mRNA were higher in cells with the G/A genotype than in
those with the G/G genotype, suggesting a close correlation between
the EGFR copy number and the steady state levels of
EGFR mRNA. Notably, concentrations of EGFR protein were
inversely correlated with the EGFR mRNA half life and steady
state levels.
The G/A mutation proved to be synonymous, which may
be due to impaired translation as opposed to protein instability.
Generally, synonymous mutations are thought to be silent. However,
cells with the G/A mutation produce less EGFR protein than those
with the G/G wild-type, thereby suggesting that the mutation
affected the translation rate. A previous study showed that a
synonymous GAA to GAG mutation resulted in a 3-fold difference in
the rate of translation due to the binding properties of tRNA to
each codon (10). Moreover, a
silent synonymous mutation in the MDR1 (multidrug resistance
1)/ABCB1 gene, which encodes P-glycoprotein, does not alter
the mRNA or protein levels, but affects protein conformation,
thereby altering the interaction between the substrate and
inhibitor (11). Thus, synonymous
mutations may affect mRNA structure and stability, the kinetics of
translation and alternative splicing (12). We found that the heterozygous
G/G→G/A mutation at codon 2607 of the EGFR gene was
correlated with gene amplification, prolongation of EGFR
mRNA half life and an increase in the steady state concentrations
of EGFR mRNA. Additionally, the mutation was inversely
correlated with the EGFR protein level, suggesting that this
mutation affects translation efficacy. Although cell lines carrying
the G/A mutation were more sensitive to gefitinib, we did not
observe a significant correlation between gefitinib IC50
and the EGFR molecular status, including the copy number and the
mRNA half life, due to the narrow range of IC50 values
in a limited number of cell lines. However, combined with our
previous findings, which demonstrated that G/A mutants exhibited a
higher sensitivity to gefitinib, the down-regulation of EGFR
protein expression, possibly due to a reduced translation efficacy,
may be closely associated with gefitinib efficacy. Since the
efficacy of gefitinib is thought to be independent of the EGFR
protein concentration, but dependent on EGFR mutations in the
tyrosine kinase domain, the structure of EGFR protein may have been
altered by the synonymous mutation. Further studies are required to
clearly determine the relationship between the efficacy of
gefitinib and the protein structure of G/A mutants of the EGFR
protein.
References
|
1
|
Eisbruch A, Blick M, Lee JS, Sacks PG and
Gutterman J: Analysis of the epidermal growth factor receptor gene
in fresh human head and neck tumors. Cancer Res. 47:3603–3605.
1987.PubMed/NCBI
|
|
2
|
Wheeler RH, Spencer S, Buchsbaum D and
Robert F: Monoclonal antibodies as potentiators of radiotherapy and
chemotherapy in the management of head and neck cancer. Curr Opin
Oncol. 11:187–190. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schlessinger J: Cell signaling by receptor
tyrosine kinases. Cell. 103:211–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taguchi T, Tsukuda M, Imagawa-Ishiguro Y,
Kato Y and Sano D: Involvement of EGFR in the response of squamous
cell carcinoma of the head and neck cell lines to gefitinib. Oncol
Rep. 19:65–71. 2008.PubMed/NCBI
|
|
5
|
Kondo N, Ishiguro Y, Kimura M, Sano D,
Fujita K, Sakakibara A, Taguchi T, Toth G, Matsuda H and Tsukuda M:
Antitumor effect of gefitinib on head and neck spuamous cell
carcinoma enhanced by trastuzumab. Oncol Rep. 20:373–378.
2008.PubMed/NCBI
|
|
6
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Helfrich BA, Raben D, Varella-Garcia M, et
al: Activating mutations in the epidermal growth factor receptor
(EGFR) tyrosine kinase inhibitor gefitinib (ZD 1839, Iressa) in
non-small cell lung cancer cell correlates with gene copy number
and EGFR mutations but not EGFR protein levels. Clin Cancer Res.
12:7117–7125. 2006. View Article : Google Scholar
|
|
9
|
Ozawa S, Kato Y, Ito S, Komori R, Shiiki
N, Tsukinoki K, Ozono S, Maehata Y, Taguchi T, Imagawa-Ishiguro Y,
Tsukuda M, Kubota E and Hata R: Restoration of BRAK/CXCL14 gene
expression by gefitinib is associated with antitumor efficacy of
the drug in head and neck squamous cell carcinoma. Cancer Sci.
100:2202–2209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sorensen MA and Pedersen S: Absolute in
vivo translation rates of individual codons in Escherichia
coli. The two glutamic acid codons GAA and GAG are translated
with a threefold difference in rate. J Mol Biol. 222:265–280. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE,
Calcagno AM, Ambudkar SV and Gottesman MM: A ‘silent’ polymorphism
in the MDR1 gene changes substrate specificity. Science.
315:525–528. 2007.
|
|
12
|
Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV
and Gottesman MM: Silent polymorphisms speak: how they affect
pharmacogenomics and the treatment of cancer. Cancer Res.
67:9609–9612. 2007. View Article : Google Scholar : PubMed/NCBI
|