Introduction
Long-term exposure to estrogen is a well-known risk
factor for the development of breast cancer (1). Estrogen signaling pathways, in
particular the mitogenic pathway, mediated by the estrogen
receptor-α (ER-α) is crucial in the development of breast cancer
stimulated by estrogen (2,3). ER-α is a ligand-activated
transcription factor comprising three independent but interacting
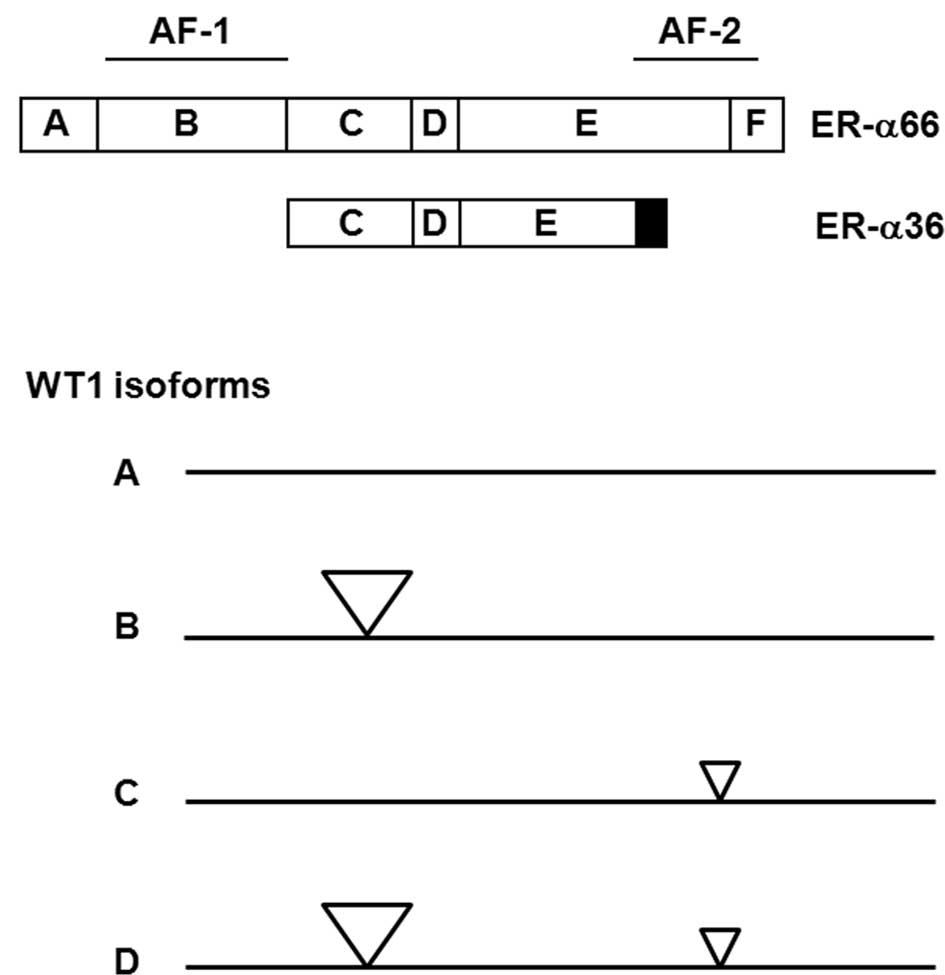
functional domains: the N-terminal A/B domain, the C or DNA-binding
domain, and the D/E/F or ligand-binding domain. The N-terminal
domain of ER-α encodes a ligand-independent activation function
(AF-1). The DNA-binding or C domain contains a two zinc-finger
structure that plays an important role in receptor dimerization and
binds to specific DNA sequences. The C-terminal E/F domain is a
ligand-binding domain that mediates ligand binding, receptor
dimerization, nuclear translocation, and a ligand-dependent
transactivation function (AF-2) (2,3).
Stimulation of the target gene expression by ER-α in response to
17β-estradiol is predominantly thought to be responsible for cell
proliferation (2).
ER-α was shown to act as a transcription factor.
However, not all of the physiological effects mediated by estrogens
are achieved through a direct effect on gene transcription. On the
other hand, a ‘non-classic’, ‘non-genomic’ or ‘membrane signaling’
pathway exists that involves cytoplasmic proteins, growth factors
and other membrane-initiated signaling pathways (4–6).
Previously, we identified and cloned a 36-kDa
variant of ER-α, i.e., ER-α36, which is mainly expressed on the
plasma membrane and mediates non-genomic estrogenic signaling
(Fig. 1A) (7,8).
ER-α36 lacks transcription activation domains AF-1 and AF-2 of the
66-kDa full-length ER-α (ER-α66), and possesses an altered
ligand-binding domain and an intact DNA-binding domain, consistent
with the fact that ER-α36 possesses no intrinsic transcriptional
activity (8). ER-α36 is
predominantly expressed on the plasma membrane and in the
cytoplasm, and mediates non-genomic estrogen signaling (8,9).
ER-α36 is generated from a promoter located in the first intron of
the ER-α66 gene (10), indicating
that ER-α36 expression is regulated differently from ER-α66,
consistent with the findings that ER-α36 is expressed in specimens
from ER-negative breast cancer patients and established ER-negative
breast cancer cells that lack ER-α66 expression (8,11,12).
Thus, the nuclear ER-α66 is mainly involved in genomic estrogen
signaling whereas extra-nuclear ER-α36 is involved in non-genomic
estrogen signaling.
Previously, the extra-nuclear ER-α36 was found to
act as a dominant-negative inhibitor of genomic estrogen signaling
through impeding the transcription activities mediated by the AF-1
and AF-2 domains of ER-α66 (8).
Recently, ER-α66 was found to suppress the promoter activity of
ER-α36 via a half estrogen response element (ERE) site located in
the 5′-flanking sequence of the ER-α36 gene (10). These findings suggest that the
genomic and non-genomic estrogen signaling pathways mediated by
ER-α66 and ER-α36 are dynamically and strictly regulated at
different levels. Dysregulated genomic and/or non-genomic estrogen
signaling may lead to various diseases including cancer. Thus, the
expression levels of ER-α66 and ER-α36 in a particular cell context
require strict coordination. However, the underlying mechanisms of
this coordination remain to be elucidated.
The Wilms’ tumor susceptibility gene, wt1, at
chromosome locus 11p13 (13–15)
encodes a C2-H2-type zinc-finger protein,
WT1. Alternative splicing results in four protein isoforms of WT1
that differ due to the presence of one 17-amino acid insert between
the transcription regulatory and DNA-binding domains, and one
3-amino-acid (KTS) insert between the third and fourth zinc fingers
(16,17). The different isoforms are referred
to as A, B, C and D, whereby the A isoform lacks both 17-amino-acid
and KTS inserts, the B isoform contains the 17-amino-acid insert
but lacks the KTS insert, the C isoform lacks the 17-amino-acid
insert but contains the KTS insert, and the D isoform contains both
inserts (Fig. 1B). Mutations of
wt1 were found to be correlated with subsets of Wilms’ tumor
(16,17), mesothelioma and ovarian tumors
(18), consistent with the role of
WT1 as a tumor suppressor. However, high levels of the wild-type
WT1 mRNA and protein have been found in leukemia (19), lung cancer (20) and breast cancer (21–23).
Breast cancer patients with tumors that highly express WT1 usually
have a lower 5-year disease-free survival rate than patients with
tumors of low WT1 expression (23),
indicating that WT1 expression is associated with aggressive
phenotype of breast cancer. However, the biological function and
underlying mechanisms of WT1 in the development of aggressive
breast tumors have yet to be investigated.
In the present study, the Wilms’ tumor suppressor
WT1 activated promoter activity of the ER-α66 gene and suppressed
ER-α36 promoter activity, suggesting that WT1 acts as a
‘coordinator’ of the genomic and non-genomic estrogen signaling
pathways through the opposite regulation of the expression of
ER-α36 and ER-α66.
Materials and methods
Cell culture and establishment of stable
cell lines
Human embryonic kidney (HEK293) cells were obtained
from American Type Culture Collection (ATCC, Manassas, VA, USA) and
maintained in DMEM and 10% fetal calf serum at 37°C in a 5%
CO2 incubator. Relatively high-passage MCF7 cells were
initially obtained from Dr Thomas F. Deuel’s laboratory at the
Scripps Research Institute. The subline of MCF7 cells used in this
study had been cultured for >75 passages and were maintained at
37°C in a 5% CO2 atmosphere in Improved Modified Eagle’s
Medium (IMEM) supplemented with 5% fetal calf serum. To establish
stable cells that express knocked-down levels of the Wilms’ tumor
suppressor, WT1, MCF7 cells were plated at a density of
1×105 cells per 60-mm dish and transfected 24 h later
with a mixture of four WT1 small hairpin (sh) RNA expressing
constructs purchased from Origene (TR300442, Rockville, MD, USA)
using the FuGene 6 transfection reagent (Roche Applied Sciences,
Indianapolis, IN, USA). The control expression vector was also
transfected into MCF7 cells to serve as a control. Following
transfection (48 h), the cells were replated and selected with 5
μg/ml of puromycin (Invitrogen Corporation, Carlsbad, CA, USA) for
two weeks. The medium was changed every three days until colonies
appeared. A number of clonal cell lines were established that
express the knocked-down levels of WT1. Two of these cell lines are
described in this study, i.e., MCF7/sh-WTl-1 and -2. More than 20
individual clones from cells transfected with the empty expression
vector were pooled and used as control MCF7/V cells.
Western blot analysis
Cells were washed three times with cold
phosphate-buffered saline (PBS) and lysed with lysis buffer [50 mM
Tris-HCl pH 8.0, 150 mM NaCl, 0.25 mM EDTA pH 8.0, 0.1% sodium
dodecyl sulfate (SDS), 1% Triton® X-100, 50 mM NaF and
the protease inhibitor cocktail from Sigma (St. Louis, MO, USA)].
Following adjustment to the same total protein content, cell
lysates were analyzed by Western blot analysis. Cell lysates (25
μg) were boiled for 5 min in SDS gel-loading buffer and separated
on a 10% SDS-PAGE gel. Following electrophoresis, the proteins were
transferred to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA,
USA). The membranes were probed with different primary antibodies,
incubated with appropriate HRP-conjugated secondary antibodies and
visualized with enhanced chemiluminescence (ECL) detection reagents
(Amersham Pharmacia Biotech., Piscataway, NJ, USA). The same
membranes were stripped and reprobed with an antibody against
β-actin (I-19) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) to
confirm equal loading.
Polyclonal anti-ER-α36 antibody was generated and
characterized as previously described (8). Anti-ER-α66 antibody (Ab-15) was
obtained from Lab Vision Products (Fremont, CA, USA). Polyclonal
anti-WT1 antibody was from Invitrogen Corporation.
Luciferase assay
HEK293 cells were transfected using FuGene 6
transfection reagent with the reporter plasmids encoding the
firefly luciferase gene driven by the 5′-flanking sequence of
ER-α66 or ER-α36 gene, ER-α66 promoter-Luc and ER-α36 promoter-Luc,
respectively. The ER-α66 promoter-Luc reporter plasmid was
purchased from Switchgear Genomics (Menlo Park, CA, USA). The
plasmid contains the DNA sequence from −748 to +324 (relative to
the major transcription initiation site) of the ER-α66 promoter
region. The ER-α36 promoter-Luc containing the 715-bp 5′-flanking
sequence of the ER-α36 gene was generated and characterized as
previously described (10).
Expression vectors containing the WT1 isoforms A, B, C and D were
previously described (24). A
cytomegalovirus-driven Renilla luciferase plasmid, pRL-CMV
(Promega, San Luis Obispo, CA, USA), was also included in the
transfection to establish transfection efficacy. Following
transfection (48 h), cell extracts were prepared, and the
luciferase activity was determined and normalized using the
Dual-Luciferase Assay System (Promega) and a TD 20/20 Luminometer
(Turner BioSystems, Inc. Sunnyvale, CA, USA) according to the
manufacturer’s instructions.
Statistical analysis
Data were summarized as the mean ± standard error
(SE) using an GraphPad InStat software program. The Tukey-Kramer
multiple comparisons test was also used. P<0.05 was considered
to be statistically significant.
Results
Knockdown of WT1 expression in MCF7 cells
alters the expression of ER-α66 and ER-α36
Recently, we found that high-passage MCF7 cells
express increased levels of the Wilms’ tumor suppressor WT1
compared to low-passage MCF7 cells (25). To determine the role played by WT1
in MCF7 cells, expression levels of WT1 were knocked down in
high-passage MCF7 cells, using the shRNA method. The clonal cell
lines MCF7/sh-WTl-1 and -2 were transfected with the WT1 shRNA
expression vectors. A cell line was generated from a mixture of
>20 clones transfected with the empty expression vector
(MCF7/V).
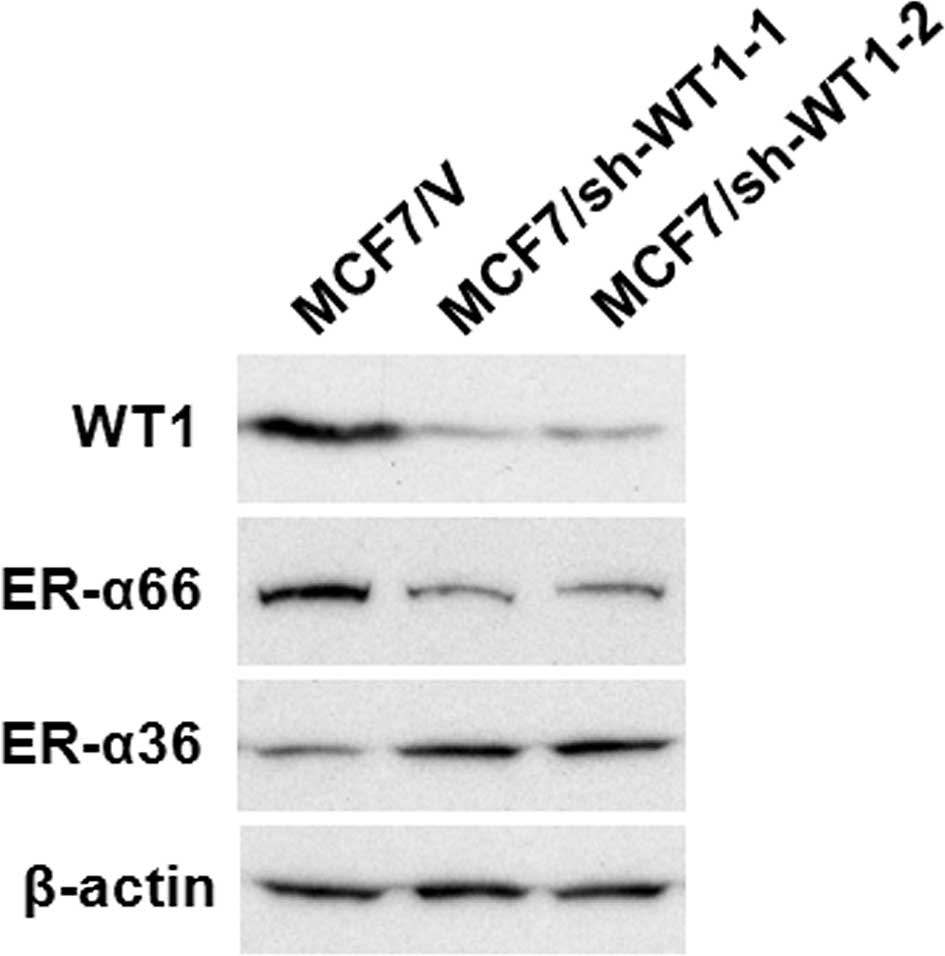
Western blot analysis using the antibody against WT1
confirmed that the WT1 protein (~52 kDa) was significantly
down-regulated in the MCF7/sh-WTl-1 and -2 cell lines compared to
the control MCF7 cells transfected with the empty vector (MCF7/V)
(Fig. 2). The expression levels of
ER-α66 were markedly decreased in the WT1 shRNA-transfected MCF7
cells, MCF7/sh-WTl-1 and -2, compared to the control (MCF7/V) cells
(Fig. 2). We also noted that ER-α36
expression was increased in the MCF7/sh-WTl-1 and -2 cells
(Fig. 2). These results suggest
that as a dual transcription regulator, WT1 modulates the promoter
activities of ER-α66 and ER-α36 oppositely.
WT1 activates the promoter activity of
ER-α66
To determine whether WT1 regulates ER-α66 promoter
activity, we performed transient co-transfection assays in HEK293
cells that express undetectable levels of WT1, ER-α66 and ER-α36.
HEK293 cells were co-transfected with a luciferase reporter gene
driven by the 5′-flanking sequence of the ER-α66 gene (−748 to
+324, relative to the major transcription initiation site) with
expression vectors encoding four different isoforms of WT1
separately to evaluate the effects of different isoforms of WT1 on
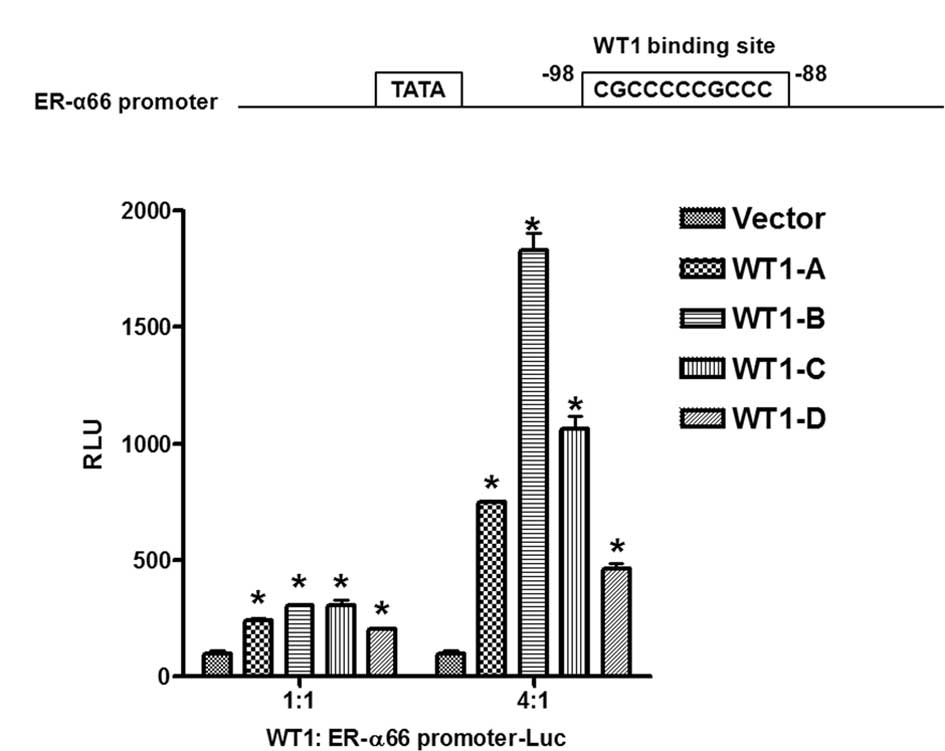
ER-α66 promoter activity. Findings showed that all four isoforms of
WT1 activated the promoter activity of ER-α66; the WT1-B isoform
exhibited the strongest activity whereas the WT1-D isoform
exbitited the weakest activity (Fig.
3B). Computer analysis of the 5′-flanking sequence of ER-α66
revealed the existence of a perfect WT1 binding site located
downstream of the TATA box (Fig.
3A). Our data thus indicated that WT1 positively regulates
ER-α66 promoter activity presumably via the WT1 binding site
located at −98 to −88 (relative to the major transcription
initiation site).
WT1 suppresses the promoter activity of
ER-α36
Recently, we cloned and characterized the
5′-flanking sequence of ER-α36 that is located in the first intron
of the ER-α66 gene (10). A
computer analysis of the promoter region of ER-α36 revealed two WT1
binding sites located both upstream and downstream of the TATA box
(Fig. 4A). We then examined whether
WT1 regulates the promoter activity of ER-α36. HEK293 cells were
co-transfected with a luciferase reporter driven by the 5′-flanking
sequence of the ER-α36 gene (−736 to +16, relative to the
transcription initiation site) with expression vectors encoding
four isoforms of WT1 to examine the effects of these WT1 isoforms
on ER-α36 promoter activity. The four isoforms of WT1 inhibited the
promoter activity of ER-α36 with different efficiency in that the
WT1-D isoform exhibited the strongest activity whereas the WT1-A
isoform exhibited the weakest activity (Fig. 4B).
Discussion
The diverse functions of estrogens are mediated by
the estrogen receptors, ER-α and ER-β, both of which play a role as
ligand-dependent transcription factors. The liganded ERs readily
form homodimers or heterodimers that interact with the palindromic
ERE in the promoter regions of estrogen responsive genes and
stimulate gene transcription (2,3).
Alternatively, ER-α may act indirectly by tethering to other
transcription factors, such as Sp1 and AP1, to modulate activities
of these transcription factors, thereby regulating downstream gene
expression (2,3).
Previously, accumulating evidence suggested a rapid
(within seconds or minutes) estrogen action that cannot be
explained by the genomic signaling pathway which usually requires a
long period of time to reach maximal gene activation (4–6,26).
This non-genomic estrogen signaling pathway cross-talks with
various signaling pathways, such as the adenylate cyclase,
cAMP-dependent signaling and the MAPK pathways (4–6,26).
Thus, the genomic and non-genomic pathways of estrogen action may
integrate with one another to achieve a complete cellular response
to estrogens.
It is well known that ER-α66 predominantly mediates
genomic estrogen signaling by regulating target gene expression,
although a previous study showed that ER-α66 is also involved in
non-genomic estrogen signaling (27). ER-α36, on the other hand, lacks
intrinsic transcription activity and mainly mediates non-genomic
estrogen signaling (8). Thus, the
expression levels of ER-α66 and ER-α36 should be dynamically and
strictly regulated in order to maintain a balance between the
genomic and non-genomic estrogen signaling pathways.
In the present study, ER-positive breast cancer MCF7
cells expressed high levels of WT1 and ER-α66, whereas MCF7 cells
with a knocked-down level of WT1 expressed a decreased level of
ER-α66, suggesting that WT1 up-regulates ER-α66 expression. We also
found that the same WT1 knocked-down MCF7 cells expressed an
increased level of ER-α36, suggesting that WT1 plays a role as a
negative regulator of ER-α36 expression. Further co-transfection
assays showed that WT directly activated the promoter activity of
the ER-α66 gene and suppressed ER-α36 promoter activity. Thus, this
study showed that WT1 plays a role as a dual transcription factor
in the regulation of the promoter activities of ER-α66 and ER-α36
oppositely.
Current evidence indicates a potentially oncogenic
role of WT1 in breast cancer (28).
WT1 expression was found in primary breast tumors (21–23),
and high levels of WT1 expression were shown to predict a poor
prognosis in breast cancer patients (23), consistent with a putative oncogenic
role of WT1. WT1 is a dual transcription regulator and that plays a
role in the activation or suppression of gene transcription
depending on the cell and promoter context (24,29–32).
Previously, we demonstrated that WT1 acts as a transcription
suppressor on promoters harboring WT1 binding sites both upstream
and downstream of the transcription initiation site. WT1 also
promotes transcription activity with WT1 binding sites located
either upstream or downstream of the transcription site (24). Our computer analysis revealed the
existence of two putative WT1 binding sites in the promoter region
of ER-α36 located both upstream and downstream of the TATA box. By
contrast, the ER-α66 promoter contained one perfect WT1 binding
site downstream of the TATA box. Consequently, WT1 functions to
oppositely regulate the promoter activities of ER-α66 and
ER-α36.
Han et al have reported that the forced
expression of WT1-B and -D isoforms in MCF7 cells down-regulated
ER-α66 expression. Additionally, the co-transfection of WT1-B and
-D isoforms moderately suppressed ER-α66 promoter activity
(33). However, in the present
study, it was noted that both WT1-B and -D isoforms up-regulated
the promoter activity of ER-α66, and knockdown of all WT1 isoforms
with shRNA down-regulated ER-α66 expression. The exact mechanisms
underlying this discrepancy have yet to be elucidated. One
possibility is that various ER-α66 promoter reporter constructs
were used that contained a different length of the 5′-flanking
sequence of the ER-α66 gene with different transcription factor
binding sites. In a recent study, the forced expression of only
WT1-B and -D isoforms was used (33). It was reported that various isoforms
of WT1 clearly affected mammary epithelial cells differently
(34). Another possibility is that
changes noted in the ratios among various isoforms of WT1 following
the forced expression of specific isoforms of WT1 may provide
different outcomes. Our results suggest that the ratios of
different WT1 isoforms expressed in mammary epithelial cells are
involved in the coordination of the genomic and non-genomic
signaling pathways by regulation of ER-α66 and ER-α36 expression
oppositely.
Acknowledgements
This study was funded by the Nebraska Tobacco
Settlement Biomedical Research Program Award (LB-595) to (Z.-Y.
Wang) and NIH grant DK070016 (Z.-Y. Wang).
References
|
1
|
Vorherr H: Breast cancer: epidemiology,
endocrinology, biochemistry, and pathobiology. Urban and
Schwarzenberg; Baltimore: 1980
|
|
2
|
Nilsson S, Makela S, Treuter E, et al:
Mechanisms of estrogen action. Physiol Rev. 81:1535–1565. 2001.
|
|
3
|
Klinge CM: Estrogen receptor interaction
with estrogen response elements. Nucleic Acids Res. 29:2905–2919.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Segars JH and Driggers PH: Estrogen action
and cytoplasmic signaling cascades. Part I: membrane-associated
signaling complexes. Trends Endocrinol Metab. 13:349–354. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Driggers PH and Segars JH: Estrogen action
and cytoplasmic signaling pathways. Part II: the role of growth
factors and phosphorylation in estrogen signaling. Trends
Endocrinol Metab. 13:422–427. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kelly MJ and Levin ER: Rapid actions of
plasma membrane estrogen receptors. Trends Endocrinol Metab.
12:152–156. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Z, Zhang X, Shen P, Loggie BW, Chang
Y and Deuel TF: Identification, cloning, and expression of human
estrogen receptor-α36, a novel variant of human estrogen
receptor-α66. Biochem Biophys Res Commun. 336:1023–1027. 2005.
|
|
8
|
Wang Z, Zhang X, Shen P, Loggie BW, Chang
Y and Deuel TF: A variant of estrogen receptor-{α}, hER-{α}36:
transduction of estrogen- and antiestrogen-dependent
membrane-initiated mitogenic signaling. Proc Natl Acad Sci USA.
103:9063–9068. 2006.
|
|
9
|
Lin SL, Yan LY, Liang XW, et al: A novel
variant of ER-α, ER-α36, mediates testosterone-stimulated ERK and
Akt activation in endometrial cancer Hec1A cells. Reprod Biol
Endocrinol. 7:1022009.
|
|
10
|
Zou Y, Ding L, Coleman M and Wang Z:
Estrogen receptor-α (ER-α) suppresses expression of its variant
ER-α 36. FEBS Lett. 583:1368–1374. 2009.
|
|
11
|
Lee LM, Cao J, Deng H, Chen P, Gatalica Z
and Wang ZY: ER-α36, a novel variant of ER-α, is expressed in
ER-positive and -negative human breast carcinomas. Anticancer Res.
28:479–483. 2008.
|
|
12
|
Shi L, Dong B, Li Z, et al: Expression of
ER-(α)36, a novel variant of estrogen receptor-(α), and resistance
to tamoxifen treatment in breast cancer. J Clin Oncol.
27:3423–3429. 2009.
|
|
13
|
Bonetta L, Kuehn SE, Huang A, et al: Wilms
tumor locus on 11p13 defined by multiple CpG island-associated
transcripts. Science. 250:994–997. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Call KM, Glaser T, Ito CY, et al:
Isolation and characterization of a zinc finger polypeptide gene at
the human chromosome 11 Wilms’ tumor locus. Cell. 60:509–520.
1990.
|
|
15
|
Gessler M, Poustka A, Cavenee W, Neve RL,
Orkin SH and Bruns GA: Homozygous deletion in Wilms tumours of a
zinc-finger gene identified by chromosome jumping. Nature.
343:774–778. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coppes MJ, Campbell CE and Williams BR:
The role of WT1 in Wilms tumorigenesis. FASEB J. 7:886–895.
1993.PubMed/NCBI
|
|
17
|
Lee SB and Haber DA: Wilms tumor and the
WT1 gene. Exp Cell Res. 264:74–99. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Little M and Wells C: A clinical overview
of WT1 gene mutations. Hum Mutat. 9:209–225. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Inoue K, Ogawa H, Sonoda Y, et al:
Aberrant overexpression of the Wilms tumor gene (WT1) in human
leukemia. Blood. 89:1405–1412. 1997.PubMed/NCBI
|
|
20
|
Oji Y, Miyoshi S, Maeda H, et al:
Overexpression of the Wilms’ tumor gene WT1 in de novo lung
cancers. Int J Cancer. 100:297–303. 2002.
|
|
21
|
Silberstein GB, Van Horn K, Strickland P,
Roberts CT Jr and Daniel CW: Altered expression of the WT1 wilms
tumor suppressor gene in human breast cancer. Proc Natl Acad Sci
USA. 94:8132–8137. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Loeb DM, Evron E, Patel CB, et al: Wilms’
tumor suppressor gene (WT1) is expressed in primary breast tumors
despite tumor-specific promoter methylation. Cancer Res.
61:921–925. 2001.
|
|
23
|
Miyoshi Y, Ando A, Egawa C, et al: High
expression of Wilms’ tumor suppressor gene predicts poor prognosis
in breast cancer patients. Clin Cancer Res. 8:1167–1171. 2002.
|
|
24
|
Wang ZY, Qiu QQ and Deuel TF: The Wilms’
tumor gene product WT1 activates or suppresses transcription
through separate functional domains. J Biol Chem. 268:9172–9175.
1993.
|
|
25
|
Wang L and Wang ZY: The Wilms’ tumor
suppressor WT1 induces estrogen-independent growth and
anti-estrogen insensitivity in ER-positive breast cancer MCF7
cells. Oncol Rep. 23:1109–1117. 2010.
|
|
26
|
Levin ER: Integration of the extranuclear
and nuclear actions of estrogen. Mol Endocrinol. 19:1951–1959.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pedram A, Razandi M and Levin ER: Nature
of functional estrogen receptors at the plasma membrane. Mol
Endocrinol. 20:1996–2009. 2006. View Article : Google Scholar
|
|
28
|
Yang L, Han Y, Suarez Saiz F and Minden
MD: A tumor suppressor and oncogene: the WT1 story. Leukemia.
21:868–876. 2007.PubMed/NCBI
|
|
29
|
Liu XW, Gong LJ, Guo LY, et al: The Wilms’
tumor gene product WT1 mediates the down-regulation of the rat
epidermal growth factor receptor by nerve growth factor in PC12
cells. J Biol Chem. 276:5068–5073. 2001.
|
|
30
|
Han Y, San-Marina S, Liu J and Minden MD:
Transcriptional activation of c-myc proto-oncogene by WT1 protein.
Oncogene. 23:6933–6941. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hewitt SM, Hamada S, McDonnell TJ,
Rauscher FJ III and Saunders GF: Regulation of the proto-oncogenes
bcl-2 and c-myc by the Wilms’ tumor suppressor gene WT1. Cancer
Res. 55:5386–5389. 1995.PubMed/NCBI
|
|
32
|
Englert C, Hou X, Maheswaran S, et al: WT1
suppresses synthesis of the epidermal growth factor receptor and
induces apoptosis. EMBO J. 14:4662–4675. 1995.PubMed/NCBI
|
|
33
|
Han Y, Yang L, Suarez-Saiz F, San-Marina
S, Cui J and Minden MD: Wilms’ tumor 1 suppressor gene mediates
antiestrogen resistance via down-regulation of estrogen receptor-α
expression in breast cancer cells. Mol Cancer Res. 6:1347–1355.
2008.
|
|
34
|
Burwell EA, McCarty GP, Simpson LA,
Thompson KA and Loeb DM: Isoforms of Wilms’ tumor suppressor gene
(WT1) have distinct effects on mammary epithelial cells. Oncogene.
26:3423–3430. 2007.
|