Introduction
The leading cause of death in breast cancer (BrCa)
patients is not the primary tumor in the breast per se, but
metastasis to distant organs. Metastasis accounts for over 90% of
deaths in BrCa patients. Millions of cells are released from the
primary tumor into the blood circulation daily, but only a small
minority of these cells survive and colonize on distant organs. The
aim of the present study was to establish the identity of these
cells. According to the current cancer stem cell hypothesis, tumors
are organized in cell hierarchies composed of tumor-initiating
cells (T-ICs), cancer cells and differentiated cells (1). T-ICs with stem cell-like properties,
so-called cancer stem cells, are not only the source of the primary
tumor; but are also responsible for tumor growth, metastasis and
possible recurrence. The CD44+CD24−/low
subpopulation in BrCa cells was reported to be enriched in BrCa
T-ICs (2). This subpopulation of
cells is capable of generating other cellular phenotypes observed
in the original tumor and initiating metastases at distant sites
from low numbers of cells (2).
However, data on the characteristics of BrCa T-ICs and their
molecular regulation is currently limited.
microRNAs (miRNAs) are small, non-coding RNAs that
negatively regulate gene expression by binding to their target
miRNAs. miRNAs regulate tumor development, prognosis and metastasis
either as oncogenes or tumor suppressors (3,4). A
burgeoning body of evidence showed that miRNAs modulate the
properties of embryonic stem (ES) and tissue stem cells in a
variety of eukaryotic organisms (5,6). A
group of miRNAs may play specialized roles in stem cell regulation,
as the Dicer (the enzyme that processes the pre-miRNAs into mature
miRNAs) knockout in mice is embryonically lethal (7), and Dicer conditional knockout mouse ES
cells grow more slowly than controls and fail to differentiate
(8).
In the present study, we investigated the
differences in miRNA expression profiling among BrCa cell lines
with various proportions of T-ICs and the potential contribution of
miRNAs to maintaining the 'stemness' of BrCa T-ICs. The results
indicate that a miRNA expression signature may help identify the
stemness of T-ICs, and that certain miRNAs may help predict
clinical features, such as metastasis, for BrCa.
Materials and methods
Cell lines and cell culture
Human BrCa cell lines MCF-7, SUM149, SUM1315 and
MDA-MB-231 were stored in the laboratory. The culture conditions
for each cell line were identical to those used in our previous
study (9).
Fluorescence-activated cell sorting
(FACS) analysis
CD24 and CD44 expression of BrCa cells was examined
by FACS analysis using anti-CD44 (FITC-conjugated) and anti-CD24
(PE-conjugated) antibodies as previously described (10). The two antibodies were obtained from
BD Biosciences (San Jose, CA, USA).
miRNA microarray analysis
A MiRVana miRNA isolation kit (Ambion, Austin, TX,
USA) was used to isolate total RNA with enriched miRNAs from cells
according to the manufacturer's instructions. miRNA microarray was
performed by LC Sciences (Houston, TX, USA; http://www.lcsciences.com) as previously described
(9). The differentially expressed
miRNAs in SUM1315 and MDA-MB-231 cells were those with a
fold-change of >2 or <−2 and P-values of the t-test <0.05
compared to MCF-7 cells.
Reverse transcription polymerase chain
reaction (RT-PCR) analyses
Total RNA with enriched miRNAs in BrCa cells was
isolated as described above. Reverse transcription (RT) was
performed using the RT primers for each individual miRNA from
Ambion according to the manufacturer's instructions. Real-time PCR
was performed using a miRVana qRT-PCR kit (Ambion) according to the
manufacturer's instructions and the protocol used previously
(11). PCR primers were purchased
from Ambion.
miRNA selection and target gene
prediction
To filter the miRNAs associated with stem cell-like
properties, a search on various literature in the PubMed database
was conducted using the keywords 'miR-xx' and 'stem cells' for each
individual miRNA. Based on the literature, miRNAs clearly involved
in stem/progenitor cell modulation were collected. Following that,
the miRGen database (http://www.diana.pcbi.upenn.edu/miRGen/v3/miRGen.html)
was used to predict target genes for selected miRNAs. The
Targets interface of the miRGen database provides access to
the union and intersection of the four widely used target
prediction programs: miRBase, miRanda, TargetScan and PicTar. To
obtain reliable target genes, the intersection function (among
miRanda, TargetScan and PicTar) was used to predict target genes
(Fig. 1). The obtained gene list
was further intersected with the target genes above the mean score
from the PicTar prediction alone to obtain the final hits for each
individual miRNA. Following this process, target genes documented
to regulate stem cell-like properties were selected from
PubMed.
Statistical analysis
The two-tailed Student's t-test was used to analyze
the statistical significance of FACS and RT-PCR data. P<0.05 was
considered to be statistically significant.
Results and Discussion
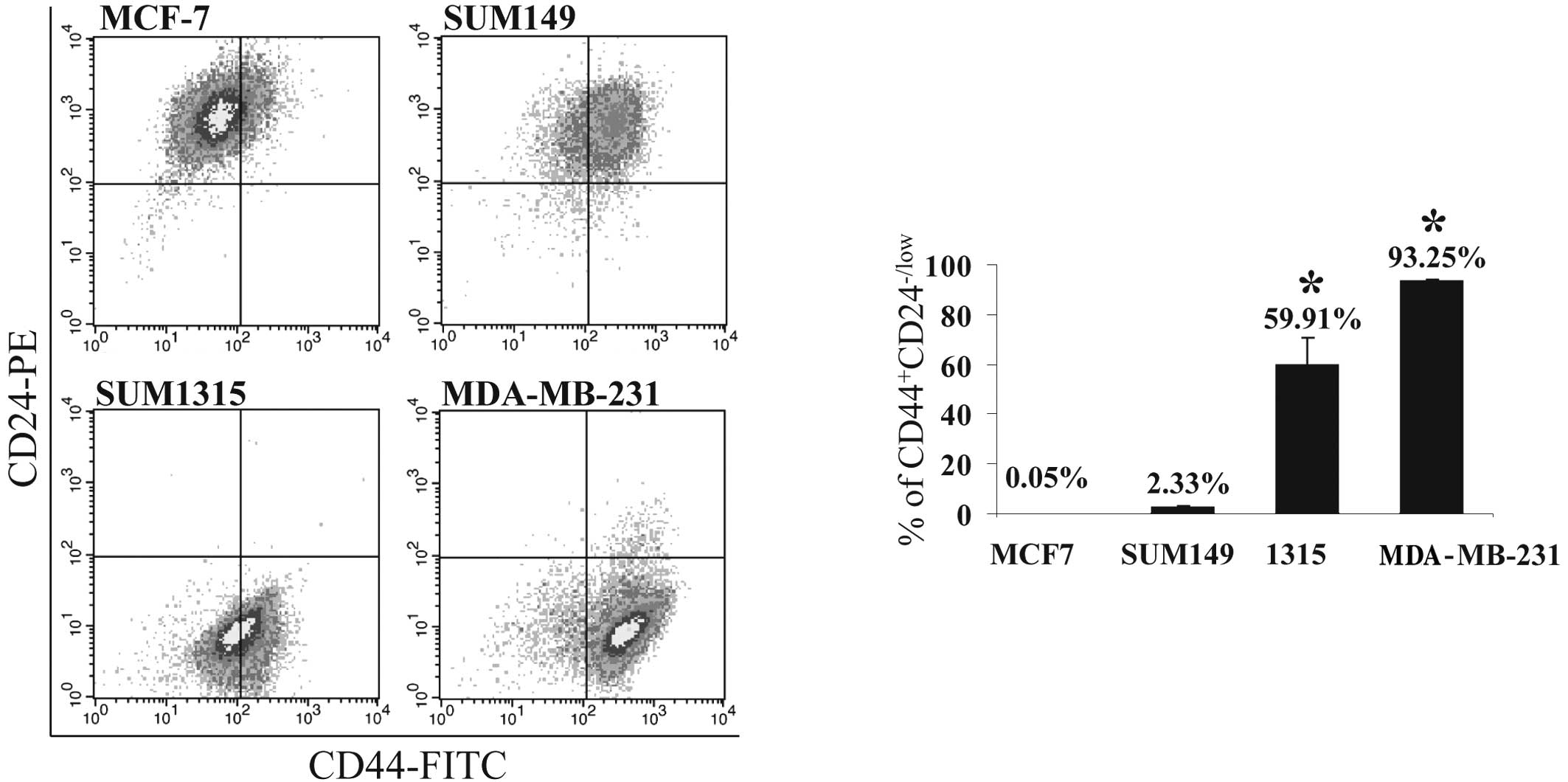
The prevalence of
CD44+CD24−/low cells in BrCa cell lines
To test the hypothesis that the prevalence of T-ICs
in a BrCa cell population is associated with the metastatic
phenotype of the BrCa cells, the proportion of the
CD44+CD24−/low subpopulation of cells was
assessed in the tumorigenic/metastatic human BrCa cell lines,
SUM1315 and MDA-MB-231, and the tumorigenic/non-metastatic human
BrCa cell lines, MCF-7 and SUM149, by FACS analysis. Consistent
with a previous report (12), the
proportion of this subpopulation is relatively higher in SUM1315
(~60%) and MDA-MB-231 cells (>90%) than in MCF-7 and SUM149 ones
(0.05 and 3%, respectively; P<0.001) (Fig. 2A and B). The greater percentage of
CD44+CD24−/low cells in SUM1315 and
MDA-MB-231 cells corresponds with the highly malignant and
metastatic phenotypes of these cell lines (13,14).
This finding is also consistent with our previous observation that
these cell lines possess enhanced propensity to metastasize
(9,13).
miRNA expression signature for
tumor-initiating properties in BrCa cells
To investigate the role of miRNAs in the maintenance
of BrCa T-ICs, total RNAs from MCF-7, SUM1315 and MDA-MB-231 cells
were used as representatives to carry out a microarray analysis of
miRNAs. The miRNA expression profiling was documented previously
(9). There are 86 and 74 miRNAs
that are differentially expressed (fold-change >2 or <−2,
P<0.05) in MDA-MB-231 and SUM1315 cells compared to MCF-7 cells,
respectively, and there are 69 common miRNAs between the 86 and 74
differentially expressed miRNAs (9). Among the 69 miRNAs common to the two
cell lines, approximately 46% (n=32) of them have been documented
to regulate the stemness of stem/progenitor cells (Table I). The expression changes of miR-22,
miR-93 and miR-203 were further validated by qRT-PCR (Fig. 3).
 | Table ImiRNAs associated with stem cell-like
properties. |
Table I
miRNAs associated with stem cell-like
properties.
| miRNA | MDA-MB-231 vs.
MCF-7 cells | SUM1315 vs. MCF-7
cells | Function
description | Representative
target genes |
|---|
| miR-375 | ↓↓↓↓↓↓↓↓ | ↓↓↓↓↓↓↓↓ | Regulates human ES
cell differentiation (15) | RLF, OTP,
INSM1 |
| miR-203 | ↓↓↓↓↓↓↓ | ↓↓↓↓↓↓↓↓ | Tumor suppressor,
and regulates stem cell differentiation (16,17) | EDN1 |
| miR-200c | ↓↓↓↓↓↓↓ | ↓↓↓↓↓↓↓ | Suppresses the
ability of normal mammary stem cells to form mammary ducts and
tumors (18) | FN1, ZFHX1B |
| miR-7 | ↓↓↓ | ↓↓↓↓ | Regulates
differentiation of germline stem cell lineage (19) | COL2A1, GLI3,
KLF4 |
| miR-98 | ↓↓ | ↓↓ | Regulates
differentiation of bronchoalveolar stem cells (20) | E2F5, HOXA9,
SEMA4C |
| miR-183 | ↓ | ↓↓ ↓ | Suppresses
expression of stem cell factors in cancer and mouse ES cells
(21) | ABCA1, LRP6,
ITGB1 |
| miR-26b | ↓↓ ↓ | ↓ | Significantly
decreased in human ES cells (22) | JAG1, NLK,
Ezh2 |
| miR-21 | ↓↓ | ↓ | Suppresses the
self-renewal of ES cells (23) | SATB1, RHOB,
PITX2 |
| miR-148b | ↓ | ↓↓ | Regulates human
mesenchymal stem cell differentiation (24) | CANX, CSF1,
DNMT1 |
| miR-93 | ↓ | ↓ | Depleted in mammary
progenitor cells (25) | LATS2, TXNIP,
ARID4B |
| miR-125a | ↓ | ↓ | Regulates neural
differentiation of stem cells (26) | TAZ, GPC4,
PLAGL2 |
| miR-106b | ↓ | ↓ | Altered expression
in bronchoalveolar stem cells (27) | ARID4B, BCL2L2,
EPHA4 |
| miR-103 | ↓ | ↓ | Modulates the
self-renewal of mesenchymal stem cells (28) | BTG2, CDK6 |
| miR-107 | ↓ | ↓ | Modulates the
self-renewal of mesenchymal stem cells (28) | OGT, RBM24,
CDK6 |
| miR-128 | ↓ | ↓ | Promotes neural
stem cell self-renewal (29) | BMI-1 |
| miR-26a | ↓ | ↓ | Modulates the
osteogenic differentiation of human adipose tissue-derived stem
cells (30) | SMAD1, HOXA5,
JAG1 |
| miR-20a | ↑ | ↑ | Differentially
expressed in developing mouse embryos, and controls differentiation
of stem cells (31) | ABCA1, BCL11B,
CD69 |
| miR-23b | ↑ | ↑ | Regulates liver
stem cell differentiation by targeting Smads (32) | PKNOX1, SRC1,
ZIC1 |
| miR-23a | ↑ | ↑ | Increased during
replication and senescence of human cord blood-derived multipotent
stem cells (33) | ELF5, SRC1 |
| miR-92b | ↑ | ↑ | Controls the G1/S
checkpoint gene p57 in human ES cells (34) | p57 |
| miR-24 | ↑ | ↑ ↑ | Inhibits endodermal
differentiation of human ES cells (35) | CDX2 |
| miR-22 | ↑ ↑ | ↑ | Highly expressed in
mammary progenitor cells (25) | LGALS1, PLAGL2,
MECP2 |
| miR-19a | ↑ ↑ | ↑ | Up-regulated by
activin A in human ES cells (36) | CD164, ARHGAP1,
WNT3 |
| miR-27a | ↑ ↑ | ↑ ↑ | Reduces the
differentiation of human mesenchymal stem cells (24) | HOXB8, APRIN,
SEMA4C |
| miR-19b | ↑ ↑ | ↑ ↑ | Down-regulated by
activin A in human ES cells (36) | NEUROD1, WNT3,
CCNT2 |
| miR-29c | ↑ ↑ ↑ | ↑ ↑ ↑ | Up-regulated in the
course of replicative senescence of mesenchymal stem cells
(37) | RLF, TRAF4,
PHC1 |
| miR-99a | ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | Differentially
expressed in human mesenchymal stem cells (38) | HOXA1 |
| miR-29a | ↑ ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | Highly expressed in
hematopoietic stem cells and down-regulated in hematopoietic
progenitors (39) | RLF, GDF8,
TRAF4 |
| miR-125b | ↑ ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | Critical for the
suppression of human glioma stem cell proliferation (40) | U251 GPC4, TAZ,
MSI1 |
| miR-100 | ↑ ↑ ↑ ↑ ↑ ↑ ↑
↑ | ↑ ↑ ↑ ↑ ↑ ↑ | Modulates
differentiation of mouse ES cells (41) | SMARCA5, HOXA1 |
| miR-222 | ↑ ↑ ↑ ↑ ↑ ↑ ↑
↑ | ↑ ↑ ↑ ↑ ↑ ↑ ↑
↑ | Highly expressed at
early developmental stages in the embryonic retina (42) | MAP3K10, PBX3 |
| miR-221 | ↑ ↑ ↑ ↑ ↑ ↑ ↑ ↑
↑ | ↑ ↑ ↑ ↑ ↑ ↑ ↑
↑ | Controls
proliferation and differentiation of CD34-positive hematopoietic
progenitor cells (43) | MAP3K10, CTCF |
A recent study of miRNA expression of mouse mammary
progenitor cells revealed a characteristic group of miRNAs: miR-22
and miR-205 are highly expressed, and miR-93 and let-7 are
decreased in progenitor cells compared to differentiated cells
(25). In agreement with this
study, the miRNA profiling data obtained in the present study
showed that miR-22 was induced and miR-93 was reduced in MDA-MB-231
and SUM1315 cells compared to MCF-7 cells (Table I and Fig. 3). This result correlates with the
high percentage of BrCa T-ICs in MDA-MB-231 and SUM1315 cells
(Fig. 2). miR-21, regulated by REST
(RE1-silencing transcription factor; also known as NRSF),
specifically suppresses the self-renewal of mouse embryonic stem
(ES) cells (23). miR-125a, along
with let-7, induces stem cell commitment by regulating the
expression of the mammalian lin-28 (a marker for pluripotency in ES
cells) (26,44) and its expression is significantly
increased during the neural differentiation of ES and
embryocarcinoma (EC) cells (26,44).
The downregulation of miR-21 and miR-125a (Table I) parallels the enriched progenitor
cells in MDA-MB-231 and SUM1315 cells compared to MCF-7 cells
(Fig. 2). miR-203 has been shown to
control the keratinocyte differentiation of epithelial progenitor
cells (16) and to be a tumor
suppressor (17). This miRNA was
depleted in a number of hematopoietic tumors due to
hypermethylation, while its re-expression is capable of reducing
tumor cell proliferation (17). Its
expression was found to be markedly reduced in MDA-MB-231 and
SUM1315 cells compared to MCF-7 cells (Table I and Fig. 3). miRNA depletion in metastatic
MDA-MB-231 and SUM1315 cells correlates with the more aggressive
phenotype of these cells compared to non-metatastic MCF-7 cells
(14).
To shed light on the potential role of
differentially expressed miRNAs in modulating tumor-initiating
features, an analysis of target gene prediction was performed using
the online miRGen database. Most miRNAs were predicted to have
numerous target genes. miRNAs such as miR-99b, miR-200c and miR-7
have tens of predicted target genes, whereas other miRNAs (e.g.,
miR-148b, miR-375 and miR-20a) have hundreds of predicted target
genes. Representative genes involved in the stemness are shown in
Table I. LATS2, a potential target
gene of miR-93, is essential for embryonic development,
proliferation control and genomic integrity (45). A possible target gene of miR-21,
SATB1, contributes to embryonic stem cell differentiation and
regulates Nanog expression (46).
As a likely target gene of miR-22, PLAGL2 restrains differentiation
in neural stem cells (47).
Although the published data on miRNA profiles in T-ICs are limited,
evidence suggests that certain miRNAs play a significant role in
the generation and maintenance of T-ICs. In C. elegans,
mutation of lin-4 and let-7 miRNAs results in the
generation of stem cell-like cells (48). Mammalian let-7 expression is
depleted in human BrCa T-ICs and significantly increased in
differentiated cells. Let-7 suppresses self-renewal by
targeting H-RAS and induces differentiation by targeting HMGA2
(10). The data from the present
study further support the hypothesis that miRNAs are involved in
the regulation of BrCa T-ICs. Future studies of the aberrantly
expressed miRNAs in BrCa T-ICs may elucidate the mechanisms
responsible for tumorigenesis, metastasis and cancer relapse, and
may identify therapeutic molecular targets for anti-cancer drug
discovery.
Taken together, these results indicate that BrCa
cells with a higher prevalence of T-ICs possess distinct miRNA
expression profiles that may contribute to the maintenance of stem
cell properties. The mechanism by which miRNAs modulate BrCa T-ICs
remains unknown. Moreover, our study suggests the potential
diagnostic value of the miRNA expression signature in BrCa,
including metastasis, although clinical evidence is required to
confirm this hypothesis.
Acknowledgements
This study was supported by grants from the 'Hundred
Talents Program', the Chinese Academy of Sciences (KZCX2-EW-404),
and the National Natural Science Foundation of China (21077128 and
20921063).
Abbreviations:
|
miRNAs
|
microRNAs
|
|
BrCa
|
breast cancer
|
|
T-ICs
|
tumor-initiating cells
|
|
ES
|
embryonic stem
|
|
EC
|
embryocarcinoma
|
|
FACS
|
fluorescence-activated cell
sorting
|
|
RT-PCR
|
reverse transcription polymerase chain
reaction
|
|
RT
|
reverse transcription
|
References
|
1
|
Shipitsin M, Campbell LL, Argani P, et al:
Molecular definition of breast tumor heterogeneity. Cancer Cell.
11:259–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ponti D, Costa A, Zaffaroni N, et al:
Isolation and in vitro propagation of tumorigenic breast
cancer cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005.PubMed/NCBI
|
|
3
|
Deng SCG, Croce CM, Coukos G and Zhang L:
Mechanisms of microRNA deregulation in human cancer. Cell Cycle.
7:2643–2646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Suh M-R, Lee Y, Kim JY, et al: Human
embryonic stem cells express a unique set of microRNAs. Dev Biol.
270:488–498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bernstein EKS, Carmell MA, Murchison EP,
Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV and Hannon GJ:
Dicer is essential for mouse development. Nat Genet. 35:215–217.
2003. View
Article : Google Scholar
|
|
8
|
Kanellopoulou C, Muljo SA, Kung AL, et al:
Dicer-deficient mouse embryonic stem cells are defective in
differentiation and centromeric silencing. Genes Dev. 19:489–501.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu S, Goldstein RH, Scepansky EM and
Rosenblatt M: Inhibition of rho-associated kinase signaling
prevents breast cancer metastasis to human bone. Cancer Res.
69:8742–8751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu S, Suragani RN, Wang F, Han A, Zhao W,
Andrews NC and Chen JJ: The function of heme-regulated eIF2alpha
kinase in murine iron homeostasis and macrophage maturation. J Clin
Invest. 117:3296–3305. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fillmore CM and Kuperwasser C: Human
breast cancer cell lines contain stem-like cells that self-renew,
give rise to phenotypically diverse progeny and survive
chemotherapy. Breast Cancer Res. 10:R252008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuperwasser C, Dessain S, Bierbaum BE, et
al: A mouse model of human breast cancer metastasis to human bone.
Cancer Res. 65:6130–6138. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kang Y, Siegel PM, Shu W, et al: A
multigenic program mediating breast cancer metastasis to bone.
Cancer Cell. 3:537–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hinton A, Afrikanova I, Wilson M, et al: A
distinct microRNA signature for definitive endoderm derived from
human embryonic stem cells. Stem Cells and Dev. 19:797–807. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lena AM, Shalom-Feuerstein R, Di Val Cervo
PR, et al: miR-203 represses 'stemness' by repressing [Delta]Np63.
Cell Death Differ. 15:1187–1195. 2008.
|
|
17
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, et al: Genetic and epigenetic silencing of microRNA-203
enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell.
13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shimono Y, Zabala M, Cho RW, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pek JW, Lim AK and Kai T:
Drosophila maelstrom ensures proper germline stem cell
lineage differentiation by repressing microRNA-7. Dev Cell.
17:417–424. 2009. View Article : Google Scholar
|
|
20
|
Mallick B, Ghosh Z and Chakrabarti J:
MicroRNome analysis unravels the molecular basis of SARS infection
in bronchoalveolar stem cells. PLoS ONE. 4:e78372009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wellner U, Schubert J, Burk UC, et al: The
EMT-activator ZEB1 promotes tumorigenicity by repressing
stemness-inhibiting microRNAs. Nat Cell Biol. 11:1487–1495. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma Y-L, Zhang P, Wang F, et al: Human
embryonic stem cells and metastatic colorectal cancer cells shared
the common endogenous human microRNA-26b. J Cell Mol Med.
View Article : Google Scholar : (E-pub ahead of
print). Accessed September 10, 2010
|
|
23
|
Singh SK, Kagalwala MN, Parker-Thornburg
J, Adams H and Majumder S: REST maintains self-renewal and
pluripotency of embryonic stem cells. Nature. 453:223–227. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schoolmeesters A, Eklund T, Leake D, et
al: Functional profiling reveals critical role for miRNA in
differentiation of human mesenchymal stem cells. PLoS ONE.
4:e56052009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ibarra I, Erlich Y, Muthuswamy SK,
Sachidanandam R and Hannon GJ: A role for microRNAs in maintenance
of mouse mammary epithelial progenitor cells. Genes Dev.
21:3238–3243. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rybak A, Fuchs H, Smirnova L, et al: A
feedback loop comprising lin-28 and let-7 controls pre-let-7
maturation during neural stem-cell commitment. Nat Cell Biol.
10:987–993. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qian S, Ding JY, Xie R, et al: MicroRNA
expression profile of bronchioalveolar stem cells from mouse lung.
Biochem Biophys Res Commun. 377:668–673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lakshmipathy U and Hart RP: Concise
review: microRNA expression in multipotent mesenchymal stromal
cells. Stem Cells. 26:356–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Godlewski J, Nowicki MO, Bronisz A, et al:
Targeting of the Bmi-1 oncogene/stem cell renewal factor by
microRNA-128 inhibits glioma proliferation and self-renewal. Cancer
Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Foshay KM and Gallicano GI: miR-17 family
miRNAs are expressed during early mammalian development and
regulate stem cell differentiation. Dev Biol. 326:431–443. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rogler CE, LeVoci L, Ader T, et al:
MicroRNA-23b cluster microRNAs regulate transforming growth
factor-beta/bone morphogenetic protein signaling and liver stem
cell differentiation by targeting Smads. Hepatology. 50:575–584.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee S, Jung J-W, Park S-B, et al: Histone
deacetylase regulates high mobility group A2-targeting microRNAs in
human cord blood-derived multipotent stem cell aging. Cell Mol Life
Sci. 68:325–336. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sengupta S, Nie J, Wagner RJ, Yang C,
Stewart R and Thomson JA: MicroRNA 92b controls the G1/S checkpoint
gene p57 in human embryonic stem cells. Stem Cells. 27:1524–1528.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tzur G, Levy A, Meiri E, et al: MicroRNA
expression patterns and function in endodermal differentiation of
human embryonic stem cells. PLoS ONE. 3:e37262008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tsai Z-Y, Singh S, Yu S-L, et al:
Identification of microRNAs regulated by activin A in human
embryonic stem cells. J Cell Biochem. 109:93–102. 2010.PubMed/NCBI
|
|
37
|
Wagner W, Horn P, Castoldi M, et al:
Replicative senescence of mesenchymal stem cells: A continuous and
organized process. PLoS ONE. 3:e22132008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang K-H, Kao A-P, Singh S, et al:
Comparative expression profiles of mRNAs and microRNAs among human
mesenchymal stem cells derived from breast, face, and abdominal
adipose tissues. Kaohsiung J Med Sci. 26:113–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Han Y-C, Park CY, Bhagat G, et al:
microRNA-29a induces aberrant self-renewal capacity in
hematopoietic progenitors, biased myeloid development, and acute
myeloid leukemia. J Exp Med. 207:475–489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi L, Zhang J, Pan T, et al: MiR-125b is
critical for the suppression of human U251 glioma stem cell
proliferation. Brain Res. 1312:120–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tarantino C, Paolella G, Cozzuto L, et al:
miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic
stem cells. FASEB J. 24:3255–3263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Decembrini S, Bressan D, Vignali R, et al:
MicroRNAs couple cell fate and developmental timing in retina. Proc
Natl Acad Sci USA. 106:21179–21184
|
|
43
|
Poliseno L, Tuccoli A, Mariani L, et al:
MicroRNAs modulate the angiogenic properties of HUVECs. Blood.
108:3068–3071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu L and Belasco JG: Micro-RNA regulation
of the mammalian lin-28 gene during neuronal differentiation of
embryonal carcinoma cells. Mol Cell Biol. 25:9198–9208. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McPherson JP, Tamblyn L, Elia A, et al:
Lats2/Kpm is required for embryonic development, proliferation
control and genomic integrity. Embo J. 23:3677–3688. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Savarese F, Dávila A, Nechanitzky R, et
al: Satb1 and Satb2 regulate embryonic stem cell differentiation
and Nanog expression. Genes Dev. 23:2625–2638. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng H, Ying H, Wiedemeyer R, et al:
PLAGL2 regulates Wnt signaling to impede differentiation in neural
stem cells and gliomas. Cancer Cell. 17:497–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Reinhart BJ, Slack FJ, Basson M, et al:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|