Contents
Introduction
Active autophagy in tumor microenvironment and
cancer cell fate
Hypoxia and anoxia
Nutrient deprivation
ECM detachment
ER stress
Autophagy induced by tumor microenvironmental
stresses and tumor metastasis
Manipulating autophagy induced by tumor
microenvironmental stresses for cancer therapy
Conclusions/perspectives
Introduction
Autophagy is an evolutionarily conserved catabolic
process in which intracellular membrane structures sequester
proteins and organelles to degrade and turn over these cytoplasmic
constituents; thus, it is essential for growth regulation and the
maintenance of homeostasis (1–3).
Autophagy is a multi-step process characterized by nucleation,
elongation and autophagosome and autolysosome formation, and is
tightly regulated by a limited number of highly conserved genes
called autophagy regulators (ATGs) (4,5).
Defective autophagy is correlated with diverse pathologies,
including neurodegeneration, liver, heart and muscle diseases,
ageing, inflammation and cancer (6).
Autophagy is activated in response to multiple
stresses during cancer progression, including hypoxia, nutrient
deprivation, extracellular matrix (ECM) detachment, endoplasmic
reticulum (ER) stress and other diverse stresses (7,8).
Autonomous proliferating cancer cells are often exposed to
conditions such as hypoxia or/and nutrient deprivation, so there
must be an alternative metabolic pathway to protect tumor cells
from these environmental stresses (9). Moreover, in order to metastasize,
tumor cells must adapt to a stressful microenvironment as they
disseminate into the systemic circulation and colonize distant
organ sites (10). Therefore, when
environmental stresses emerge, tumor cells are able to catabolize
existing cytoplasmic components to provide essential ingredients to
maintain survival by autophagy (11).
Autophagy facilitates cellular survival by enabling
cancer cells to grow under stressful conditions. The enhancement of
autophagy leads to degradation of proteins and organelles to
provide amino acids, fatty acids and nucleotides for reuse
(12). It is increasingly
appreciated that autophagy provides cancer cells with certain
selective advantages in response to various stresses in the primary
tumor microenvironment as well as the microenvironment during
dissemination and metastasis (13).
Paradoxically, however, in certain cases autophagy also contributes
to the death of cancer cells by scavenging damaged oxidative
organelles (14). In this review,
we argue that understanding the net effect of autophagy on enabling
cells to cope with diverse stresses of the microenvironment, and
thereby controlling the fate of cancer cells and metastasis, may
develop new therapeutic strategies based on the regulation of
autophagy.
Active autophagy in tumor microenvironment
and cancer cell fate
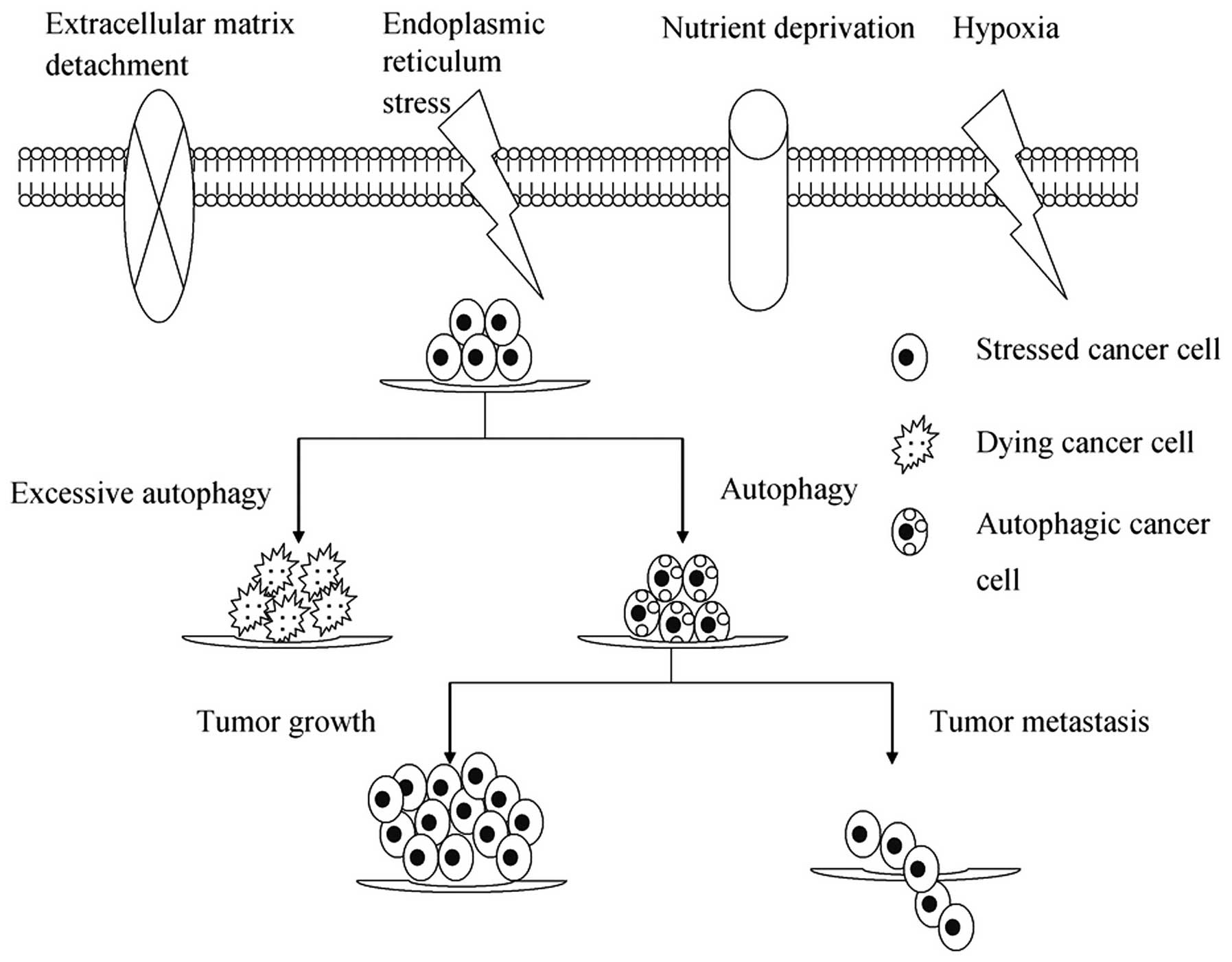
Microenvironmental stresses, as a result of either
insufficient oxygen/nutrient supply or increased energetic demands
of rapidly dividing tumor cells, induce autophagy as an alternative
source of energy and metabolites to ensure that cell growth is
appropriate to the environmental conditions (15). Increasing evidence suggests that
autophagy constitutes a major protective mechanism that allows
cells to survive in response to multiple stresses, including
hypoxia, nutrient deprivation, ECM detachment and ER and other
stresses (15–17). However, if microenvironmental
stresses persist, excessive autophagy may ultimately lead to
autophagic cell death, termed type II-programmed cell death
(Fig. 1).
Hypoxia and anoxia
Hypoxia and anoxia (with oxygen concentrations
<3% and <0.1%, respectively) induce autophagy through a
variety of different mechanisms (18). Enhanced autophagy is frequently
observed in hypoxic regions of solid tumors caused by inadequate
vascularization and contributes to cell survival (19). These hypoxic regions are considered
to be associated with altered cellular metabolism and poor
prognosis. The main transcription factors mediating the hypoxic
response are hypoxia-inducible factors (HIFs), which modulate tumor
cell metabolism, angiogenesis, growth and metastasis (20). Bcl-2/adenovirus E1B 19
kDa-interacting protein (BNIP3), a BH3-only protein, is a
downstream target of HIF-1α and has been shown to induce autophagy
by disrupting the Beclin 1-Bcl-2 complex and releasing Beclin 1 in
response to a hypoxic microenvironment (21,22).
BNIP3L (BNIP3-like protein, also known as NIX), another
HIF-1-induced target, is also important for targeting the
mitochondria to autophagosomes for clearance (23). Further study has revealed hypoxia-
and oxidative stress-mediated activation of the HIF-1α and NFκB
pathway in fibroblasts, thereby driving the autophagic flux to
promote tumor cell survival (24).
HIF-2 is also a potent regulator of chondrocyte autophagy and this
protein acts as a brake to the stimulatory function of HIF-1
(25). Recently, the epidermal
growth factor receptor antibody cetuximab was found to induce
autophagy in cancer cells by downregulating HIF-1α and Bcl-2 and
activating the Beclin 1/hVps34 complex (26). In addition, several distinct oxygen
sensing pathways that regulate the cellular response to hypoxia
have been defined, including activation of the unfolded protein
response (UPR), inhibition of the mammalian target of rapamycin
(mTOR) kinase signaling pathway and activation of AMP-responsive
protein kinase (AMPK), which are all associated with the induction
of autophagy (Fig. 2) (27,28).
Although hypoxia-driven tumor metabolism and autophagy have been
demonstrated, a more detailed mechanism of the interaction between
autophagy and a hypoxic tumor microenvironment remains to be
determined.
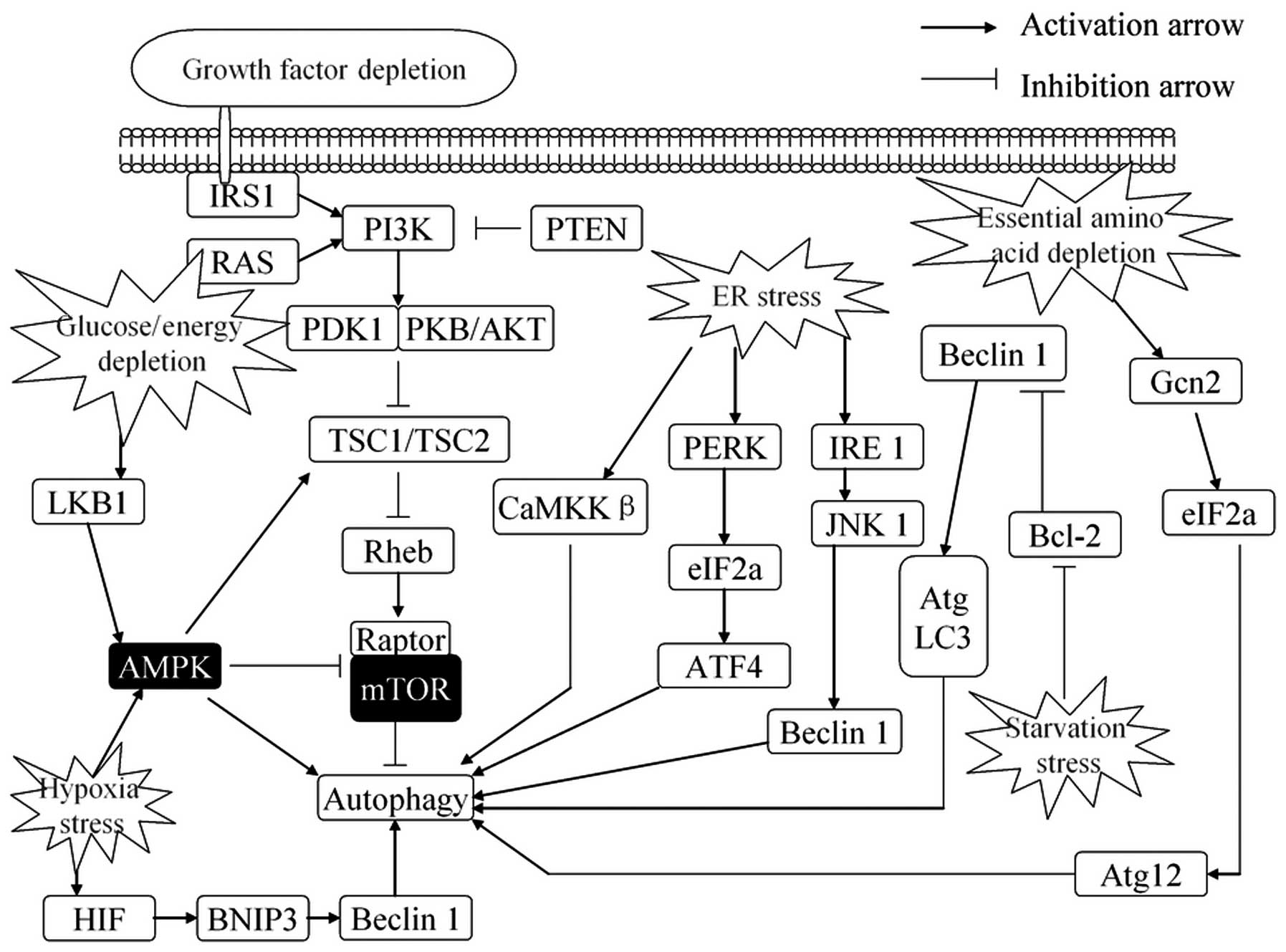 | Figure 2.Regulation of autophagy in response
to stress. Autophagy is activated in response to multiple stresses
during cancer progression, including nutrient deprivation, ER
stress, hypoxia, glucose/energy depletion and other diverse
stresses. ER stress stimulates autophagy through the PERK-eIF2α
pathway, IRE1-JNK1 pathway and Ca2+ release. Growth
factors, through AKT-dependent and ERK-dependent phosphorylation,
suppress autophagy. Depletion of nutrients or energy (amino acids,
glucose, energy or serum) induces autophagy by activating the AMPK
pathway or promoting upregulate transcription of certain autophagy
genes. Autophagy is also induced by hypoxia that signals via AMPK
to inhibit mTOR activity or disrupt the Bcl-2-Beclin 1 interaction
and activate Beclin 1. Conversely, autophagy is inhibited by
increased growth factor signaling through the activation of the
Class I group of PI3-kinases and Akt to promote mTOR activity. ER,
endoplasmic reticulum; ERK, extracellular signal-regulated kinase;
AMPK, AMP-responsive protein kinase; ATF4, activating transcription
factor 4; mTOR, mammalian target of rapamycin; BNIP3,
Bcl-2/adenovirus E1B 19 kDa-interacting protein; ATG, autophagy
regulator. |
Nutrient deprivation
Proliferating cancer cells require continuous access
to resources that sustain intracellular energy and nutrient levels,
but the tumor microenvironment is not sufficient to supply these
essential ingredients for cancer cell survival (29). Under these conditions, cancer cells
are likely to encounter a shortage of nutrients; therefore, cancer
cells must seek alternative metabolic processes to cope with this
stress and maintain their survival. Studies have shown that
autophagy plays a critical role in protecting cells against a
shortage of nutrients by removing damaged substrates for recycling,
but the exact mechanism by which cancer cells obtain energy sources
under conditions in which their external nutrient supply is
extremely limited remains unclear (30,31).
Nutrient (including amino acids and glucose)
depletion is the most potent known physiological inducer of
autophagy. Ammonia, generated from glutamine deamination in
mitochondria, was found to function as an autocrineand/or
paracrine-acting stimulator of autophagic flux (32). Autophagosomes were actively induced
and promptly consumed in colorectal cancer cells under amino acid-
and glucose-deprived conditions, which may contribute to the
survival of the cancer cells in their microenvironment (29). Glucose deprivation may cause
oxidative stress and stimulate autophagy (33). mTOR and AMPK have been best
characterized as critical signaling pathways regulating nutrient
deprivation-induced autophagy (Fig.
2) (25,34). Autophagy is also triggered to
protect cancer cells from nutrient deprivation by activation of
AMPK (35). A previous study has
suggested that ubiquilins also accelerate autophagosome maturation
and promote cell survival during nutrient starvation (36). The cellular amino acids, especially
branched chain amino acids, are a crucial upstream component for
the functional activation of mTORC1. The absence of amino acids
induces autophagy through the regulation of mTOR activity (Fig. 2) (37). In addition to amino acids, cells
must also be supplied with glucose to maintain a constant supply of
ATP; during a lack of glucose, autophagy is often activated to
maintain intracellular energy homeostasis (38,39).
Moreover, it has been reported that the receptor for advanced
glycation end products (RAGE) sustains autophagy and limits
apoptosis by inhibiting mTOR, resulting in the promotion of
pancreatic tumor cell survival (40). Overall, autophagy constitutes a
major protective mechanism that allows cells to survive nutrient
deprivation.
ECM detachment
Integrin-mediated attachment of epithelial cells to
the ECM is vital for cell growth and survival (41). The loss of ECM attachment leads to
apoptosis, termed anoikis (42).
However, previous studies have shown that a lack of appropriate
matrix contact also robustly induces autophagy to promote cell
survival, either during early carcinoma formation or in the later
stages of dissemination and metastasis (43,44).
Moreover, ECM components modulate autophagy and mitigate its role
in cell survival. In HeLa cells, the mechanism by which this occurs
has been shown to be dependent on the adhesion of the cells to
collagen I or IV (45). In a
three-dimensional (3D) culture system using MCF10A mammary
epithelial cells grown in low ECM attachment conditions, autophagy
was rapidly induced to enhance cell survival during anoikis
(46). Although the intracellular
signals linking ECM detachment to autophagy remain unclear, the
results suggest that autophagy may be a previously unrecognized
mechanism which enhances the survival of tumor cells lacking proper
ECM contact.
ER stress
The ER is an organelle responsible for crucial
biosynthetic and signaling functions in eukaryotic cells (47). Dysfunction of ER or ER stress may
result from various disturbances, including hypoxia and oxidative
stress, which elicit a cellular stress response known as the UPR
(48). The UPR initially serves as
an adaptive mechanism to maintain ER homeostasis. However, severe
or prolonged ER stress also switches the cytoprotective functions
of UPR and autophagy into cell death, usually by activating
intrinsic apoptosis (49).
It has been recognized that in order to clear the
accumulation of terminally misfolded protein aggregates that cannot
be degraded by the proteasome, the UPR may upregulate the autophagy
machinery (50). Activating
transcription factor 4 (ATF4) has been shown to facilitate
autophagy through direct binding to a cyclic AMP response element
binding site in response to ER stress (51). Activation of AMPK by atorvastatin
enhances p21 expression and ER stress response, leading to
autophagy, which promotes the survival of cancer cells (52). Autophagy may also eliminate a
specific type of misfolded procollagen and play a protective role
in cell survival against ER stress (53). By contrast, persistent ER stress
also induces cell death by activating apoptosis. Cannabinoid action
induces autophagy-mediated cell death through stimulation of ER
stress in human glioma cells (54).
Moreover, the ER stress activates radiation-induced autophagy by
PERK-eIF2α in caspase-3/7-deficient cells, which promotes
radiosensitivity in vitro and in vivo(55). It has been demonstrated that ER
stress-induced cell death was mediated by autophagy (56), which was partly attributed to the
inactivation of AKT/TSC/mTOR (Fig.
2). As discussed above, it is clear that ER stress and
autophagy are capable of activating prosurvival mechanisms as well
as lethal programs, but the specific mechanisms linking UPR to
autophagy during ER stress remain poorly understood.
Autophagy induced by tumor
microenvironmental stresses and tumor metastasis
Tumor microenvironmental stresses have recently
gained much attention as a critical determinant of tumor
progression since autophagy is often induced as a major protective
mechanism that allows cells to survive in response to these
stresses. In addition, some clinical evidence suggests that
autophagy is used as a survival strategy by established tumors to
promote tumor progression.
Autophagy may promote metastasis by enhancing tumor
cell fitness in response to microenvironmental stresses. Pancreatic
cancer remains a devastating and poorly understood malignant cancer
and hypoxia in pancreatic cancers is known to increase malignant
potential. In the peripheral area of pancreatic cancer tissue, high
expression of LC3, a key component of autophagy, is correlated with
poor overall survival and a shorter disease-free period (57). Recent study has also suggested that
high expression of the autophagy-related Beclin 1 protein predicts
poorer overall survival, progression-free survival and distant
metastasis-free survival for nasopharyngeal carcinoma patients
(58). The microtubule-associated
protein 1 light chain 3 (LC3A) is an essential component of the
autophagic vacuoles and LC3A immunohistochemistry renders three
patterns of autophagic expression in breast carcinomas: diffuse
cytoplasmic, perinuclear and ‘stone-like’ intracellular structures
(SLS). Perinuclear LC3A accumulation in colorectal tumour cells is
a marker of good prognosis, while high SLS counts were associated
with metastases and poor prognosis (59). Phospho-enriched protein in
astrocytes (PEA-15) is a 15-kDa phosphoprotein that induces
autophagy in human ovarian cancer cells and is associated with
prolonged overall survival (60).
γ-aminobutyric acid type A (GABAA) receptor-associated protein
(GABARAP), the mammalian homolog of yeast Atg8, is involved in
autophagosome formation during autophagy and is a new independent
prognostic marker for colorectal carcinoma and the overexpression
of this protein is associated with poor differentiation as well as
shortened overall survival in colorectal cancers (61).
Conversely, autophagy may also inhibit metastasis.
Beclin 1 and LC3, crucial genes for autophagy, are altered in
several types of human cancer. A higher level of Beclin 1
expression is strongly associated with longer survival of colon
cancer patients with stage IIIB disease (62). Autophagy-active Beclin 1 has also
been shown to be significantly correlated with the survival of
non-Hodgkin lymphoma patients (63). Moreover, Beclin 1 and LC3
significantly decrease with melanoma progression (64). Beclin 1 may play a role in the
inhibition of the development of breast cancer and this inhibition
may be due to an interaction with Bcl-2 protein and inactivation of
PI3K/PKB signaling pathway (65,66).
The high expression level of Beclin 1 protein has been demonstrated
to be positively correlated with apoptosis and negatively with cell
proliferation in gliomas (67).
Beclin 1 defects caused by the overexpression of Bcl-xL may
facilitate tumor malignant differentiation, which results in a more
aggressive cancer cell phenotype and poor prognosis of
hepatocellular carcinoma (68). Low
Beclin 1 expression is associated with worse overall survival and
progression-free survival in extranodal natural killer T-cell
lymphoma (69).
Although these proteins have been used to detect and
measure levels of autophagy in human tumor samples, few may be
universally and accurately applied for autophagy detection in
clinical samples. Consequently, there is a rapidly growing need for
exploiting ‘gold standard’ for methods and better markers to
monitor autophagic activity (70).
Manipulating autophagy induced by tumor
microenvironmental stresses for cancer therapy
As discussed above, cancer cells gain survival and
proliferation advantages by autophagy to cope with
micro-environmental stresses. Despite the determination of the
survival-promoting role of autophagy, it is also well recognized
that elevated and/or prolonged autophagy may result in cell death.
Therefore, inhibiting autophagy induced by tumor microenvironmental
stresses or enhancing excessive microenvironmental stresses to give
rise to autophagic cell death may be a promising strategy for
cancer therapy. Based on the correlation between microenvironmental
stresses and autophagy, certain chemotherapeutic agents and
antineoplastic therapies have been reported as an adjuvant therapy
for cancer, including acid sphingomyelinase (71), thiazolidinediones (72), tetraspanin (73), bortezomib (74), Δ(9)-tetrahydrocannabinol (54), etformin (75), 2-deoxyglucose (76) and the arginine deiminase ADI-PEG20
(77). However, this therapy has
not been further explored for clinical application. In order to
accelerate this clinical application, large-scale and multicenter
collaboration are necessary.
Conclusions/perspectives
Autophagy is a catabolic adaptive process usually
activated in response to adverse microenvironmental stresses which
may have either a beneficial or detrimental cellular effect,
depending on the response to environmental stresses (78,79).
Currently, it is becoming clear that autophagy is a survival
pathway that enables tumor cells to survive under stressful
conditions, including hypoxia, nutrient deprivation, ECM detachment
and ER stress. By contrast, prolonged activation of autophagy may
lead to cell death by cellular self-degradation (80–82).
The tumor environment is a complex and highly
dynamic environment, playing a central role in controlling tumor
cell behavior and metastasis formation (83). Reduced levels of oxygen and
nutrients and malfunction of ECM and ER are critical parameters
modulating the tumor microenvironment. As discussed above,
abnormality in the tumor microenvironment induces autophagy to aid
the maintenance of cancer cell viability and promote cancer cell
metastasis under these stressful conditions. However, in certain
cases autophagy also contributes to cancer cell death and inhibits
metastasis. Based on the functional correlation between
microenvironmental stresses and autophagy, a number of new cancer
therapeutics have been exploited, but certain limitations prevent
widespread clinical application. First, the question of whether we
should try to enhance or inhibit autophagy in cancer treatment is
not straightforward since it is unclear how autophagic cell death
is distinguished from autophagy during cell survival. The
engulfment receptor Draper was found to be the first factor that
distinguishes autophagy associated with cell death from that
associated with cell survival (84). This finding is especially critical
since numerous current cancer therapeutics activate or inhibit
autophagy, although Draper has not been applied to cancer research.
Second, to maximize the potential to be applied for more stringent
clinical study, characteristics of methods and better markers to
monitor autophagic activity may need to be examined. Third,
published studies concerning antineoplastic therapies based on the
correlation between the autophagy and tumor microenvironment are
short of high-level clinical evidence. Large-scale and multicenter
collaborations are necessary in the future. Finally, the molecular
mechanisms that underlie autophagy induced by multiple tumor
microenvironmental stresses and cancer metastasis remain to be
determined.
Abbreviations:
|
ATGs
|
autophagy regulators
|
|
AMPK
|
AMP-responsive protein kinase
|
|
ATF4
|
activating transcription factor 4
|
|
BNIP3
|
Bcl-2/adenovirus E1B 19
kDa-interacting protein
|
|
BNIP3L
|
BNIP3-like protein
|
|
ECM
|
extracellular matrix
|
|
HIFs
|
hypoxia-inducible factors
|
|
mTOR
|
mammalian target of rapamycin
|
|
SLS
|
‘stone-like’ intracellular
structures
|
|
UPR
|
unfolded protein response
|
References
|
1.
|
Klionsky DJ: Autophagy: from phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Hippert MM, O’Toole PS and Thorburn A:
Autophagy in cancer: good, bad, or both? Cancer Res. 66:9349–9351.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Høyer-Hansen M and Jäättelä M: Autophagy:
an emerging target for cancer therapy. Autophagy. 4:574–580.
2008.
|
|
4.
|
Chen N and Debnath J: Autophagy and
tumorigenesis. FEBS Lett. 584:1427–1435. 2010. View Article : Google Scholar
|
|
5.
|
Tsuchihara K, Fujii S and Esumi H:
Autophagy and cancer: dynamism of the metabolism of tumor cells and
tissues. Cancer Lett. 278:130–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Bao XH, Naomoto Y, Hao HF, Watanabe N,
Sakurama K, Noma K, et al: Autophagy: Can it become a potential
therapeutic target? Int J Mol Med. 25:493–503. 2010.PubMed/NCBI
|
|
7.
|
Kondo Y, Kanzawa T, Sawaya R and Kondo S:
The role of autophagy in cancer development and response to
therapy. Nat Rev Cancer. 5:726–734. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Yang Z and Klionsky DJ: An overview of the
molecular mechanism of autophagy. Curr Top Microbiol Immunol.
335:1–32. 2009.PubMed/NCBI
|
|
9.
|
Vousden KH and Ryan KM: p53 and
metabolism. Nat Rev Cancer. 9:691–700. 2009. View Article : Google Scholar
|
|
10.
|
Kenific CM, Thorburn A and Debnath J:
Autophagy and metastasis: another double-edged sword. Curr Opin
Cell Biol. 22:241–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Lum JJ, DeBerardinis RJ and Thompson CB:
Autophagy in metazoans: cell survival in the land of plenty. Nat
Rev Mol Cell Biol. 6:439–448. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Apel A, Zentgraf H, Büchler MW and Herr I:
Autophagy-A double-edged sword in oncology. Int J Cancer.
125:991–995. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Roy S and Debnath J: Autophagy and
tumorigenesis. Semin Immunopathol. 32:383–396. 2010. View Article : Google Scholar
|
|
14.
|
Brech A, Ahlquist T, Lothe RA and Stenmark
H: Autophagy in tumour suppression and promotion. Mol Oncol.
3:366–375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wang RC and Levine B: Autophagy in
cellular growth control. FEBS Lett. 584:1417–1426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Altman BJ and Rathmell JC: Autophagy: not
good OR bad, but good AND bad. Autophagy. 5:569–570. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Rosenfeldt MT and Ryan KM: The role of
autophagy in tumour development and cancer therapy. Expert Rev Mol
Med. 11:e362009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Kroemer G, Mariño G and Levine B:
Autophagy and the integrated stress response. Mol Cell. 40:280–293.
2010. View Article : Google Scholar
|
|
19.
|
Degenhardt K, Mathew R, Beaudoin B, Bray
K, Anderson D, Chen G, et al: Autophagy promotes tumor cell
survival and restricts necrosis, inflammation, and tumorigenesis.
Cancer Cell. 10:51–64. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Bertout JA, Patel SA and Simon MC: The
impact of O2 availability on human cancer. Nat Rev
Cancer. 8:967–975. 2008.
|
|
21.
|
Mazure NM and Pouysségur J:
Hypoxia-induced autophagy: cell death or cell survival. Curr Opin
Cell Biol. 22:177–180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Mazure NM and Pouysségur J: Atypical
BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia.
Autophagy. 5:868–869. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Sandoval H, Thiagarajan P, Dasgupta SK,
Schumacher A, Prchal JT, Chen M and Wang J: Essential role for Nix
in autophagic maturation of erythroid cells. Nature. 454:232–235.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Martinez-Outschoorn UE, Trimmer C, Lin Z,
Whitaker-Menezes D, Chiavarina B, Zhou J, et al: Autophagy in
cancer associated fibroblasts promotes tumor cell survival: Role of
hypoxia, HIF1 induction and NFκB activation in the tumor stromal
microenvironment. Cell Cycle. 9:3515–3533. 2010.PubMed/NCBI
|
|
25.
|
Srinivas V, Bohensky J, Zahm AM and
Shapiro IM: Autophagy in mineralizing tissues: microenvironmental
perspectives. Cell Cycle. 8:391–393. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Li X and Fan Z: The epidermal growth
factor receptor antibody cetuximab induces autophagy in cancer
cells by downregulating HIF-1alpha and Bcl-2 and activating the
beclin 1/hVps34 complex. Cancer Res. 70:5942–5952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Rouschop KM and Wouters BG: Regulation of
autophagy through multiple independent hypoxic signaling pathways.
Curr Mol Med. 9:417–424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Pouysségur J, Dayan F and Mazure NM:
Hypoxia signalling in cancer and approaches to enforce tumour
regression. Nature. 441:437–443. 2006.PubMed/NCBI
|
|
29.
|
Sato K, Tsuchihara K, Fujii S, Sugiyama M,
Goya T, Atomi Y, et al: Autophagy is activated in colorectal cancer
cells and contributes to the tolerance to nutrient deprivation.
Cancer Res. 67:9677–9684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Moreau K, Luo S and Rubinsztein DC:
Cytoprotective roles for autophagy. Curr Opin Cell Biol.
22:206–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Jin S and White E: Role of autophagy in
cancer: management of metabolic stress. Autophagy. 3:28–31. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Eng CH and Abraham RT: Glutaminolysis
yields a metabolic by-product that stimulates autophagy. Autophagy.
6:968–970. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Marambio P, Toro B, Sanhueza C, Troncoso
R, Parra V, Verdejo H, et al: Glucose deprivation causes oxidative
stress and stimulates aggresome formation and autophagy in cultured
cardiac myocytes. Biochim Biophys Acta. 1802:509–518. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Neufeld TP: TOR-dependent control of
autophagy: biting the hand that feeds. Curr Opin Cell Biol.
22:157–168. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Lum JJ, DeBerardinis RJ and Thompson CB:
Autophagy in metazoans: cell survival in the land of plenty. Nat
Rev Mol Cell Biol. 6:439–448. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
N’Diaye EN, Debnath J and Brown EJ:
Ubiquilins accelerate autophagosome maturation and promote cell
survival during nutrient starvation. Autophagy. 5:573–575.
2009.PubMed/NCBI
|
|
37.
|
Liao XH, Majithia A, Huang X and Kimmel
AR: Growth control via TOR kinase signaling, an intracellular
sensor of amino acid and energy availability, with crosstalk
potential to proline metabolism. Amino Acids. 35:761–770. 2008.
View Article : Google Scholar
|
|
38.
|
Kumar SH and Rangarajan A: Simian virus 40
small T antigen activates AMPK and triggers autophagy to protect
cancer cells from nutrient deprivation. J Virol. 83:8565–8574.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Hardie DG: AMP-activated/SNF1 protein
kinases: conserved guardians of cellular energy. Nat Rev Mol Cell
Biol. 8:774–785. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kang R, Tang D, Schapiro NE, Livesey KM,
Farkas A, Loughran P, et al: The receptor for advanced glycation
end products (RAGE) sustains autophagy and limits apoptosis,
promoting pancreatic tumor cell survival. Cell Death Differ.
17:666–676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Miranti CK and Brugge JS: Sensing the
environment: a historical perspective on integrin signal
transduction. Nat Cell Biol. 4:E83–E90. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Gilmore AP: Anoikis. Cell Death Differ.
12(Suppl 2): 1473–1477. 2005. View Article : Google Scholar
|
|
43.
|
Debnath J: Detachment-induced autophagy
during anoikis and lumen formation in epithelial acini. Autophagy.
4:351–353. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Lock R and Debnath J: Extracellular matrix
regulation of autophagy. Curr Opin Cell Biol. 20:583–588. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Tuloup-Minguez V, Greffard A, Codogno P
and Botti J: Regulation of autophagy by extracellular matrix
glycoproteins in HeLa cells. Autophagy. 77:27–39. 2011. View Article : Google Scholar
|
|
46.
|
Debnath J, Mills KR, Collins NL, Reginato
MJ, Muthuswamy SK and Brugge JS: The role of apoptosis in creating
and maintaining luminal space within normal and oncogene-expressing
mammary acini. Cell. 111:29–40. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Inagi R: Endoplasmic reticulum stress as a
progression factor for kidney injury. Curr Opin Pharmacol.
10:156–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Kaushik S, Singh R and Cuervo AM:
Autophagic pathways and metabolic stress. Diabetes Obes Metab.
12:4–14. 2010. View Article : Google Scholar
|
|
49.
|
Verfaillie T, Salazar M, Velasco G and
Agostinis P: Linking ER Stress to Autophagy: Potential Implications
for Cancer Therapy. Int J Cell Biol. 17:9305–9309. 2010.PubMed/NCBI
|
|
50.
|
Ogata M, Hino S, Saito A, Morikawa K,
Kondo S, Kanemoto S, et al: Autophagy is activated for cell
survival after endoplasmic reticulum stress. Mol Cell Biol.
26:9220–9231. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Rzymski T, Milani M, Pike L, Buffa F,
Mellor HR, Winchester L, et al: Regulation of autophagy by ATF4 in
response to severe hypoxia. Oncogene. 29:4424–4435. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Yang PM, Liu YL, Lin YC, Shun CT, Wu MS
and Chen CC: Inhibition of autophagy enhances anticancer effects of
atorvastatin in digestive malignancies. Cancer Res. 70:7699–7709.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Ishida Y and Nagata K: Autophagy
eliminates a specific species of misfolded procollagen and plays a
protective role in cell survival against ER stress. Autophagy.
5:1217–1219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Salazar M, Carracedo A, Salanueva IJ,
Hernández-Tiedra S, Lorente M, Egia A, et al: Cannabinoid action
induces autophagy-mediated cell death through stimulation of ER
stress in human glioma cells. J Clin Invest. 119:1359–1372. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Kim KW, Moretti L, Mitchell LR, Jung DK
and Lu B: Endoplasmic reticulum stress mediates radiation-induced
autophagy by perk-eIF2alpha in caspase-3/7-deficient cells.
Oncogene. 29:3241–3251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Qin L, Wang Z, Tao L and Wang Y: ER stress
negatively regulates AKT/TSC/mTOR pathway to enhance autophagy.
Autophagy. 6:239–247. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Fujii S, Mitsunaga S, Yamazaki M, Hasebe
T, Ishii G, Kojima M, et al: Autophagy is activated in pancreatic
cancer cells and correlates with poor patient outcome. Cancer Sci.
99:1813–1819. 2008.PubMed/NCBI
|
|
58.
|
Wan XB, Fan XJ, Chen MY, Xiang J, Huang
PY, Guo L, et al: Elevated Beclin 1 expression is correlated with
HIF-1alpha in predicting poor prognosis of nasopharyngeal
carcinoma. Autophagy. 6:395–404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Giatromanolaki A, Koukourakis MI, Harris
AL, Polychronidis A, Gatter KC and Sivridis E: Prognostic relevance
of light chain 3 (LC3A) autophagy patterns in colorectal
adenocarcinomas. J Clin Pathol. 63:867–872. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Bartholomeusz C, Rosen D, Wei C, Kazansky
A, Yamasaki F, Takahashi T, et al: PEA-15 induces autophagy in
human ovarian cancer cells and is associated with prolonged overall
survival. Cancer Res. 68:9302–9310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Miao Y, Zhang Y, Chen Y, Chen L and Wang
F: GABARAP is overexpressed in colorectal carcinoma and correlates
with shortened patient survival. Hepatogastroenterology.
57:257–261. 2010.PubMed/NCBI
|
|
62.
|
Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan
DS, et al: The expression of beclin 1 is associated with favorable
prognosis in stage IIIB colon cancers. Autophagy. 5:303–306. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Nicotra G, Mercalli F, Peracchio C,
Castino R, Follo C, Valente G and Isidoro C: Autophagy-active
beclin-1 correlates with favourable clinical outcome in non-Hodgkin
lymphomas. Mod Pathol. 23:937–950. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Miracco C, Cevenini G, Franchi A, Luzi P,
Cosci E, Mourmouras V, et al: Beclin 1 and LC3 autophagic gene
expression in cutaneous melanocytic lesions. Hum Pathol.
41:503–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Won KY, Kim GY, Kim YW, Song JY and Lim
SJ: Clinicopathologic correlation of beclin-1 and bcl-2 expression
in human breast cancer. Hum Pathol. 41:107–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Duan ZL, Peng ZL and Wang ZH: Expression
and involved signal transduction pathway of autophagy gene Beclin 1
in epithelial ovarian cancer. Sichuan Da Xue Xue Bao Yi Xue Ban.
38:239–242. 2007.(In Chinese).
|
|
67.
|
Pirtoli L, Cevenini G, Tini P, Vannini M,
Oliveri G, Marsili S, et al: The prognostic role of Beclin 1
protein expression in high-grade gliomas. Autophagy. 5:930–936.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai
Z, et al: Association of autophagy defect with a malignant
phenotype and poor prognosis of hepatocellular carcinoma. Cancer
Res. 68:9167–9175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Huang JJ, Li HR, Huang Y, Jiang WQ, Xu RH,
Huang HQ, et al: Beclin 1 expression: a predictor of prognosis in
patients with extranodal natural killer T-cell lymphoma, nasal
type. Autophagy. 6:777–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy research. Cell. 140:313–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Smith EL and Schuchman EH: Acid
sphingomyelinase over-expression enhances the antineoplastic
effects of irradiation in vitro and in vivo. Mol Ther.
16:1565–1571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Wei S, Kulp SK and Chen CS: Energy
restriction as an antitumor target of thiazolidinediones. J Biol
Chem. 285:9780–9791. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Zismanov V, Lishner M, Tartakover-Matalon
S, Radnay J, Shapiro H and Drucker L: Tetraspanin-induced death of
myeloma cell lines is autophagic and involves increased UPR
signaling. Br J Cancer. 101:1402–1409. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Fels DR, Ye J, Segan AT, Kridel SJ,
Spiotto M, Olson M, et al: Preferential cytotoxicity of bortezomib
toward hypoxic tumor cells via overactivation of endoplasmic
reticulum stress pathways. Cancer Res. 68:9323–9330. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Buzzai M, Jones RG, Amaravadi RK, Lum JJ,
DeBerardinis RJ, Zhao F, et al: Systemic treatment with the
antidiabetic drug metformin selectively impairs p53-deficient tumor
cell growth. Cancer Res. 67:6745–6752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Ben Sahra I, Laurent K, Giuliano S,
Larbret F, Ponzio G, Gounon P, et al: Targeting cancer cell
metabolism: the combination of metformin and 2-deoxyglucose induces
p53-dependent apoptosis in prostate cancer cells. Cancer Res.
70:2465–2475. 2010.
|
|
77.
|
Kim RH, Coates JM, Bowles TL, McNerney GP,
Sutcliffe J, Jung JU, et al: Arginine deiminase as a novel therapy
for prostate cancer induces autophagy and caspase-independent
apoptosis. Cancer Res. 69:700–708. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar
|
|
79.
|
Zhu K, Dunner K Jr and McConkey DJ:
Proteasome inhibitors activate autophagy as a cytoprotective
response in human prostate cancer cells. Oncogene. 29:451–462.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Dalby KN, Tekedereli I, Lopez-Berestein G
and Ozpolat B: Targeting the prodeath and prosurvival functions of
autophagy as novel therapeutic strategies in cancer. Autophagy.
6:322–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81.
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82.
|
Scherz-Shouval R, Weidberg H, Gonen C,
Wilder S, Elazar Z and Oren M: p53-dependent regulation of
autophagy protein LC3 supports cancer cell survival under prolonged
starvation. Proc Natl Acad Sci USA. 107:18511–18516. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
83.
|
Chouaib S, Kieda C, Benlalam H, Noman MZ,
Mami-Chouaib F and Rüegg C: Endothelial cells as key determinants
of the tumor microenvironment: interaction with tumor cells,
extracellular matrix and immune killer cells. Crit Rev Immunol.
30:529–545. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84.
|
McPhee CK, Logan MA, Freeman MR and
Baehrecke EH: Activation of autophagy during cell death requires
the engulfment receptor Draper. Nature. 465:1093–1096. 2010.
View Article : Google Scholar : PubMed/NCBI
|