Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors of the liver and the third most common
cause of mortality from cancer in eastern Asia. More than 85% of
HCC is caused by the hepatitis B virus (HBV) or C virus (HCV)
infection (1–3). Other causes are exposure to aflatoxin
B1 (4), vinyl chloride (5) and tobacco (6), as well as chronic ethanol ingestion
(7). Many of these factors are
known causes of chronic hepatitis (CH) and liver cirrhosis, which
represent a pre-neoplastic condition of HCC. HCC is often far
advanced and may have multiple lesions at the time of diagnosis.
Curative resection cannot be expected in cases with extrahepatic
metastases. In cases without extrahepatic metastases, curative
resection could potentially be performed; however, postoperative
recurrence and intrahepatic metastases occur frequently, and the
postoperative 5-year survival rate is reported to be 30–40%
(2).
microRNAs (miRNAs) are short (19–25 nucleotides)
noncoding single-stranded RNA molecules, which are cleaved from
70–100 nucleotide miRNA precursors. miRNAs regulate gene expression
either at the transcriptional or translational level, based on
specific binding to the complementary sequence in the coding or
noncoding region of mRNA transcripts. Recent findings, based on
microarray analysis of global miRNA expression profiles in cancer
tissues, have revealed that miRNA profiles discriminate
malignancies of the breast (8),
lung (8,9), pancreas (8,10) and
liver (11–17) from their counterparts.
Expression profiling of miRNA in HCC was first
reported by Murakami et al (11). They comprehensively analyzed miRNA
expression in HCC and non-tumor tissues and compared expression
patterns in tumor and non-tumor tissues, in three differentiation
levels and also between chronic hepatitis and liver cirrhosis
(11). Since then, various groups
have profiled miRNA expression in HCC and surrounding non-tumor
tissues (12–17). Laderio et al performed miRNA
profiling in HCC patients with various clinical features including
normal liver samples (14).
However, in their comparison, not all of the HCC samples were
accompanied by their surrounding non-tumor tissues. Ura et
al compared miRNA expression between HBV- and HCV-related HCC
(16). Although they used normal
liver tissue samples, all patients analyzed had the virus
infection. In this way, there is no study which comprehensively
compared the expression of miRNAs in HCC patients with various
clinical features using tumor and surrounding non-tumor tissues. In
this study, we profiled miRNA expressions in tumor and non-tumor
tissues from 40 HCC patients with heterogeneous pathogenesis. We
also investigated 6 surrounding non-tumor tissues from patients
with metastatic liver cancer. These were used as normal liver
samples. To identify miRNAs specific to each disease state, we
compared the expression of miRNAs in various combinations.
Materials and methods
Clinical specimens
Operative specimens of primary HCC and metastatic
liver cancer tissues were obtained with informed consent from 40
and 6 patients, respectively, at the Department of
Hepato-Biliary-Pancreatic Surgery at Tokyo Medical and Dental
University Hospital between November 2005 and May 2008 (18). This research project was approved by
the local ethics committee and all samples were obtained with the
informed consent of the patients. The clinical findings were
reviewed and analyzed from the patients’ medical records, and are
shown in Table I. Of the 40
patients with HCC, 12 were infected with HBV, 12 with HCV, and 16
were not infected with HBV or HCV. In addition, 6 normal liver
tissue samples obtained during surgery for metastatic liver cancer
originating in the colon were used as control samples. All
specimens were immediately frozen in liquid nitrogen and then
stored at −80°C for RNA analysis.
 | Table I.Clinical characteristics of 40 HCC
patients and 6 patients with metastatic liver cancer. |
Table I.
Clinical characteristics of 40 HCC
patients and 6 patients with metastatic liver cancer.
| Patient | Age | Gender | Cancer type | Virus |
|---|
| L18 | 70 | M | HCC | HBV |
| L23 | 57 | M | HCC | HBV |
| L34 | 57 | M | HCC | HBV |
| L64 | 48 | F | HCC | HBV |
| L76 | 70 | F | HCC | HBV |
| L84 | 76 | F | HCC | HBV |
| L85 | 51 | M | HCC | HBV |
| L87 | 46 | M | HCC | HBV |
| L116 | 68 | M | HCC | HBV |
| L127 | 40 | M | HCC | HBV |
| L144 | 78 | M | HCC | HBV |
| L154 | 66 | F | HCC | HBV |
| L42 | 66 | M | HCC | HCV |
| L50 | 55 | F | HCC | HCV |
| L63 | 68 | M | HCC | HCV |
| L71 | 75 | M | HCC | HCV |
| L72 | 53 | M | HCC | HCV |
| L77 | 73 | M | HCC | HCV |
| L119 | 63 | F | HCC | HCV |
| L120 | 71 | F | HCC | HCV |
| L124 | 43 | F | HCC | HCV |
| L130 | 63 | M | HCC | HCV |
| L132 | 67 | M | HCC | HCV |
| L155 | 77 | M | HCC | HCV |
| L57 | 58 | M | HCC | - |
| L58 | 73 | M | HCC | - |
| L73 | 85 | M | HCC | - |
| L88 | 72 | M | HCC | - |
| L90 | 65 | M | HCC | - |
| L97 | 80 | M | HCC | - |
| L98 | 75 | M | HCC | - |
| L100 | 77 | M | HCC | - |
| L102 | 69 | M | HCC | - |
| L104 | 64 | F | HCC | - |
| L112 | 78 | M | HCC | - |
| L117 | 47 | M | HCC | - |
| L131 | 76 | M | HCC | - |
| L139 | 73 | M | HCC | - |
| L160 | 56 | M | HCC | - |
| L165 | 50 | F | HCC | - |
| C62 | 56 | M | Metastatic liver
cancer | |
| C81 | 70 | M | Metastatic liver
cancer | |
| C114 | 54 | M | Metastatic liver
cancer | |
| C126 | 59 | M | Metastatic liver
cancer | |
| C132 | 65 | F | Metastatic liver
cancer | |
| C145 | 76 | M | Metastatic liver
cancer | |
RNA isolation
Small RNA with a miRNA-rich fraction was extracted
from tissue specimens using the miRNeasy Mini kit (Qiagen,
Valencia, CA, USA) according to the manufacturer’s instructions.
RNAs were quantified with NanoDrop ND-1000. The integrity of the
obtained RNA was assessed using Agilent Bioanalyzer RNA 6000 Nano
Assay (Agilent Technologies, Palo Alto, CA, USA). All samples had
an RNA integrity number (RIN) ≥4.0. The extracted RNAs were then
analyzed by miRNA microarray using 3D-Gene (Toray Industries,
Tokyo, Japan). The microarrays were scanned using GenePix
4000B.
Statistical analysis
The obtained microarray datasets were background
corrected and used for statistical analyses using R statistical
software. The Wilcoxon rank-sum test for paired data was performed
to estimate the significance of miRNA gene expression differences
between tumor and non-tumor tissues in 1) the 12 patients with HBV
infection, 2) the 12 patients with HCV infection, 3) the 16
patients without virus infection, 4) the 24 patients with HBV or
HCV infection, and 5) all 40 patients (see Fig. 1). In addition, an unpaired version
of the Wilcoxon rank-sum test was used to compare the expression of
miRNAs in the 6 normal liver samples with those in 6) the tumor and
7) non-tumor tissues in the 12 patients with HBV infection, 8) the
tumor and 9) non-tumor tissues in the 12 patients with HCV
infection, 10) the tumor and 11) non-tumor tissues in the 16
patients without virus infection, and 12) the tumor and 13)
non-tumor tissues in all 40 patients (Figs. 2 and 3). MiRNAs with a fold change >1.5 and a
p-value <0.05 were deemed as differentially expressed.
Many miRNAs have been reported to be involved in
cancer. In addition to the previously mentioned miRNA profiling
studies (11–17), numerous reviews and papers (19–56)
were found through a literature survey on miRNA and cancer. It
should be noted that we deemed a miRNA to be previously reported
only when the exact name of the miRNA was found in a study. In what
follows, a miRNA whose association with cancer has been previously
reported is called as a previously reported miRNA. Such miRNAs may
be classified into two types depending on whether they show the
same or opposite tendencies to the previous reports. For certain
miRNAs, it is difficult to determine whether their expressions were
reportedly up- or downregulated due to inconsistent reports and
lack of available information on the change direction. In the
latter case, such miRNAs were described simply as cancer
markers.
Results
Table II indicates
the differentially expressed miRNAs between the tumor tissues in
all HCC patients and the 6 normal liver samples (Combination 12).
Thirty miRNAs were downregulated in the tumor tissues while 18 were
upregulated. The double-underlined miRNAs have been previously
reported to be involved in HCC while the miRNAs with a single
underline have a reported association with other types of cancer.
Most of the 18 upregulated miRNAs were known to be involved in HCC
and other types of cancer. The expression of miR-15b is reported to
be upregulated in HCC (23).
Overexpression of miR-18a contributes to HCC cell proliferation in
females through the targeting of the estrogen receptor (21). miR-21 is known to be highly
overexpressed in HCC, and its inhibition in cultured HCC cells
increases the expression of PTEN, a direct target of miR-21,
as well as tumor cell proliferation, migration and invasion
(20). The overexpression of miR-96
is reported in HBV-related HCC (14,21).
miR-222 is a HCC biomarker which is irrespective of viral
association (21). miR-224 is
overexpressed in HCC when compared to a benign hepatocellular
tumor, hepatocellular adenoma (14), and is involved in the inhibition of
apoptosis in HCC cells (21). In
contrast to the upregulated miRNAs, only two downregulated miRNAs
are previously reported. miR-101 is involved in the inhibition of
apoptosis and its downregulation contributes to the spread of HCC
(21).
 | Table II.Differentially expressed miRNAs among
the tumor tissues in all HCC patients and the 6 normal liver
samples. |
Table II.
Differentially expressed miRNAs among
the tumor tissues in all HCC patients and the 6 normal liver
samples.
| miRNA | Fold change | P-value |
|---|
| m-2116* | 0.048 | <0.001 |
| m-545* | 0.077 | 0.012 |
| m-581 | 0.101 | 0.004 |
| m-2114 | 0.104 | 0.012 |
| m-129* | 0.127 | 0.012 |
| m-296-5p | 0.161 | <0.001 |
| m-1267 | 0.168 | 0.008 |
| m-191* | 0.178 | 0.001 |
| m-222* | 0.188 | 0.025 |
| m-1249 | 0.197 | <0.001 |
| m-18b* | 0.208 | <0.001 |
| m-92a-2* | 0.233 | 0.040 |
| m-625* | 0.233 | 0.001 |
| m-934 | 0.237 | 0.002 |
| m-1225-5p | 0.239 | 0.026 |
| m-1913 | 0.284 | 0.001 |
| m-1238 | 0.286 | 0.002 |
| m-557 | 0.288 | 0.001 |
| m-940 | 0.292 | 0.003 |
| m-1228 | 0.293 | 0.002 |
| m-133b | 0.315 | 0.010 |
| let-7f-1* | 0.324 | 0.002 |
| m-1225-3p | 0.364 | 0.028 |
| m-802 | 0.369 | 0.030 |
| m-1224-3p | 0.377 | 0.012 |
| m-144 | 0.382 | 0.006 |
| m-101 | 0.427 | 0.034 |
| m-486-5p | 0.551 | 0.038 |
| m-29c | 0.573 | 0.047 |
| m-422a | 0.576 | 0.004 |
| m-1979 | 2.71 | 0.009 |
| m-140-3p | 2.83 | 0.023 |
| let-7i | 3.59 | 0.017 |
| m-151-3p | 3.78 | 0.005 |
| m-130b | 3.80 | 0.031 |
| m-425 | 4.86 | 0.033 |
| m-25 | 6.55 | <0.001 |
| m-21 | 7.44 | 0.005 |
| m-18a | 9.05 | 0.024 |
| m-221 | 10.9 | 0.004 |
| m-452 | 11.0 | 0.017 |
| m-222 | 14.2 | 0.026 |
| m-15b | 18.3 | 0.016 |
| m-224 | 82.4 | 0.006 |
| m-34b* | 134 | 0.032 |
| m-374b | 148 | 0.039 |
| m-96 | 181 | 0.032 |
| m-216a | 263 | 0.039 |
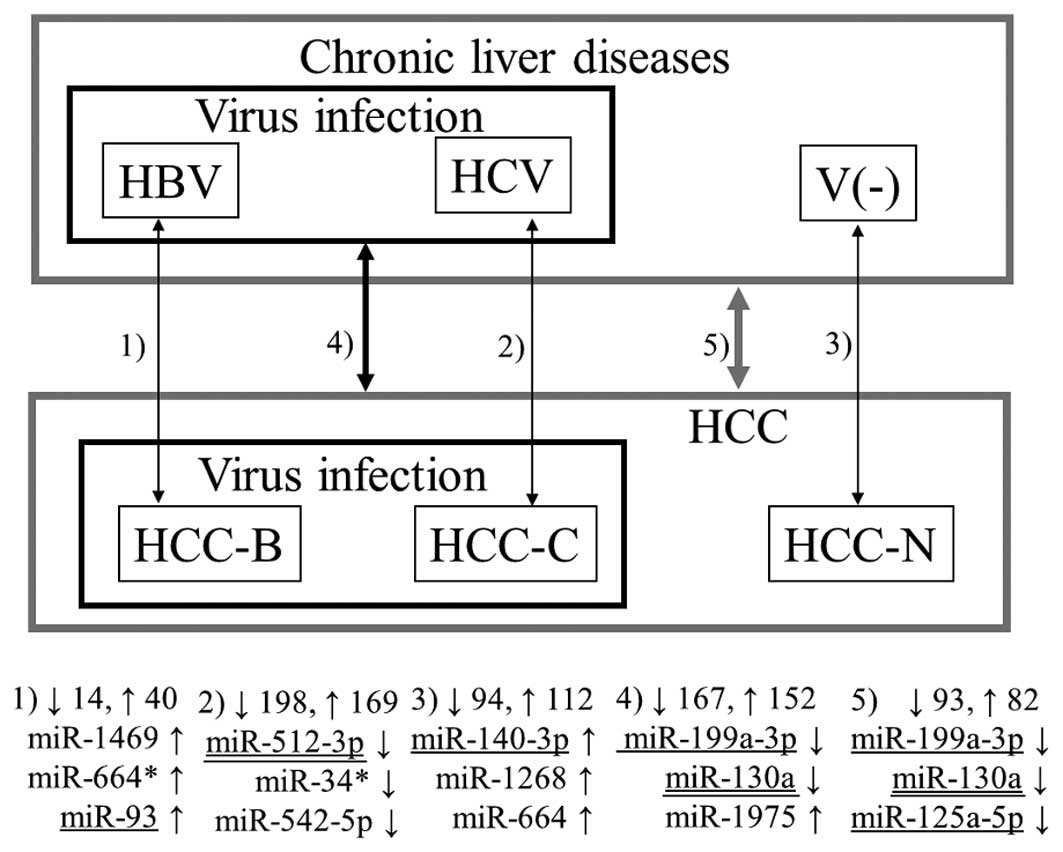
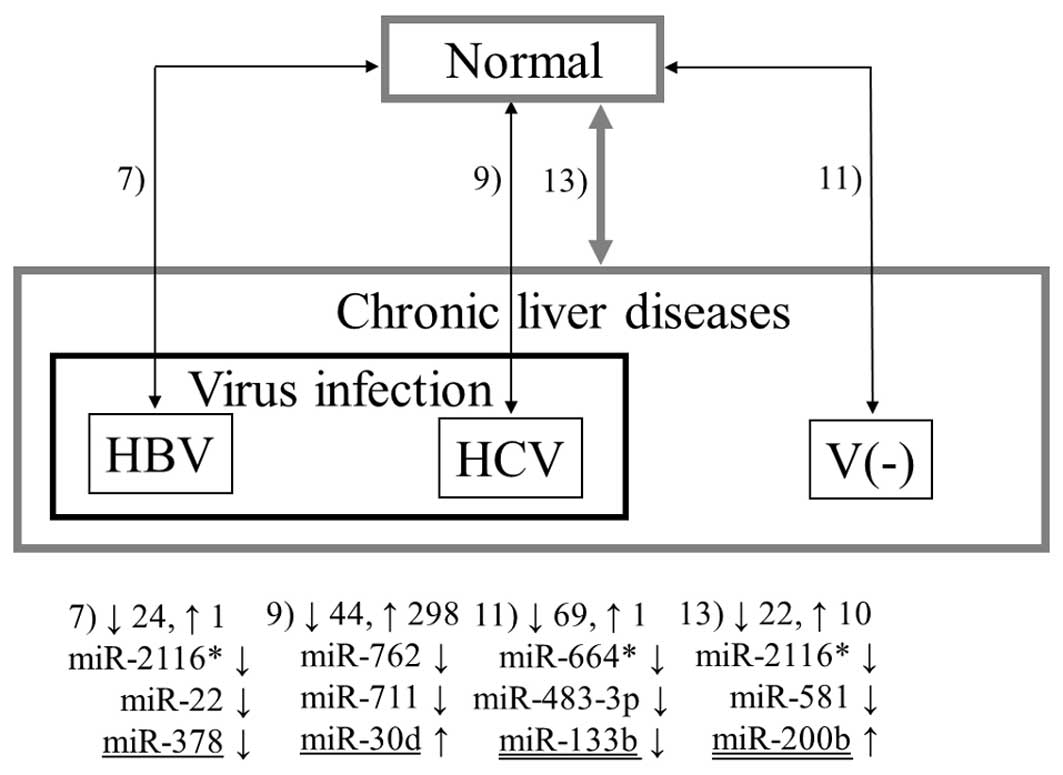
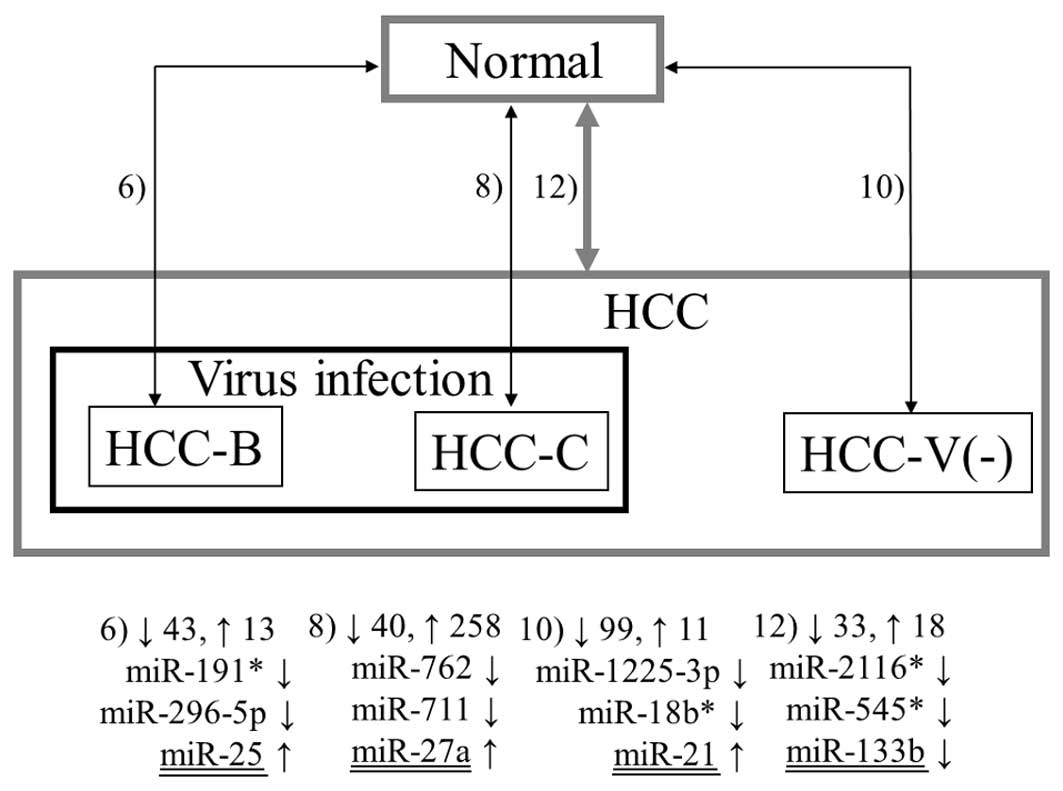
Figs. 1–3 summarize the results from all 13
combinations described in Materials and methods. The numbers 1–13
denote the aforementioned combinations. The numbers beside the
upwards and downwards-pointing arrows indicate the numbers of up-
and downregulated miRNAs, respectively. Three representative miRNAs
in each combination are also shown. Here, ‘representative’ means
the miRNAs with the first and second lowest p-values and the
previously reported miRNA with the lowest p-value among the
previously reported ones (underlined in the figures in the same way
as in Table II). If the previously
reported miRNA was the same as either of the first two, the miRNA
with the third lowest p-value is shown in the figure. In all five
combinations comparing tumor and non-tumor tissues, previously
reported miRNAs demonstrated the lowest p-values (Fig. 1). Notably, the three representative
miRNAs for HCC with HBV infection (Combination 1) and HCC without
virus infection (Combination 3) do not include any miRNA with
previous association with HCC. Conversely, many novel miRNAs as
well as previously reported miRNAs were detected by the unpaired
Wilcoxon rank-sum test (Figs. 2 and
3).
If a miRNA is deemed to be upregulated in the tumor
tissue when comparing tumor and non-tumor tissue, and also
downregulated in the non-tumor tissues when comparing normal liver
samples and non-tumor tissues, these results indicate that the
miRNA was actually downregulated in the non-tumor tissue and thus
it was detected as upregulated in the tumor when comparing the
tumor and non-tumor tissues. To investigate such detailed changes,
we compared lists of the differentially expressed miRNAs from the
13 combinations. Table III shows
the results. For example, Normal↑ denotes that these miRNAs were
downregulated in the tumor and non-tumor tissues compared with the
normal liver. Similarly, Tumor↑ denotes that these miRNAs were
upregulated in the tumor tissues only and their expression was not
altered in the non-tumor tissues. HCV infection↑ indicates
upregulation in the tumor and non-tumor tissues in patients with
HCV infection while HCV↑ represents upregulation in the non-tumor
tissue of the patients with HCV. V(−) denotes patients without
virus infection. The expression levels of many miRNAs were altered
in HCV-positive patients whereas a relatively small number of
miRNAs were detected in patients with HBV and without virus
infection.
 | Table III.Differentially expressed miRNAs
detected in various conditions. |
Table III.
Differentially expressed miRNAs
detected in various conditions.
| Expression
change | Total detected | Detected
miRNAs |
|---|
| Normal↑ | 8 | miR-18b*,
miR-296-5p, miR-557, miR-581, miR-625*, miR-1228, miR-1249,
miR-2116* |
| Tumor↑ | 1 | miR-183 |
| Tumor↓ | 5 | miR-29c, miR-129*, miR-146b-3p,
miR-448, miR-486-5p |
| HCV infection↑ | 125 | let-7a, let-7c, miR-22, miR-23a, miR-93, miR-100, miR-106b, miR-126, miR-135a, miR-210 |
| HCV infection↓ | 7 | miR-143, miR-548q, miR-670,
miR-711, miR-718, miR-759, miR-762 |
| HCC-C↑ | 68 | miR-31, miR-96, miR-103-2*, miR-130b*,
miR-132,
miR-134, miR-138,
miR-184, miR-301a,
miR-372 |
| HCC-C↑,
V(-)_T↓ | 24 | let-7a2, miR-21*,
miR-25*, miR-33a*, miR-92b*, miR-127-3p, miR-149, miR-330-5p,
miR-338-3p, miR-517* |
| HCV↑ | 94 | miR-10a, miR-101, miR-130a, miR-146a, miR-155, miR-181b, miR-195, miR-197, miR-200a, miR-200b |
| HCV↑, V(-)_N↓ | 10 | miR-30b*, miR-187*,
miR-188-5p, miR-193b, miR-199a-5p, miR-542-5p, miR-574-5p, miR-658,
miR-720, miR-1287 |
| HCV↓ | 7 | let-7b*,
miR-139-3p, miR-220c, miR-616*, miR-708, miR-767-3p, miR-1911 |
| HBV infection↑ | 1 | miR-422a |
| HCC-B↑ | 2 | miR-605,
miR-1909 |
| V(-)↑ | 1 | miR-217 |
| V(-)↓ | 5 | miR-92a-2*,
miR-361-3p, miR-513a-5p, miR-1234, miR-1914* |
| V(-)_T↑ | 1 | miR-216a |
| V(-)_T↓ | 10 | miR-133a,
miR-199b-5p, miR-338-5p, miR-491-3p, miR-509-3-5p, miR-543,
miR-627, miR-887, miR-921, miR-1247 |
| V(-)_N↓ | 6 | miR-664,
miR-671-3p, miR-1246, miR-1268, miR-1280, miR-1978 |
Discussion
In this study, miRNA expression was profiled in
tumor and surrounding non-tumor tissues from 40 HCC patients with
various clinical features. We first performed pairwise comparisons
of miRNA expression between the tumor and non-tumor tissues. We
also profiled miRNA expression in non-tumor tissue samples from 6
patients with metastatic liver cancer. These non-tumor samples were
considered as normal liver and compared with the results from each
of the tumor and non-tumor tissues in patients with HBV and HCV or
without infection. These comprehensive comparisons provided
valuable insight into the alteration of miRNAs in the tumor and
non-tumor tissues. For example, when a ratio of 1.5 is obtained
from a comparison of the miRNA expression level in tumor and
non-tumor tissues, it cannot be proven whether the miRNA expression
was increased in the tumor tissue or decreased in the non-tumor
tissue, since both cases will produce the same results. Employing
normal liver samples enables this distinction.
As shown in Fig. 2,
miR-30d, miR-124, miR-200b and miR-378 demonstrated significant
changes in expression in the non-tumor tissues in HCC patients
compared with normal liver samples in Combinations 7, 9, 11 and 13.
In all combinations except 9, only a few previously detected miRNAs
were observed: let-7e, let-7f, miR-98 and miR-144. Notably, none of
them demonstrated the same tendency observed in previous studies.
In Combination 9, more than 80 of the 342 miRNAs detected were
previously reported, and 21 and 24 of them demonstrated the same
and opposite tendencies observed in previous reports, respectively.
These results suggest that these miRNAs may play a role in the very
early stages of carcinogenesis in HCC and some may have different
roles in HCC and chronic liver diseases.
As shown in Table
III, miR-2116* was significantly downregulated in HCC as well
as the surrounding non-tumor tissues compared with the normal liver
samples in all possible combinations; thus, miR-2116* may have some
influence upon carcinogenesis of HCC. Although this miRNA has no
previously reported correlation with cancer, its predicted target
genes involve genes associated with cancer. The top four predicted
targets with the strongest prediction scores (57) were ZDHHC11, MTCH2, DIRC2 and
PEA15 (as of January 2012 according to www.microrna.org). The gene copy number of
ZDHHC11 was altered in nearly half of the patients with
non-small cell lung cancer (58)
and bladder cancer (59).
MTCH2 is a gene exhibiting highly restricted levels of gene
expression variation in tumor tissues compared to non-malignant
tissues (60). Bodmer et al
identified DIRC2 as a familial renal cell
carcinoma-associated gene (61).
The expression level of PEA15 was used for grading the
malignancy of astrocytic tumors (62). These facts indicate that the
predicted targets play roles in cancer. The expression of miR-2116*
was altered in the non-tumor tissues when compared to the normal
liver samples, and accordingly, as discussed earlier, this miRNA
may also play a role in the early stages of carcinogenesis. Other
miRNAs which were consistently downregulated in the tumor and
non-tumor tissues are miR-18b*, miR-296-5p, miR-557, miR-581,
miR-625*, miR-1228 and miR-1249. Similar to miR-2116*, the
predicted target genes of miR-1249 involved numerous genes whose
association with cancer has been reported.
The results strongly suggest that the above miRNAs
are novel biomarker candidates for chronic liver diseases. Some are
novel biomarker candidates for HCC irrespective of virus infection
and HCC with HCV, as shown in Table
III. In this way, through miRNA expression profiling, we
identified various pathogenesis-related miRNAs which may be used as
biomarkers for a specific disease state, although further
investigation is required.
Acknowledgements
This study was funded by the
Scientific Research Grant (No. 20510184), Science and Technology
Promotion Adjustment Expenses (No. 08005234), and the Integrated
Database Project, from the Ministry of Education, Culture, Sports,
Science and Technology of Japan.
References
|
1.
|
HB El-SeragKL RudolphHepatocellular
carcinoma: epidemiology and molecular
carcinogenesisGastroenterology13225572576200710.1053/j.gastro.2007.04.06117570226
|
|
2.
|
H BlumHepatocellular carcinoma: therapy
and preventionWorld J Gastroenterol1173917400200516437707
|
|
3.
|
M CrampHBV + HCV = HCC?Gut451681691999
|
|
4.
|
Y SoiniSC ChiaWP BennettAn
aflatoxin-associated mutational hotspot at codon 249 in the p53
tumor suppressor gene occurs in hepatocellular carcinomas from
MexicoCarcinogenesis1710071012199610.1093/carcin/17.5.10078640905
|
|
5.
|
P BoffettaL MatisaneKA MundtLD
DellMeta-analysis of studies of occupational exposure to vinyl
chloride in relation to cancer mortalityScand J Work Environ
Health29220229200310.5271/sjweh.72512828392
|
|
6.
|
H TsukumaT HiyamaA OshimaA case-control
study of hepatocellular carcinoma in Osaka, JapanInt J
Cancer45231236199510.1002/ijc.2910450205
|
|
7.
|
F DonatoA TaggerU GelattiAlcohol and
hepatocellular carcinoma: the effect of lifetime intake and
hepatitis virus infections in men and womenAm J
Epidemiol155323333200210.1093/aje/155.4.32311836196
|
|
8.
|
S VoliniaGA CalinCG LiuA microRNA
expression signature of human solid tumors defines cancer gene
targetsProc Natl Acad Sci
USA10322572261200610.1073/pnas.051056510316461460
|
|
9.
|
N YanaiharaN CaplenE BowmanUnique micro
RNA molecular profiles in lung cancer diagnosis and prognosisCancer
Cell9189198200610.1016/j.ccr.2006.01.02516530703
|
|
10.
|
EJ LeeY GusevJ JiangExpression profiling
identifies microRNA signature in pancreatic cancerInt J
Cancer12010461054200710.1002/ijc.2239417149698
|
|
11.
|
Y MurakamiT YasudaK SaigoComprehensive
analysis of microRNA expression patterns in hepatocellular
carcinoma and non-tumorous
tissuesOncogene2525372545200610.1038/sj.onc.120928316331254
|
|
12.
|
A BudhuHL JiaM ForguesIdentification of
metastasis-related microRNAs in hepatocellular
carcinomaHepatology47897907200810.1002/hep.2216018176954
|
|
13.
|
H VarnhortU DrebberF SchulzeMicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinomaHepatology4712231232200810.1002/hep.2215818307259
|
|
14.
|
Y LaderioG CouchyC BalabaudMicroRNA
profiling in hepatocellular tumor is associated with clinical
features and oncogene/tumor suppressors gene
mutationsHepatology4719551963200810.1002/hep.2225618433021
|
|
15.
|
W LiL XieX HeDiagnostic and prognositic
implications of microRNAs in human hepatocellular carcinomaInt J
Cancer12316161622200810.1002/ijc.2369318649363
|
|
16.
|
S UraM HondaT YamashitaDifferential
microRNA expression between hapatitis B and hepatitis C leading
disease progression to hepatocellular
carcinomaHepatology4910981112200910.1002/hep.2274919173277
|
|
17.
|
S ToffaninY HoshidaA
LachenmayerMicroRNA-based classification of hepatocellular
carcinoma and oncogenic role of
miR-517aGastoenterology14016181628201110.1053/j.gastro.2011.02.00921324318
|
|
18.
|
M YasenH MizushimaK MogushiExpression of
Aurora B and alterative variant forms in hepatocellular carcinoma
and adjacent tissueCancer
Sci100472480200910.1111/j.1349-7006.2008.01068.x19134008
|
|
19.
|
L GramantieriF FornariE CallegariMicroRNA
involvement in hepatocellular carcinomaJ Cell Mol
Med1221892204200810.1111/j.1582-4934.2008.00533.x19120703
|
|
20.
|
RN AravalliCJ SteerNK CressmanMolecular
mechanisms of hepatocellular
carcinomaHepatology4820472063200810.1002/hep.22580
|
|
21.
|
J JiXW WangNew kids on the blockCancer
Biol Therapy816831690200910.4161/cbt.8.18.8898
|
|
22.
|
L LiangCM WongQ YingMicroRNA-125b
suppressed human liver cancer cell proliferation and metastasis by
directly targeting oncogene
LIN28BHepatology5217311740201010.1002/hep.2390420827722
|
|
23.
|
M OsakiF TakeshitaT OchiyaMicroRNAs as
biomarkers and therapeutic drugs in human
cancerBiomarkers13658670200810.1080/1354750080264657219096960
|
|
24.
|
J TakamizawaH KonishiK YanagisawaReduced
expression of the let-7 micro RNAs in human lung cancers in
association with shortened postoperative survivalCancer
Res6437533756200410.1158/0008-5472.CAN-04-063715172979
|
|
25.
|
M FabbriR GarzonA CimminoMicroRNA-29
family reverts aberrant methylation in lung cancer by targeting DNA
methyltransferases 3A and 3BProc Natl Acad Sci
USA1041580515810200710.1073/pnas.070762810417890317
|
|
26.
|
GT BommerI GerinY Fengp53-mediated
activation of miRNA34 candidate tumor-suppressor genesCurr
Biol1712981307200710.1016/j.cub.2007.06.06817656095
|
|
27.
|
M CrawfordE BrawnerK BattleMicroRNA-126
inhibits invasion in non-small cell lung carcinoma cell
linesBiochem Biophys Res
Commun373607612200810.1016/j.bbrc.2008.06.09018602365
|
|
28.
|
Y HayashitaH OsadaY TatematsuA
polycistronic microRNA cluster, miR-17-92, is overexpressed in
human lung cancers and enhances cell proliferationCancer
Res6596289632200510.1158/0008-5472.CAN-05-235216266980
|
|
29.
|
S VoliniaGA CalinCG LiuA microRNA
expression signature of human solid tumors defines cancer gene
targetsProc Natl Acad Sci
USA10322572261200610.1073/pnas.051056510316461460
|
|
30.
|
M GarofaloC QuintavalleG Di LevaMicroRNA
signatures of TRAIL resistance in human non-small cell lung
cancerOncogene2738453855200810.1038/onc.2008.618246122
|
|
31.
|
N YanaiharaN CaplenE BowmanUnique microRNA
molecular profiles in lung cancer diagnosis and prognosisCancer
Cell9189198200610.1016/j.ccr.2006.01.02516530703
|
|
32.
|
Z HuJ ChenT TianGenetic variants of miRNA
sequences and non-small cell lung cancer survivalJ Clin
Invest11826002608200818521189
|
|
33.
|
SL YuHY ChenGC ChangMicroRNA signature
predicts survival and relapse in lung cancerCancer
Cell134857200810.1016/j.ccr.2007.12.00818167339
|
|
34.
|
A MarkouEG TsarouchaL KaklamanisM FotinouV
GeogouliasES LianidouPrognostic value of mature microRNA-21 and
microRNA-205 overexpression in non-small cell lung cancer by
quantitative real-time RT-PCRClin
Chem5416961704200810.1373/clinchem.2007.10174118719201
|
|
35.
|
LX YanXF HuangQ ShaoMicroRNA miR-21
overexpression in human breast cancer is associated with advanced
clinical stage, lymph node metastasis and patient poor
prognosisRNA1423482360200810.1261/rna.103480818812439
|
|
36.
|
SH ChanCW WuAF LiCW ChiWC LinmiR-21
microRNA expression in human gastric carcinomas and its clinical
associationAnticancer Res28907911200818507035
|
|
37.
|
T SchepelerJT ReinertMS
OstenfeldDiagnostic and prognostic microRNAs in stage II colon
cancerCancer
Res6864166424200810.1158/0008-5472.CAN-07-611018676867
|
|
38.
|
G ChildsM FazzariG KungLow-level
expression of microRNAs let-7d and miR-205 are prognostic markers
of head and neck squamous cell carcinomaAm J
Pathol174736745200910.2353/ajpath.2009.08073119179615
|
|
39.
|
C RoldoE MissiagliaJP HaganMicroRNA
expression abnormalities in pancreatic endocrine and acinar tumors
are associated with distinctive pathologic features and clinical
behaviorJ Clin Oncol2446774684200610.1200/JCO.2005.05.5194
|
|
40.
|
G MarcucciMD RadmacherK MaharryMicroRNA
expression in cytogenetically normal acute myeloid leukemiaN Engl J
Med35819191928200810.1056/NEJMoa07425618450603
|
|
41.
|
GA CalinM FerracinA CimminoA microRNA
signature associated with prognosis and progression in chronic
lymphocytic leukemiaN Engl J
Med35317931801200510.1056/NEJMoa05099516251535
|
|
42.
|
L LuD KatsarosIA de la LongraisO SochircaH
YuHypermethylation of let-7a-3 in epithelial ovarian cancer is
associated with low insulin-like growth factor-II expression and
favorable prognosisCancer
Res671011710122200710.1158/0008-5472.CAN-07-254417974952
|
|
43.
|
Y GuoZ ChenL ZhangDistinctive microRNA
profiles relating to patient survival in esophageal squamous cell
carcinomaCancer
Res682633200810.1158/0008-5472.CAN-06-441818172293
|
|
44.
|
SS ChimTK ShingEC HungDetection and
characterization of placental microRNAs in material plasmaClin
Chem54482490200810.1373/clinchem.2007.09797218218722
|
|
45.
|
S GiladE MeiriY YogevSerum microRNAs are
promising biomarkersPLoS
One3e3148200810.1371/journal.pone.000314818773077
|
|
46.
|
PS MitchelRA ParkinEM KrohCirculating
microRNAs as stable blood-based markers for cancer detectionProc
Natl Acad Sci
USA1051051310518200810.1073/pnas.080454910518663219
|
|
47.
|
CH LawrieCd CooperE BallabioJ ChiD
TramontiCS HattonAberrant expression of microRNA biosynthetic
pathways components is a common feature of haematological
malignancyBr J Hematol141672675200819298586
|
|
48.
|
R DiazJ SilvaJM GarciaDeregulated
expression of miR-106a predicts survival in human colon cancer
patientsGenes Chromosomes
Cancer47794802200810.1002/gcc.2058018521848
|
|
49.
|
K ScheeO FodstadK FlatmarkMicroRNAs as
biomarkers in colorectal cancerAm J
Patho17715921599201010.2353/ajpath.2010.10002420829435
|
|
50.
|
WKK WuCW LeeCH ChoD FanK WuJ YuJJY
SungMicroRNA dysregulation in gastric cancer: a new player enters
the gameOncogene2957615671201010.1038/onc.2010.35220802530
|
|
51.
|
J ZavadilH YeZ LiuProfiling and functional
analyses of microRNAs and their target gene products in human
uterine leiomyomasPLoS
One5e12362201010.1371/journal.pone.001236220808773
|
|
52.
|
MV IorioC PiovanCM CroceInterplay between
microRNAs and the epigenetic machinery: an intricate networkBoichim
Biophys Acta1799694701201010.1016/j.bbagrm.2010.05.00520493980
|
|
53.
|
N ValeriI VanniniF FaniniF CaloreB AdairM
FabbriEpigenetics, miRNAs, and human cancer: a new chapter in human
gene regulationMamm
Genome20573580200910.1007/s00335-009-9206-519697081
|
|
54.
|
A SchaeferM JungG KristiansenMicroRNAs and
cancer: current state and future perspectives in urologic
oncologyUrologic
Oncol28413201010.1016/j.urolonc.2008.10.02119117772
|
|
55.
|
R GarzonG MarcucciCM CroceTargeting
microRNAs in cancer: rationale, strategies and challengesNat Rev
Drug Discov9775789201010.1038/nrd317920885409
|
|
56.
|
GA CalinCG LiuC SevignaniMicroRNA
profiling reveals distinct signatures in B cell chronic lymphocytic
leukemiasProc Natl Acad Sci
USA1011175511760200410.1073/pnas.040443210115284443
|
|
57.
|
D BetelA KoppalP AngiusC SanderC
LeslieComprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sitesGenome
Biol11R90201010.1186/gb-2010-11-8-r9020799968
|
|
58.
|
JU KangSH KooKC KwonJW ParkJM KimGain at
chromosomal region 5p15.33, containing TERT, is the most frequent
genetic event in early stages of non-small cell lung cancerCancer
Genet
Cytogenet82111200810.1016/j.cancergencyto.2007.12.00418328944
|
|
59.
|
Y YamamotoY ChochiH MatuyamaGain of
5p15.33 is associated with progression of bladder
cancerOncology72132138200710.1159/000111132
|
|
60.
|
K YuK GanesanLK TanA precisely regulated
gene expression cassette potentially modulates metastasis and
survival in multiple solid cancersPLoS
Genet4e1000129200810.1371/journal.pgen.100012918636107
|
|
61.
|
D BodmerM EleveldE Kater-BaatsDisruption
of a novel MFS transporter gene, DIRC2, by a familial renal cell
carcinoma-associated t(2;3)(q35;q21)Hum Mol
Genet11641649200210.1093/hmg/11.6.64111912179
|
|
62.
|
Y WatanabeF YamasakiY KajiwaraExpression
of phosphoprotein enriched in astrocytes 15 kDa (PEA-15) in
astrocytic tumors: a novel approach of correlating malignancy grade
and prognosisJ
Neurooncol100449457201010.1007/s11060-010-0201-120455002
|