Introduction
Despite recent improvements in diagnosis, lung
cancer is a major cause of mortality from malignant diseases due to
its high incidence, malignant behavior and lack of major
advancements in treatment strategy (1). Although there have been advances in
understanding the biology of lung cancer and introduction of new
chemotherapeutic agents for treatment, the 5-year survival rate
remains less than 15% (2).
Recently, progression in understanding oncogenic kinase signaling
pathways has provided more successful targets for developing
effective therapeutic strategies (3), which may improve the outcome of lung
cancer.
A potential therapeutic target is the mammalian
target of rapamysin (mTOR) pathway, which plays a central role in
regulating cell functions, including proliferation, growth,
survival, mobility and angiogenesis (4,5).
Dysregulation of the mTOR pathway has been reported in lung cancers
(6,7). A member of the small G protein family,
RagD, which encodes a recently discovered activator of the
mTOR pathway (8), was significantly
upregulated in cells expressing mutant NRF2 (9). It has been demonstrated that mutations
of the NRF2 gene (NFE2L2) are associated with primary
lung cancer (10–13). It has also been revealed that
patients with lung tumors containing the NRF2 gene mutation
display a poorer prognosis compared to patients with non-mutant
tumors (11,12). Additionally, NRF2 gene
somatic mutation is more common in lung squamous cell carcinomas
(11).
Although we have revealed the NRF2 gene
mutation status in lung cancer (11), the correlation between NRF2
gene mutation and RagD expression status in lung cancer has
not been reported. To determine the RagD mRNA expression
status, we performed quantitative real-time polymerase chain
reaction (qPCR) using a LightCycler (Roche Diagnostics GmbH,
Mannheim, Germany). The findings were compared to the
clinicopathological features of lung squamous cell carcinomas.
Patients and methods
Patients
The study group included 90 lung squamous cell
carcinoma patients who had undergone surgery at the Department of
Oncology, Immunology and Surgery, Nagoya City University Graduate
School of Medical Sciences, Nagoya, Japan. All tumor samples were
immediately frozen and stored at −80°C until assayed. Written
informed consent was obtained from each patient prior to the study.
The study was approved by the Institutional Review Board of Nagaya
City University Graduate School of Medicine.
The clinical and pathological characteristics of the
90 lung squamous cell carcinoma patients are shown in Table I. Among the 90 patients, 83 were
male and the mean age was 66.8 years (range, 49–80 years). A total
of 30 patients had lymph node metastasis and 47 cases were
pathological stage I, 19 were stage II and 24 were stage III. All
patient samples were sequenced for the NRF2 gene (11) and 14 cases were positive for the
NRF2 gene mutation.
 | Table I.Clinicopathological data of 90 lung
squamous cell carcinoma patients. |
Table I.
Clinicopathological data of 90 lung
squamous cell carcinoma patients.
| | RagD gene
status
|
|---|
|
Characteristics | Number of patients
(%) | RagD/β-actin
mRNA levels | P-value |
|---|
| Age | | | |
| Mean ± SD
(years) | 66.8±8.0 | | |
| ≤65 | 37 (41.1) | 1.532±2.477 | 0.1683 |
| >65 | 53 (58.9) | 2.294±2.617 | |
| Gender | | | |
| Male | 83 (92.2) | 2.070±2.648 | 0.2622 |
| Female | 7 (7.8) | 0.929±1.011 | |
| Pathological
stage | | | |
| I | 47 (52.2) | 1.357±1.560 | 0.0039a |
| II | 19 (21.1) | 1.979±2.559 | |
| III | 24 (26.7) | 3.204±3.623 | |
| Lymph node
metastasis | | | |
| N0 | 60 (66.7) | 1.810±2.106 | NS |
| N1 | 14 (15.6) | 2.000±3.116 | |
| N2 | 16 (17.8) | 2.606±3.593 | |
| BI status | | | |
| <400 | 4 (4.4) | 1.625±0.624 | 0.7789 |
| ≥400 | 86 (95.6) | 1.998±2.630 | |
|
Differentiation | | | |
| Well | 22 (24.4) | 2.214±2.938 | NS |
| Moderate | 45 (50.0) | 2.204±2.696 | |
| Poor | 21 (23.3) | 1.367±1.921 | |
| NRF2 mutation | | | |
| Mutant | 14 (15.6) | 3.107±3.633 | 0.0747 |
| Wild-type | 76 (84.4) | 1.774±2.301 | |
PCR assays for NRF2
Total RNA was extracted from lung cancer tissues
using an Isogen kit (Nippon Gene Co., Ltd., Tokyo, Japan) according
to the manufacturer’s instructions. RNA concentration was
determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop
Technologies Inc., Rockland, DE, USA). Approximately 5 cases were
excluded from each assay due to the an insufficient number of tumor
cells to effectively extract tumor RNA. RNA (1 μg) was
reverse transcribed using a First-Strand cDNA synthesis kit with
0.5 μg oligo(dT)16 (Roche Diagnostics GmbH)
according to the manufacturer’s instructions. The reaction mixture
was incubated at 25°C for 15 min, 42°C for 60 min, 99°C for 5 min
and then at 4°C for 5 min. The cDNA concentration was also
determined using a NanoDrop ND-1000 Spectrophotometer.
Approximately 200 ng of each cDNA was used for PCR analysis. To
ensure the accuracy of mRNA extraction and reverse transcription,
all samples were subjected to PCR amplification using a
β-actin primers kit (Nihon Gene Research Laboratory, Miyagi,
Japan) and a LightCycler FastStart DNA Master HybProbe kit (Roche
Diagnostics GmbH). The RT-PCR assay reactions were performed using
a LightCycler FastStart DNA Master SYBR Green I kit (Roche
Diagnostics GmbH) in a 20 μl reaction volume. The primer
sequences for the RagD gene were as follows: forward,
5′-GACAAAGTTCCTGGCTCTCG-3′ and reverse, 5′-AGCACTCTAGGGGTCCCATT-3′
(210 bp). Cycling conditions consisted of an initial denaturation
period at 95°C for 10 min, followed by 40 cycles at 95°C for 10
sec, 62°C for 10 sec and 72°C for 9 sec.
Statistical analysis
Statistical analyses were conducted using the
Mann-Whitney U test for unpaired samples and the Wilcoxon’s singed
rank test for paired samples. Linear relationships between
variables were determined by means of simple linear regression.
Correlation coefficients were determined by rank correlation using
the Spearman’s rho test and Chi-squared test. The overall survival
of lung cancer patients was examined using the Kaplan-Meier
analysis, and differences were examined using the log-rank test.
The Stat-View software package (Abacus Concepts Inc., Berkeley, CA,
USA) was used for all statistical analyses and P<0.05 was
considered to indicate a statistically significant difference.
Results
NRF2 gene mutation in lung cancer
Previously, we investigated the NRF2 gene
mutation status in the N-terminal domain by direct sequencing
(11). A total of 291 non-small
cell lung cancer (NSCLC) patients, including 148 lung squamous cell
carcinoma patients, were investigated and 16 were identified to
express NRF2 gene mutations. All of the mutations were
identified in male patients with lung squamous cell carcinomas.
RagD mRNA levels in lung cancer
patients
In this study, we investigated 90 lung squamous cell
carcinoma patients, including 14 NRF2 mutant patients, in
order to examine their RagD/β-actin levels (Table I). We revealed that the mean
RagD/β-actin level in lung cancer tissues was 2.138±2.698
and did not correlate with age (R2=0.17; P=0.2487).
Additionally, RagD/β-actin mRNA levels were not correlated
with age (≤65 vs. >65 years; P= 0.1683), Brinkman index (<400
vs. ≥400; P= 0.7789), lymph node metastasis, tumor invasion status
or pathological differentiation status. RagD/β-actin mRNA
level was correlated with pathological stage, and there was a
tendency towards higher RagD/β-actin mRNA level in
higher pathological stages (stage I, 1.357±1.560; stage II,
1.979±2.599; stage III, 3.204±3.623). RagD/β-actin mRNA
level was significantly higher in stage III cases compared to stage
I cases (P=0.0039). In addition, significantly higher levels of
RagD/β-actin mRNA were demonstrated in NRF2 mutant
cases (3.107±3.633) compared to NRF2 wild-type cases
(1.774±2.301) (P=0.0747).
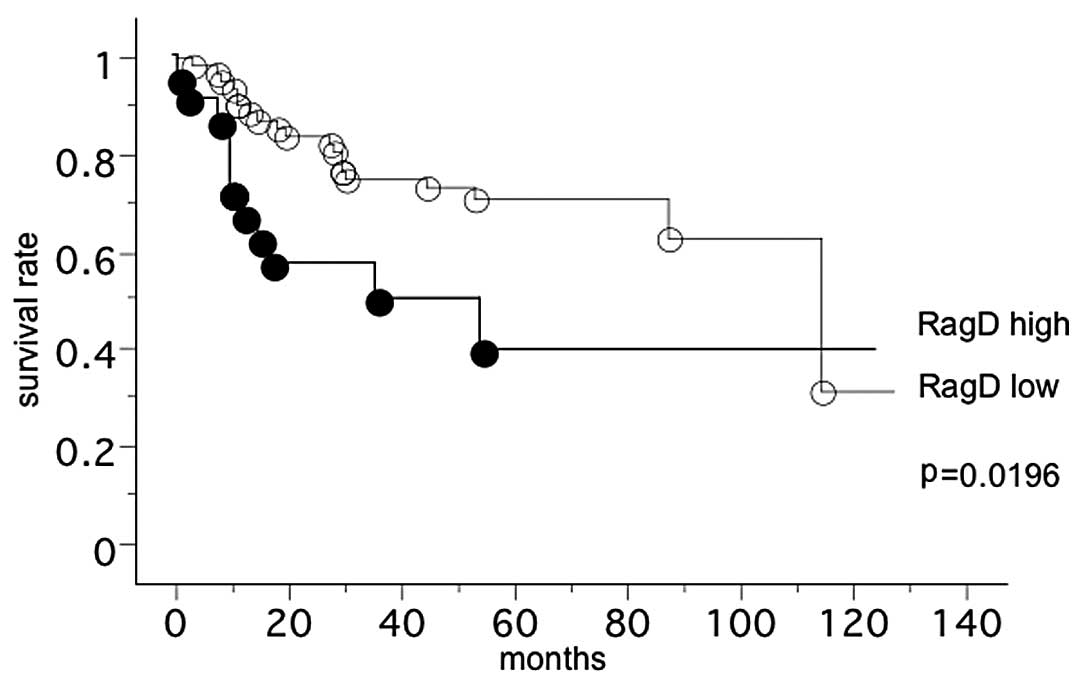
The overall survival of 90 lung squamous cell
carcinoma patients, with follow-up until December 31, 2010, was
studied in reference to the RagD/β-actin mRNA level. The
survival of patients with high RagD/β-actin mRNA levels
(11/21 mortalities; mean survival, 33.6 months) was significantly
less compared to patients with low RagD/β-actin mRNA levels
(19/68 mortalities; mean survival, 85.0 months) (log-rank test, P=
0.0196) (Fig. 1). However,
multivariate analysis demonstrated RagD mRNA was not an
independent prognostic factor.
Discussion
In this study, we identified that RagD mRNA
levels were correlated with advanced stage lung squamous cell
carcinomas. We also demonstrated that high RagD mRNA levels
correlated with poor prognosis using univariate analysis. Although
the sample size was small, there was a tendency towards higher
RagD mRNA levels in NRF2 mutant lung squamous cell
carcinoma patients.
RagD is a member of the small G protein
family gene, which encodes a recently discovered positive regulator
of the mTOR pathway (8,14,15),
and is upregulated in NRF2 mutant cell lines. It has been
demonstrated that RagD knockdown reduces the activation of
mTOR signaling and NRF2 down-regulation reduces RagD
expression. Therefore, RagD plays an important role in the
proliferation of NRF2-mutant cancer cells (9). However, the putative promoter region
of the RagD gene contains no ARE sequence, and chromatin
immunoprecipitate sequence analyses has revealed that RagD
is not a direct target of NRF2 (9). An additional regulatory mediator may
link the NRF2 gene and RagD.
The NRF2 gene is a master transcriptional
activator of genes encoding a number of cytoprotective enzymes that
are induced in response to environmental and endogenously derived
oxidative/electrophilic agents (16–18). A
previous study demonstrated that RNAi-mediated silencing of
NRF2 gene expression in NSCLC inhibited tumor growth
(19). A NRF2 gene promoter
polymorphism has been identified and was suggested to correlate
with carcinogenesis (20). The
correlation between NRF2 mutations and RagD mRNA
levels of lung squamous cell carcinomas suggests a role of
NRF2 in tumor growth. Constitutive expression of NRF2
may provide a survival advantage to invasive and metastatic cancer
cells. NFR2 may adapt these cancer cells to the
microenvironment by increasing chemoresistance under hypoxic
conditions (21,22).
Higher RagD mRNA levels were correlated with
poor prognosis, however, this may be due to the correlation with
pathological stages. Our previous study demonstrated that mutant
NRF2 had poor prognosis (11), which confirmed results from other
experiments (10,12). In addition, previous studies
revealed that mTOR expression was a prognostic biomarker for poor
survival of lung cancers (6,7,23).
A longer follow-up period and larger cohort are required to analyze
RagD expression as a prognostic biomarker for lung
cancers.
Acknowledgements
The authors thank Mrs Miki Mochizuki
for her excellent technical assistance. This study was supported by
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (JSPS) (Nos. 23659674, 21390394 and 21591820)
and a grant for cancer research of Program for Developing the
Supporting System for Upgrading the Education and Research (2009)
from the Ministry of Education, Culture, Sports, Science and
Technology of Japan.
References
|
1.
|
RJ GinsbergMK KrisJG ArmstrongCancer of
the lungPrinciples and Practice of OncologyVT DeVita JrS HellmanSA
Rosenberg4th editionLippincottPhiladelphia6736821993
|
|
2.
|
A JemalR SiegelE WardCancer statistics,
2009Ca Cancer J Clin59225249200910.3322/caac.20006
|
|
3.
|
X ZhouM TanV Stone HawthorneActivation of
the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2
overexpression predicts tumor progression in breast cancersClin
Cancer Res1067796788200410.1158/1078-0432.CCR-04-011215501954
|
|
4.
|
MA BjornstiPJ HoughtonThe TOR pathway:
target for cancer therapyNat Rev
Cancer4335348200410.1038/nrc1362
|
|
5.
|
EB BordersC BivonaPJ MedinaMammalian
target of rapamycin: biological function and target for novel
anticancer agentsAm J Health Syst
Pharm6720952106201010.2146/ajhp10002021116000
|
|
6.
|
D LiuY HuangB ChenActivation of mammalian
target of rapamycin pathway confers adverse outcome in nonsmall
cell lung
carcinomaCancer11737633773201110.1002/cncr.2595921387259
|
|
7.
|
K GatelyB Al-AlaoT DhillonOverexpression
of the mammalian target of rapamycin (mTOR) and angioinvasion are
poor prognostic factors in early stage NSCLC: a verification
studyLung
Cancer75217222201210.1016/j.lungcan.2011.06.01221802763
|
|
8.
|
Y SancakTR PetersonYD ShaulThe Rag GTPases
bind raptor and mediate amino acid signaling to
mTORC1Science32014961501200810.1126/science.115753518497260
|
|
9.
|
T ShibataS SaitoA KokubuGlobal downstream
pathway analysis reveals a dependence of oncogenic NF-E2-related
factor 2 mutation on the mTOR growth signaling pathwayCancer
Res7090959105201010.1158/0008-5472.CAN-10-038421062981
|
|
10.
|
LM SolisC BehrensW DongNrf2 and Keap1
abnormalities in non-small cell lung carcinoma and association with
clinicopathologic featuresClin Cancer
Res1637433753201010.1158/1078-0432.CCR-09-335220534738
|
|
11.
|
H SasakiY HikosakaK OkudaNFE2L2 gene
mutation in male Japanese squamous cell carcinoma of the lungJ
Thorac Oncol5786789201010.1097/JTO.0b013e3181db3dd320421815
|
|
12.
|
T ShibataT OhtaKI TongCancer related
mutations in Nrf2 impair its recognition by Keap1-Clu3 E3 ligase
and promote malignancyProc Natl Acad Sci
USA1051356813573200810.1073/pnas.080626810518757741
|
|
13.
|
Y HuY JuD LinMutation of the Nrf2 gene in
non-small cell lung cancerMol Biol RepOct12011(Epub ahead of
print).
|
|
14.
|
XM MaJ BlenisMolecular mechanisms of
mTOR-mediated translational controlNat Rev Mol Cell
Biol10307318200910.1038/nrm267219339977
|
|
15.
|
E KimP Goraksha-HicksL Liregulation of
TORC1 by Rag GTPases in nutrient responseNat Cell
Biol10935945200810.1038/ncb175318604198
|
|
16.
|
K ItohT ChibaS TakahashiAn Nef2/small Maf
heterodimer mediates the induction of phase II detoxifying enzyme
genes through antioxidant response elementsBiochem Biophys Res
Commun236313322199710.1006/bbrc.1997.69439240432
|
|
17.
|
TH RushmoreAN KongPharmacogenomics,
regulation and signaling pathways of phase I and II detoxifying
enzymesCurr Drug
Metab3481490200210.2174/138920002333717112369894
|
|
18.
|
T NgyyenCS YangCB PickettThe pathways and
molecular mechanisms regulating Nrf2 activation in response to
chemical stressFree Radic Biol
Med37433441200410.1016/j.freeradbiomed.2004.04.03315256215
|
|
19.
|
A SinghS Boldin-AdamskyRK
ThimmulappaRNAi-mediated silencing of nuclear factor
erythroid-2-related factor 2 gene expression in non-small cell lung
cancer inhibits tumor growth and increases efficacy of
chemotherapyCancer
Res6879757984200810.1158/0008-5472.CAN-08-140118829555
|
|
20.
|
T ArisawaT TaharaT ShibataNrf2 gene
promoter polymorphism and gastric
carcinogenesisHepatogastroenterology55750754200818613447
|
|
21.
|
GL SemenzaTargeting HIF-1 for cancer
therapyNat Rev Cancer3721732200310.1038/nrc1187
|
|
22.
|
J ZhouT ScmidS SchnitzerTumor hypoxia and
cancer progressionCancer
Lett2371021200610.1016/j.canlet.2005.05.028
|
|
23.
|
L WangW YueL ZhangmTOR and PTEN expression
in non-small cell lung cancer: analysis by real-time fluorescence
quantitative polymerase chain reaction and immunohistochemistrySurg
TodayNov302011(Epub ahead of print).
|