Introduction
Gallbladder carcinoma (GBC) is a relatively uncommon
neoplasm in the majority of countries and its incidence rate shows
marked geographic and ethnic variation. It is up to three times
more common in females compared with males in almost all
populations. The highest incidences in the world are among women
from Chile (27/100,000), Poland (14/100,000), India (10/100,000),
Japan (7/100,000) and Israel (5/100,000). In the United States and
the United Kingdom, the incidence is <2/100,000 (1–3). GBC
is a highly lethal disease since it is usually diagnosed at an
advanced stage (4). The 5-year
survival rate of patients with GBC is ∼10–30% despite surgical
resection (5–7). Moreover, the majority of patients have
frequent recurrences following surgery and unsatisfactory results
following chemotherapy or radiotherapy (8). Several prognostic models have been
designed to identify patients with a high risk of disease
progression following cholecystectomy, and features such as grade,
depth of wall infiltration and lymph node metastasis have been
determined to be classic clinicopathological prognostic factors
(9). In addition, certain molecular
biomarkers have been identified including cyclooxygenase-2 (Cox2)
and hypoxia-inducible factor (HIF) (10–21).
However, the majority of these molecular and genetic factors are
not markedly associated with GBC. Therefore, it would be useful to
identify new molecular markers, that may be associated with
prognosis and used as therapeutic targets.
Carboxyl-terminus of heat shock protein
70-interacting protein (CHIP) is a well-described U-box-type E3
ubiqutin ligase that induces ubiquitination and proteasomal
degradation of its substrates, which include several tumor-related
proteins (22–26). CHIP participates in the degradation
of p53, a common tumor suppressor protein frequently mutated in
cancers (27,28). The stress-dependent death
domain-associated protein (Daxx)-CHIP interaction also suppresses
p53 apoptotic pathways (29).
Through the formation of the heat shock protein 70
(HSP70)/CHIP/apoptosis signal-regulating kinase 1 (ASK1) complex,
HSP70 promotes ASK1 proteasomal degradation and prevents tumor
necrosis factor-α (TNF-α)-induced cell apoptosis (30). In addition, CHIP is a negative
regulator of forkhead class O1 transcription factor (FoxO1)
activity through ubiquitin-mediated degradation, thus promoting
cell survival (31). Xu et
al demonstrated that CHIP contributes to the tumorigenesis of
malignant gliomas by regulating survivin (32). These data suggest that CHIP may be
significant in cancer by regulating tumor-related proteins.
However, the clinical relevance of CHIP in GBC has not been
investigated. The present study analyzed the expression of CHIP in
tumor specimens from patients who underwent surgical treatment of
GBC and investigated the association between CHIP expression and
clinicopathological features, as well as patient survival.
Materials and methods
Patients and tumor samples
Tumor samples from 78 consecutive patients who
underwent cholecystectomy for GBC at the Chungnam National
University Hospital from 1999 to 2010 were investigated.
Clinicopathological data were obtained by reviewing medical
records. The patient population included 38 males and 40 females
who ranged in age from 25 to 87 years (median, 68 years). All
tumors were diagnosed as adenocarcinoma and defined as primary
tumors arising from the gallbladder. The T classification was
defined according to 2002 American Joint Committee on Cancer
criteria. Tumor samples were collected from tissue blocks used for
routine pathological examination. All patients signed informed
consent for therapy, as well as for subsequent tissue studies,
which had received prior approval by the local ethics
committee.
Histological grading
The GBC specimens were examined by routine
hematoxylin and eosin staining. The specimens were graded into
well- (G1), moderately (G2) and poorly differentiated (G3) and
undifferentiated (G4) adenocarcinoma according to the World Health
Organization classification. Eight cases were well differentiated,
46 were moderately differentiated, 21 were poorly differentiated
and three were undifferentiated.
Tissue microarray (TMA) construction
TMAs were constructed from archival, original
formalin-fixed, paraffin-embedded tissue blocks from 78 patients
with GBC. For each tumor, a representative tumor area was carefully
selected from a hematoxylin and eosin stained section of a donor
block. Each case was represented by two 2-mm-diameter core
cylinders from tumors which were obtained using an automated tissue
array (UNITMA, Seoul, Korea). TMA blocks containing a total of 156
cylinders were constructed from these samples.
Immunohistochemistry (IHC)
The expression of CHIP was analyzed by IHC on
paraffin-embedded tissue sections from the GBC samples. Sections
(3-μm thick) from the paraffin blocks were used for IHC with the
rabbit EnVision-HRP detection system (Dako, Carpinteria, CA, USA).
A polyclonal rabbit CHIP antibody (AP6413a; ABGENT, San Diego CA,
USA) was used for IHC. Following deparaffinization and antigen
retrieval with a pressure cooker in 10 mM sodium citrate buffer (pH
6.0) at full power for 4 min, the tissue sections were treated with
3% hydrogen peroxide for 10 min. The primary antibody was diluted
(1:100) with background-reducing diluents (Dako) and incubated
overnight at 4°C in a humid chamber. The slides were then incubated
with the EnVision reagent for 30 min, sequentially incubated with
DAB chromogen for 5 min, counterstained with Meyer’s hematoxylin
and mounted. Careful rinsing with several changes of 0.3% Tween-20
in TBS buffer was performed between each step. For the IHC negative
control, the primary antibody was excluded.
Evaluation of immunostaining
The level of CHIP expression in each sample was
evaluated by two independent pathologists (J.M. Kim and M.R. Kim)
who were blinded to the patients’ clinicopathological details. The
IHC staining was categorized according to a scoring method in which
the tumors were classified into four grades based on the staining
intensity: (0, no staining; +1, low staining intensity; +2,
intermediate staining intensity; and +3, high staining intensity).
In cases of heterogeneous staining within the samples, the higher
score was selected if >50% of the cells exhibited the higher
staining intensity. For all patients, the scores from the two tumor
cores from the same patient were averaged to obtain a mean score.
Cases with staining intensity scores of 0, +1 and +2 were included
in the CHIP low-expression group (LEG), whereas those with staining
intensity scores of +3 were included in the CHIP high-expression
group (HEG) for all analyses.
Statistical analysis
Group comparisons of categorical variables were
evaluated using the χ2 test or the linear-by-linear
association test. Cancer-specific survival was defined from the
date of surgery to the date of mortality by GBC. Survival curves
were plotted using the Kaplan-Meier method and analyzed using the
log-rank test. Cox’s proportional hazards model was used to
identify prognostic factors for survival. For all analyses,
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed using the SPSS
version 17.0 statistical software (SPSS Inc., Chicago, IL,
USA).
Results
Immunohistochemical analysis of CHIP
expression in GBC
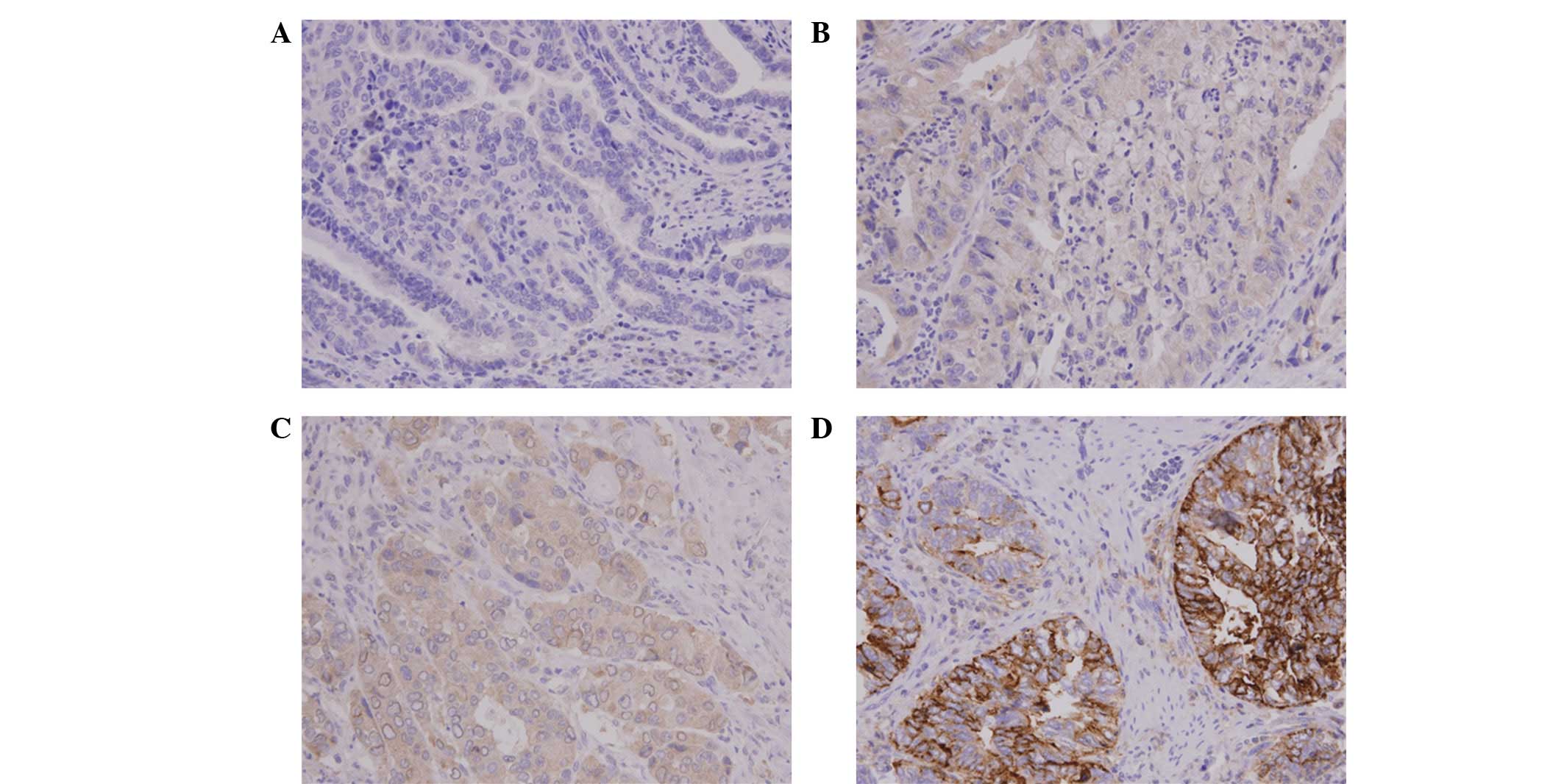
CHIP expression was analyzed by IHC analysis of the
tumor specimens obtained from 78 patients with GBC. CHIP staining
varied by intensity and location. It was predominantly located in
the cytoplasm of the GBC cells, but also appeared in the nucleus or
cell membrane in certain cells (Fig.
1). Next, the CHIP expression levels were analyzed by
determining the intensity of the positively stained tumor cells. A
total of 21 cases (26.9%) had +3 staining intensity (CHIP-HEG) and
57 cases had a lower staining intensity (CHIP-LEG), specifically +2
(23 cases), +1 (31 cases) or 0 (3 cases).
Correlation between CHIP expression and
cliniocopathological factors
The correlations between CHIP expression and various
clinicopathological factors known to affect the prognosis of
patients with GBC were analyzed. The results are presented in
Table I. No significant differences
were observed with regard to age and gender between the CHIP-LEG
and CHIP-HEG patients. In addition, other clinicopathological
factors, such as pathological T stage, lymph node and distant
metastases, stage, differentiation, perineural invasion and
lymphatic invasion were not significantly associated with CHIP
expression.
 | Table IAssociation of CHIP expression with
clinicopathological characteristics of gallbladder carcinoma. |
Table I
Association of CHIP expression with
clinicopathological characteristics of gallbladder carcinoma.
| | CHIP
| |
|---|
| Variable | Total n=78 | LEG (n=57) % | HEG (n=21) % | P-value |
|---|
| Age (years) | | | | 0.676a |
| <65 | 22 | 17 (29.8) | 5 (23.8) | |
| ≥65 | 56 | 40 (70.2) | 16 (76.2) | |
| Gender | | | | 0.157a |
| Male | 38 | 25 (43.9) | 13 (61.9) | |
| Female | 40 | 32 (56.1) | 8 (38.1) | |
| Pathological T
stage | | | | 0.675b |
| 1 | 15 | 9 (15.8) | 6 (28.6) | |
| 2 | 35 | 28 (49.1) | 7 (33.3) | |
| 3 | 25 | 18 (31.6) | 7 (33.3) | |
| 4 | 3 | 2 (3.5) | 1 (4.8) | |
| Nodal metastasis | | | | 0.155a |
| Absent | 56 | 38 (66.7) | 18 (85.7) | |
| Present | 22 | 19 (33.3) | 3 (14.3) | |
| Distant
metastasis | | | | 0.559a |
| Absent | 75 | 54 (94.7%) | 21 (100) | |
| Present | 3 | 3 (5.3%) | 0 (0) | |
| Stage | | | | 0.623a |
| I | 41 | 29 (50.9) | 12 (57.1) | |
| II–IV | 37 | 28 (49.1) | 9 (42.9) | |
|
Differentiation | | | | 0.432b |
| G1 | 8 | 4 (7.0) | 4 (19.0) | |
| G2 | 46 | 36 (63.2) | 10 (47.6) | |
| G3 | 21 | 14 (24.6) | 7 (33.3) | |
| G4 | 3 | 3 (5.3) | 0 (0) | |
| Perineural
invasion | | | | 0.596a |
| Absent | 37 | 26 (45.6) | 11 (52.4) | |
| Present | 41 | 31 (54.4) | 10 (47.6) | |
| Lymphatic
invasion | | | | 0.353a |
| Absent | 27 | 18 (31.6) | 9 (42.9) | |
| Present | 51 | 39 (68.4) | 12 (57.1) | |
Correlation between CHIP expression and
survival
To determine the clinical utility of CHIP expression
with regard to the GBC prognosis, the association between CHIP
expression and patient survival was investigated. Survival curves
according to CHIP expression are shown in Fig. 2. The median cancer-specific survival
rates were 8.0 months (range, 1–34 months) and 13.0 months (range,
1–110 months) in patients with CHIP-LEG and -HEG tumors,
respectively (P=0.023). Next, univariate analyses were performed to
estimate the clinical significance of various parameters that may
affect survival in patients with GBC. As shown in Table II, pathological T stage (P=0.002),
stage (P=0.002), differentiation (P=0.047), lymphatic invasion
(P=0.027) and CHIP expression (P=0.029) were statistically
significant risk factors affecting the cancer-specific survival of
patients with GBC. To determine the independent prognostic effects
of these various factors, multivariate analyses using Cox’s
proportional hazards model were performed. The model revealed that
none of these factors were independent risk factors for predicting
short-term cancer-specific survival, although CHIP expression
(hazard ratio, 2.221; 95% confidence interval, 0.984–5.016;
P=0.055) was close to being a significant independent risk factor
for predicting patient survival (Table
III).
 | Table IIUnivariate analysis of the
association of prognosis with clinicopatholocal parameters and CHIP
expression in patients with gallbladder carcinoma. |
Table II
Univariate analysis of the
association of prognosis with clinicopatholocal parameters and CHIP
expression in patients with gallbladder carcinoma.
| Variables | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Age (years, ≥65 vs.
<60) | 2.064 | 0.896–4.756 | 0.089 |
| Gender (male vs.
female) | 1.579 | 0.799–3.118 | 0.188 |
| Pathological T
stage (T3/4 vs. T1/2) | 3.028 | 1.525–6.014 | 0.002 |
| Nodal metastasis
(yes vs. no) | 1.618 | 0.762–3.434 | 0.210 |
| Distant metastasis
(yes vs. no) | 1.965 | 0.461–8.368 | 0.361 |
| Stage (II–IV vs.
I) | 3.212 | 1.552–6.648 | 0.002 |
| Differentiation
(G3/4 vs. G1/2) | 1.990 | 1.008–3.928 | 0.047 |
| Perineural invasion
(yes vs. no) | 1.878 | 0.872–4.043 | 0.107 |
| Lymphatic invasion
(yes vs. no) | 2.938 | 1.133–7.613 | 0.027 |
| CHIP (HEG vs.
LEG) | 2.373 | 1.093–5.152 | 0.029 |
 | Table IIIMultivariate analysis of the
association of prognosis with clinicopathological parameters and
CHIP expression in patients with gallbladder carcinoma. |
Table III
Multivariate analysis of the
association of prognosis with clinicopathological parameters and
CHIP expression in patients with gallbladder carcinoma.
| Variables | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Pathological T
stage (T3/4 vs. T1/2) | 1.421 | 0.406–4.974 | 0.582 |
| Stage (II–IV vs.
I) | 2.075 | 0.542–7.951 | 0.287 |
| Differentiation
(G3/4 vs. G1/2) | 1.499 | 0.714–3.149 | 0.285 |
| Lymphatic invasion
(yes vs. no) | 1.681 | 0.581–4.861 | 0.338 |
| CHIP (HEG vs.
LEG) | 2.221 | 0.984–5.016 | 0.055 |
Discussion
The present study is the first to demonstrate that
CHIP expression varied among GBC samples and that there was a
significant difference in cancer-specific survival between CHIP-LEG
and CHIP-HEG groups. Since CHIP was not associated with other
clinicopathological prognostic parameters, such as pathological T
stage, nodal and distant metastases, stage, differentiation,
perineural invasion and lymphatic invasion, it may be an ideal
complementary molecular marker for predicting patient outcomes in
GBC. Univariate analyses clearly demonstrated that CHIP expression
was a statistically significant risk factor for the cancer-specific
survival of patients with GBC and multivariate analyses showed that
it was close to being an independent risk factor. The statistically
significant effect of CHIP expression was more significant than
that of the various clinicopathological parameters that are widely
used at present, suggesting that CHIP expression may be a useful
marker for predicting patient survival.
Several prognostic molecular markers for GBC have
been described previously. Mutations in p53 and v-Ki-ras2 Kirsten
rat sarcoma viral oncogene homolog (K-ras), cycle-related proteins
p16 and p21, Cox2, vascular endothelial growth factor (VEGF),
c-erb-B2 (HER-2/neu), inducible nitric oxide synthase (iNOS) and
adhesion molecules (E-cadherin, β-catenin, CD54 and CD44) have been
studied (10–20). We previously reported that L1 cell
adhesion molecule expression is a novel independent prognostic
factor that indicates a poor prognosis for patients with
gallbladder carcinoma (21). The
deregulation of E3 ligases contributes to cancer development and
their overexpression is often associated with poor prognosis, as
has been shown in studies of inhibitor of apoptosis protein
(IAP)-family genes (33), murine
double minute 2 (Mdm2) (34),
Casitas B-lineage lymphoma (CBL)-family proteins (35) and anaphase promoting complex (APC)
(36). Li et al reported
that the overexpression of Skp2, an Skp1-Cullin-F-box protein (SCF)
ubiquitin ligase-related protein, was significantly correlated with
unfavorable clinicopathological parameters and short-term survival
(37). Furthermore, certain E3
ubiquitin ligases have emerged as therapeutic targets for cancer
(38,39). In the present study, it was observed
that CHIP, a member of the E3 ubiquitin ligase family, was
associated with poor prognosis for GBC. These results provide
further support for E3 ligases as biological markers for GBC.
The pathogenic mechanism of CHIP expression in human
malignancy is not yet clear and a number of studies have suggested
that CHIP may have opposing roles in different cancers (22–26,40).
CHIP suppresses tumor progression in human breast cancer by
inhibiting oncogenic pathways and CHIP levels are negatively
correlated with the malignancy of human breast tumor tissues. The
anchorage-independent growth and invasiveness of CHIP-knockout
cells is significantly elevated due to the increased expression of
B-cell CLL/lymphoma 2 (Bcl2), protein kinase B (Akt)1, small
mothers against decapentaplegic (Smad) and Twist, a transcription
factor. Proteomic analysis identified the transcriptional
co-activator steroid receptor coactivator 3 (SRC-3) as a direct
target for ubiquitylation and degradation by CHIP (40). Another study noted that the
overexpression of CHIP inhibited the lung cancer cell growth and
invasion mediated by Met (the receptor for hepatocyte growth
factor) (26). By contrast, Xu
et al(32) showed that CHIP
contributed to the tumorigenesis of human gliomas by regulating
survivin. The authors also observed that CHIP expression in glioma
samples was associated with tumor grades, with more marked staining
in high-grade gliomas compared with low-grade gliomas. A knockdown
of CHIP expression suppressed the proliferation and colony
formation of glioma cells, while the overexpression of CHIP
resulted in enhanced proliferation and colony formation in
vitro. An intratumoral injection of CHIP RNA interference
(RNAi) lentivirus significantly delayed tumor growth and was
associated with decreased mRNA and protein levels of survivin in a
nude mouse xenograft model, while CHIP overexpression resulted in
enhanced tumor growth and increased the mRNA and protein levels of
survivin in vivo(32).
Collectively, whether CHIP contributes to tumor progression or
tumor suppression in various human cancers remains unclear,
suggesting the necessity of further extensive investigation of its
role in tumorigenesis.
In conclusion, the present results indicate that, as
a member of the E3 ubiquitin ligase family, CHIP was differentially
expressed in GBC and higher expression levels were associated with
poor prognosis in patients who were surgically treated for GBC,
suggesting that it may be a useful molecular marker in GBC.
However, the present study was limited by its small sample size and
retrospective nature. Further prospective investigations with a
large number of patients would allow an improved understanding of
the important role of CHIP in GBC progression. Molecular studies
are also required to elucidate the pathogenic mechanism of CHIP’s
involvement in GBC.
Acknowledgements
This study was financially supported
by the research fund of Chungnam National University in 2011.
References
|
1
|
Diehl AK: Epidemiology of gallbladder
cancer: a synthesis of recent data. J Natl Cancer Inst.
65:1209–1214. 1980.PubMed/NCBI
|
|
2
|
Randi G, Franceschi S and La Vecchia C:
Gallbladder cancer worldwide: geographical distribution and risk
factors. Int J Cancer. 118:1591–1602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller G and Jarnagin WR: Gallbladder
carcinoma. Euro J Surg Oncol. 34:306–312. 2008. View Article : Google Scholar
|
|
4
|
Gourgiotis S, Kocher HM, Solaini L,
Yarollahi A, Tsiambas E and Salemis NS: Gallbladder cancer. Am J
Surg. 196:252–264. 2008. View Article : Google Scholar
|
|
5
|
Balachandran P, Agarwal S, Krishnani N, et
al: Predictors of long-term survival in patients with gallbladder
cancer. J Gastrointest Surg. 10:848–854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varga M, Obrist P, Schneeberger S, et al:
Overexpression of epithelial cell adhesion molecule antigen in
gallbladder carcinoma is an independent marker for poor survival.
Clin Cancer Res. 10:3131–3136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park JS, Yoon DS, Kim KS, et al: Analysis
of prognostic factors after curative resection for gallbladder
carcinoma. Korean J Gastroenterol. 48:32–36. 2006.(In Korean).
|
|
8
|
Malka D, Boige V, Dromain C, Debaere T,
Pocard M and Ducreux M: Biliary tract neoplasms: update 2003. Curr
Opin Oncol. 16:364–371. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roa I, de Aretxabala X, Araya JC, et al:
Morphological prognostic elements in gallbladder cancer. Rev Med
Chil. 130:387–395. 2002.(In Spanish).
|
|
10
|
Roa I, Melo A, Roa J, Araya J, Villaseca M
and de Aretxabala X: P53 gene mutation in gallbladder cancer. Rev
Med Chil. 128:251–258. 2000.(In Spanish).
|
|
11
|
Itoi T, Watanabe H, Ajioka Y, et al: APC,
K ras codon 12 mutations and p53 gene expression in carcinoma and
adenoma of the gall bladder suggest two genetic pathways in gall
bladder carcinogenesis. Pathol Int. 46:333–340. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Puhalla H, Wrba F, Kandioler D, et al:
Expression of p21(Wafl/Cip1), p57(Kip2) and HER2/neu in patients
with gall-bladder cancer. Anticancer Res. 27:1679–1684.
2007.PubMed/NCBI
|
|
13
|
Shi YZ, Hui AM, Li X, Takayama T and
Makuuchi M: Overexpression of retinoblastoma protein predicts
decreased survival and correlates with loss of p16INK4 protein in
gall-bladder carcinomas. Clin Cancer Res. 6:4096–4100.
2000.PubMed/NCBI
|
|
14
|
Zhi YH, Liu RS, Song MM, et al:
Cyclooxygenase-2 promotes angiogenesis by increasing vascular
endothelial growth factor and predicts prognosis in gallbladder
carcinoma. World J Gastroenterol. 11:3724–3728. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giatromanolaki A, Sivridis E, Simopoulos
C, et al: Hypoxia inducible factors 1alpha and 2alpha are
associated with VEGF expression and angiogenesis in gallbladder
carcinomas. J Surg Oncol. 94:242–247. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim YW, Huh SH, Park YK, Yoon TY, Lee SM
and Hong SH: Expression of the c-erb-B2 and p53 protein in
gallbladder carcinomas. Oncol Rep. 8:1127–1132. 2001.PubMed/NCBI
|
|
17
|
Zhang M, Pan JW, Ren TR, Zhu YF, Han YJ
and Kühnel W: Correlated expression of inducible nitric oxide
synthase and P53, Bax in benign and malignant diseased gallbladder.
Ann Anat. 185:549–554. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roa I, Ibacache G, Melo A, et al:
Subserous gallbladder carcinoma: expression of cadherine-catenine
complex. Rev Med Chil. 130:1349–1357. 2002.(In Spanish).
|
|
19
|
Chang HJ, Jee CD and Kim WH: Mutation and
altered expression of beta-catenin during gallbladder
carcinogenesis. Am J Surg Pathol. 26:758–766. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi YL, Xuan YH, Shin YK, et al: An
immunohistochemical study of the expression of adhesion molecules
in gallbladder lesions. J Histochem Cytochem. 52:591–601. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Choi SY, Jo YS, Huang SM, et al: L1 cell
adhesion molecule as a novel independent poor prognostic factor in
gallbladder carcinoma. Hum Pathol. 42:1476–1483. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Connell P, Ballinger CA, Jiang J, et al:
The co-chaperone CHIP regulates protein triage decisions mediated
by heat-shock proteins. Nat Cell Biol. 3:93–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Demand J, Alberti S, Patterson C and
Höhfeld J: Cooperation of a ubiquitin domain protein and an E3
ubiquitin ligase during chaperone/proteasome coupling. Curr Biol.
11:1569–1577. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou P, Fernandes N, Dodge IL, et al:
ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol
Chem. 278:13829–13837. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo W, Zhong J, Chang R, Hu H, Pandey A
and Semenza GL: Hsp70 and CHIP selectively mediate ubiquitination
and degradation of hypoxia-inducible factor (HIF)-1α but not
HIF-2α. J Biol Chem. 285:3651–3663. 2010.PubMed/NCBI
|
|
26
|
Jang KW, Lee JE, Kim SY, et al: The
C-terminus of Hsp70-interacting protein promotes Met receptor
degradation. J Thorac Oncol. 6:679–687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Esser C, Scheffner M and Höhfeld J: The
chaperone-associated ubiquitin ligase CHIP is able to target p53
for proteasomal degradation. J Biol Chem. 280:27443–27448. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tripathi V, Ali A, Bhat R and Pati U: CHIP
chaperones wild type p53 tumor suppressor protein. J Biol Chem.
282:28441–28454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McDonough H, Charles PC, Hilliard EG, et
al: Stress-dependent Daxx-CHIP interaction suppresses the p53
apoptotic program. J Biol Chem. 284:20649–20659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao Y, Han C, Huang H, et al: Heat shock
protein 70 together with its co-chaperone CHIP inhibits TNF-alpha
induced apoptosis by promoting proteasomal degradation of apoptosis
signal-regulating kinase1. Apoptosis. 15:822–833. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li F, Xie P, Fan Y, et al: C terminus of
Hsc70-interacting protein promotes smooth muscle cell proliferation
and survival through ubiquitin-mediated degradation of FoxO1. J
Biol Chem. 284:20090–20098. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu T, Zhou Q, Zhou J, et al: Carboxyl
terminus of Hsp70-interacting protein (CHIP) contributes to human
glioma oncogenesis. Cancer Sci. 102:959–966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Salvesen GS and Duckett CS: IAP proteins:
blocking the road to death’s door. Nat Rev Mol Cell Biol.
3:401–410. 2002.
|
|
34
|
Haupt Y, Maya R, Kazaz A and Oren M: Mdm2
promotes the rapid degradation of p53. Nature. 387:296–299. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duan L, Miura Y, Dimri M, et al:
Cbl-mediated ubiquitinylation is required for lysosomal sorting of
epidermal growth factor receptor but is dispensable for
endocytosis. J Biol Chem. 278:28950–28960. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harper JW, Burton JL and Solomon MJ: The
anaphase-promoting complex: it’s not just for mitosis any more.
Genes Dev. 16:2179–2206. 2002.
|
|
37
|
Li SH, Li CF, Sung MT, et al: Skp2 is an
independent prognosticator of gallbladder carcinoma among
p27Kip1-interacting cell cycle regulators: an immunohistochemical
study of 62 cases by tissue microarray. Mod Pathol. 20:497–507.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lakshmanan M, Bughani U, Duraisamy S,
Diwan M, Dastidar S and Ray A: Molecular targeting of E3 ligases -
a therapeutic approach for cancer. Expert Opin Ther Targets.
12:855–870. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Y: E3 ubiquitin ligases as cancer
targets and biomarkers. Neoplasia. 8:645–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kajiro M, Hirota R, Nakajima Y, et al: The
ubiquitin ligase CHIP acts as an upstream regulator of oncogenic
pathways. Nat Cell Biol. 11:312–319. 2009. View Article : Google Scholar : PubMed/NCBI
|