Introduction
Thyroid cancer is the most frequently occurring
endocrine malignancy. Since this cancer often afflicts young
adults, thyroid cancer represents a challenging clinical problem.
Thyroid carcinomas are derived from follicular epithelial cells and
have a broad spectrum of neoplastic phenotypes. These phenotypes
are well-differentiated thyroid carcinoma, including papillary
thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC),
poorly differentiated thyroid carcinoma, representing ∼5% of
thyroid cancers, and the rare but always rapidly lethal anaplastic
thyroid carcinoma (ATC) (1).
Although thyroid neoplasms may be diagnosed from fine needle
aspirates, differentiating the more frequently occurring follicular
adenoma from malignant lesions remains challenging. There are
currently no markers to separate these groups; therefore the
majority of patients are subjected to surgery and radiotherapy. PTC
is associated with mutations in RET, BRAF and RAS, while FTC
exhibits either RAS mutations or PPARg gene rearrangements
(2–6). The search for genetic alterations for
the identification of malignancy has low sensitivity, since
numerous cancer samples do not bear any of these genetic
alterations, and low specificity, since benign adenoma shares
genetic lesions (RAS, PPARg and RET/PTC) with cancer (7). Immunohistochemical studies of thyroid
cancer have allowed the development of potential molecular
diagnostic tools (8). Galectin-3
(Gal-3) has received significant attention and is considered to be
the most accurate stand-alone marker for differentiated thyroid
cancer diagnosis. Gal-3 is highly expressed in thyroid cancer, but
not in normal thyroid tissue and infrequently in benign thyroid
lesions (9). In a large scale
study, Gal-3 was reported to have a sensitivity of 83%, specificity
of 92% and accuracy of 95% (10).
Despite initial enthusiasm, extensive experience with Gal-3 as a
potential marker of malignancy by immunocytochemistry has failed to
provide clear evidence of superior diagnostic accuracy compared
with traditional cytology (11).
Clearly, there is a need for additional markers in order to
accurately diagnose malignant thyroid lesions and avoid patients
undergoing unnecessary and potentially harmful therapies.
The DNA methylation state of several tumor
suppressor genes has been proposed to be an advantageous marker of
malignancy in various tumor types (12,13).
Aberrant hypermethylation and the consequent silencing of tumor
suppressor genes have been frequently observed in thyroid cancer
(14–16) but failed as a selective molecular
marker in thyroid tumorigenesis. However, at present the
possibility that genes that are silent in the normal thyroid but
specifically activated in thyroid malignancy are subject to
cancer-specific epigenetic alterations, has not yet been
investigated.
The present study addressed the hypothesis that the
DNA methylation state of Gal-3 gene may be associated with
malignancy in thyroid neoplasias.
Materials and methods
Tissue samples
Neoplastic and normal human thyroid tissues were
obtained from surgical specimens and immediately frozen in liquid
nitrogen. Thyroid samples were collected at the Service
d’Anatomo-Pathologie (Centre Hospitalier Lyon Sud, Pierre Benite,
France). The study was approved by the ethics committee of the
University of Naples, Naples, Italy.
DNA extraction from tissues
Genomic DNA was extracted for each sample from a
portion of liquid nitrogen-pulverized tissue and was prepared using
a QIAamp DNAMini kit (Qiagen, Hilden, Germany), following the
manufacturer’s instructions.
Bisulfite treatment
The sodium bisulfite conversion was performed using
an EZ DNA Methylation kit (Zymo Research, Irvine, CA, USA). The
manufacturer’s instructions were followed by using 2 mg of genomic
DNA and eluting in 30 ml of H2O.
MassARRAY methylation analysis
MassCLEAVE biochemistry was performed as described
previously (17,18). Mass spectra were acquired using a
MassARRAY Compact matrix-assisted laser
desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometer
(Sequenom, San Diego, CA, USA) and spectra methylation ratios were
generated by the Epityper software version 1.0 (Sequenom).
The primers used in the present analysis were: Gal3m
forward, 5′-aggaagagagTTTATTTAGGTGATTTTG GAGAGGG-3′; and Gal3m
reverse, 5′-cagtaatacgact
cactatagggagaaggctAAAAACAAAACACAAACTATAAAA CTCTC-3′. For reverse
primer, an additional T7 promoter tag for in vivo
transcription was added, as well as a 10-mer tag on the forward
primer to adjust for melting temperature differences. Sequences of
these tags are indicated in lower case. The presence of CpG islands
in the genomic region analyzed was assessed using the CGplot
software (http://www.ebi.ac.uk/emboss/cpgplot/).
Pyrosequencing methylation analysis
Quantitative DNA methylation analysis was performed
using a Pyrosequencing PSQ 96MA (Biotage AB, Uppsala, Sweden)
following the manufacturer’s instructions. The reactions were
assayed on the PSQ 96MA using the provided SNP analysis software.
The primers used for the PCR reactions were: LGalS3 forward,
5′-GGTTCGGGGAGAGGATTGGT-3′; and LGalS3 reverse,
5′-ATAACTCCAAACCTCAAATACTCC-3′ (5′-biotinylated). Amplifications
were performed using the protocols developed previously (19,20).
The sequencing primer (LGalS3S1) was 5′-AGGATTGGTTGGGTAG-3′. The
target CpGs were evaluated by anlalyzing the resulting pyrograms.
Analysis of a non-CpG cytosine was used as an internal control for
the completeness of the bisulphite treatment.
Statistical analysis
The statistical significance of differences between
the groups was assessed by the Student’s t-test. Data were
expressed as the mean ± standard deviation (SD). The variance on
the variable among the groups was calculated by Levene’s test for
the equality of variance. All experiments were repeated at least
three times. The methylation score was the sum of the methylation
value for each CpG site (nucleotide positions +134, +137, +142,
+147 and +156) of each patient. P<0.05 was considered to
indicate statistically significant differences.
Databases
The Gal-3 (LGALS3) gene sequences were retrieved by
the Ensembl database accession number, ENST0000025430.
Results
Gal-3 gene methylation analysis
DNA methylation analysis of the Gal-3 promoter
region was performed on genomic DNA extracted from human tissue
samples derived from 42 tumors (39 papillary and 3 anaplastic
carcinomas), 3 goiters and 5 normal thyroid tissues obtained from
surgical specimens. The map of the human Gal-3 gene, including the
relative positions of the analyzed CpG sites, is shown in Fig. 1A. To investigate the methylation
state of a Gal-3 genomic region encompassing the transcriptional
start site, two independent quantitative DNA methylation analysis
techniques were performed. Mass spectrometry-based methylation
analysis (MassARRAY) and pyrosequencing technology were used to
assess the precise degree of methylation of each CpG site. First, a
large genomic region from −89 to +408, which included the
transcriptional start site and 65 CpG sites, was analyzed by
MassARRAY. The results of duplicate experiments indicated that, in
this genomic region, the majority of the CpG sites were
unmethylated or slightly methylated (data not shown) in the
neoplastic and non-neoplastic tissues, with the exception of a
small region, including 5 CpG sites, localized downstream of the
transcriptional start site, where differential methylation was
detected. However, since MALDI-TOF analysis did not allow the
determination of the methylation degree of each single CpG site in
this region, focused DNA methylation analysis using a different
technology was performed. Pyrosequencing analysis was then
performed with the aim of quantitatively evaluating the methylation
state of each of the 5 CpG sites (nucleotide positions +134, +137,
+142, +147 and +156). The pyrosequencing assays were performed
using the primers indicated in Fig.
1A, covering the region from +104 to +196, and the results were
plotted on a histogram showing the methylation degree of each CpG
site in each tissue sample (Fig.
1B). Marked differential methylation was observed between the
neoplastic and non-neoplastic groups. A low methylation degree
(0–28%) at the 5 CpG sites was observed in all the neoplastic
samples, including anaplastic and papillary carcinomas, while a
high methylation degree (up to 80%) was present at a minimum of 2
out of the 5 analyzed CpG sites in the non-neoplastic tissues.
Notably, the identity of the hypermethylated CpG sites in the
non-neoplastic tissues was variable among the samples. This
suggested that the methylation state of the whole region
(+134/+156), rather than the methylation state of single CpG site,
is associated with the sample groups. Thus, statistical analysis of
the data was performed.
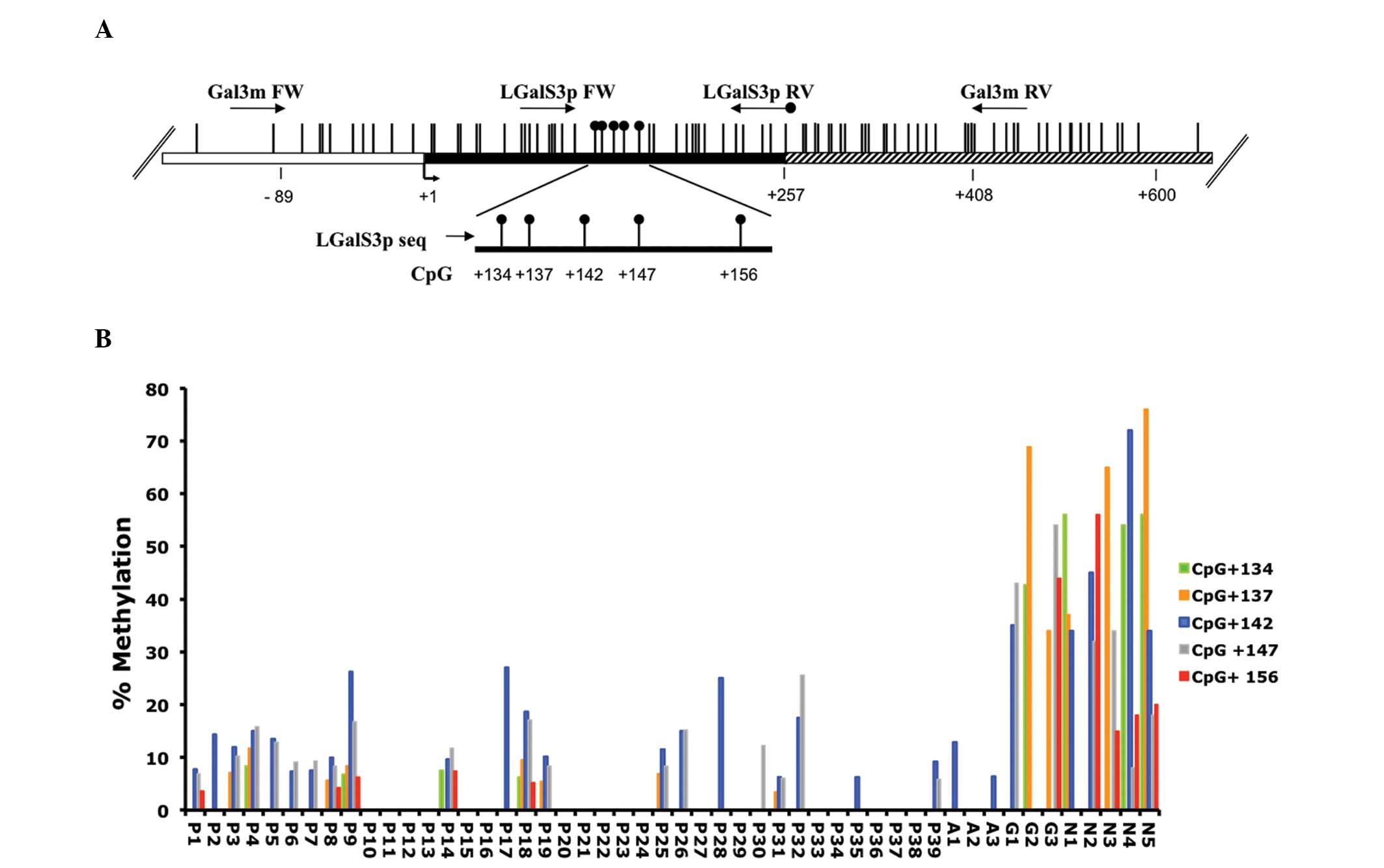 | Figure 1.Gal-3 gene methylation analysis. (A)
Structure of the human Gal-3 promoter gene. The transcriptional
start site (+1) is indicated by an arrow. The regulatory upstream
region (white box), exons (black) and first intron (striped box)
are indicated. Vertical bars represent the relative positions of
each CpG site. The primer positions used for MALDI-TOF and
pyrosequencing analysis are indicated by arrows (Gal3m FW/Gal3m RV
and LGalS3p FW/LGalSp RV biotinylated). Black circles represent the
CpG sites analyzed by pyrosequencing (CpG +134, +137, +142, +147
and +156). (B) Histogram representing the percentage of methylation
of each CpG analyzed in each sample. P1-P39, papillary thyroid
carcinoma; A1-A3, anaplastic thyroid carcinoma; G1-G3, thyroid
goiter; N1-N5, normal thyroid; Gal-3, galectin-3; MALDI-TOF,
matrix-assisted laser desorption/ionization-time-of-flight. |
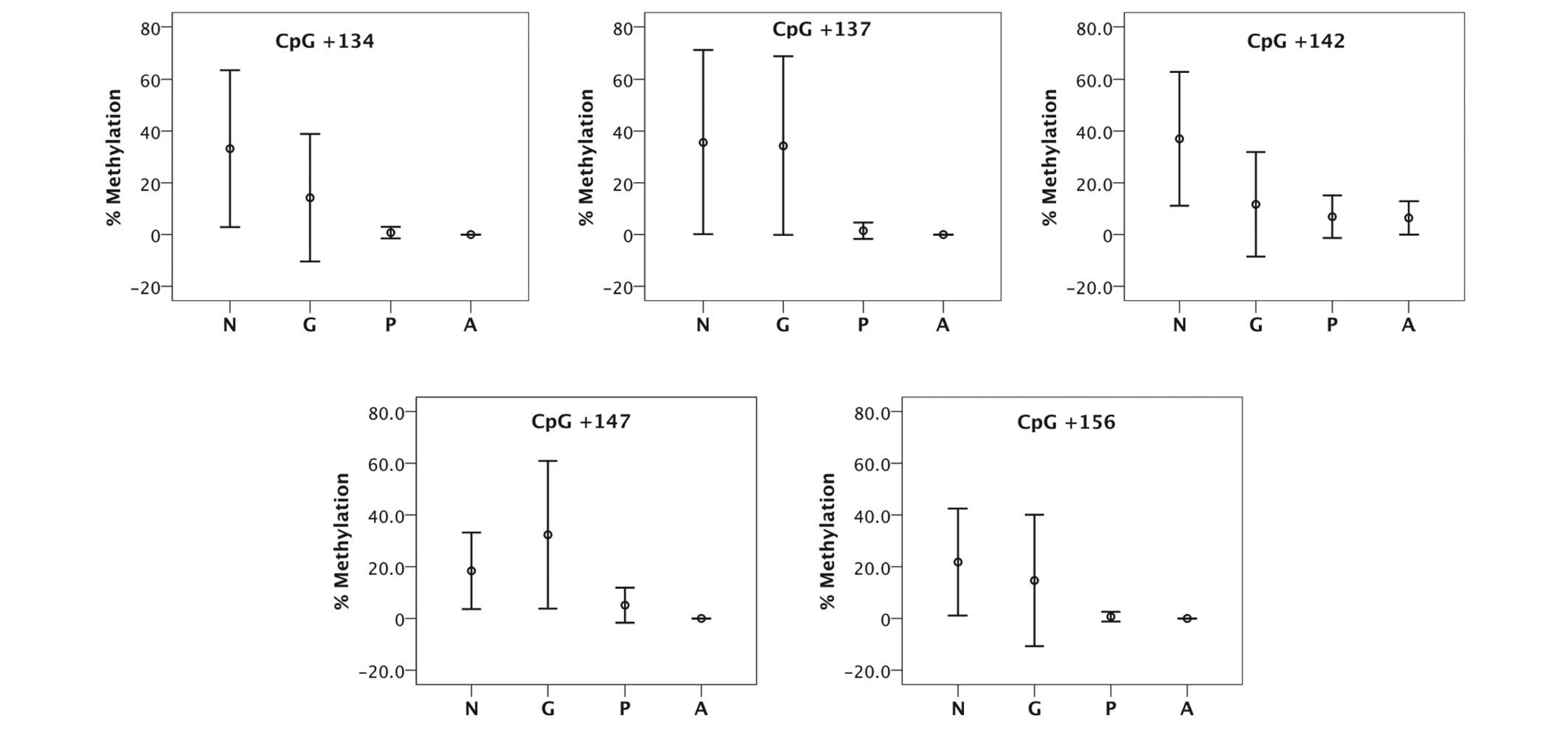
Statistical analysis
The associations between the methylation degree of
each of the 5 CpG sites and tumor types were analyzed. In the first
analysis, each methylated region was compared among the various
thyroid tissues. Although the difference in the percentage of
methylation was higher in the normal thyroid and goiter than the
papillary and anaplastic thyroid carcinoma, the high variability of
the normal thyroid and goiter tissues makes these results poor in
terms of statistical significance and reproducibility (Fig. 2).
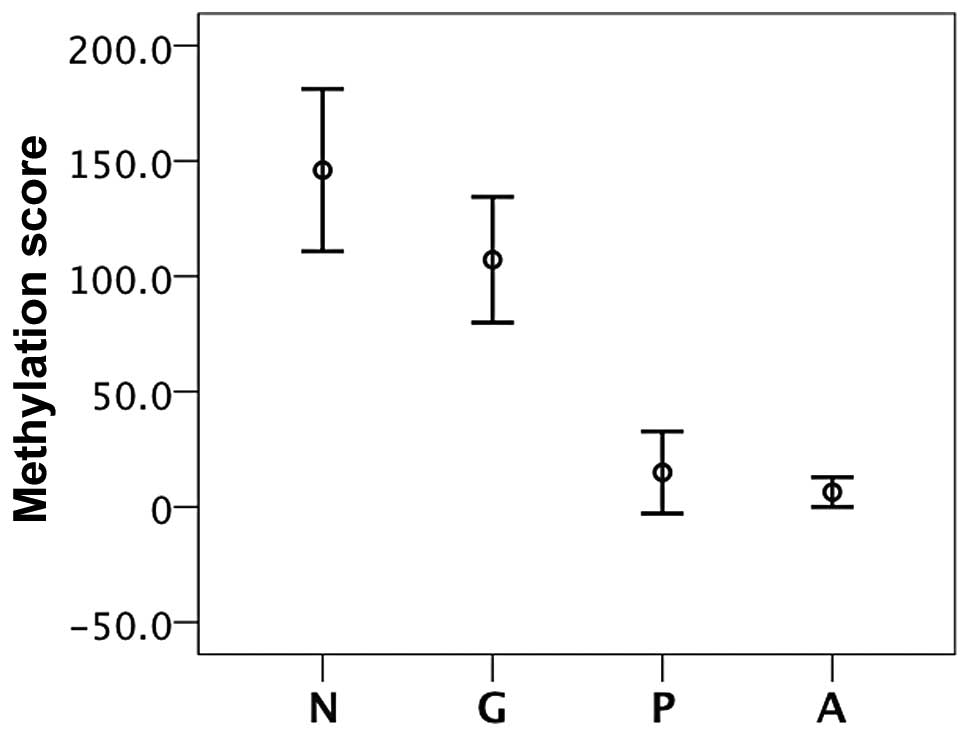
To obtain robust and reproducible results, a new
variable named ‘methylation score’ was then created by the addition
of the methylation values of the CpG sites lying in the
differentially methylated region (+134, +137, +142, +147 and +156)
for each patient. Statistical analysis was performed by considering
the average methylation score of the CpG sites. The results, shown
in Fig. 3, indicate that the
average DNA methylation degree at the 5 CpG sites was significantly
lower (P<0.004) in the anaplastic and papillary thyroid
carcinomas compared with either the goiters or normal thyroid
tissues. Moreover, a reduction of data variability was observed.
The use of the methylation score, which includes more CpG
methylated sites, may be a reliable diagnostic tool for
distinguishing the cancer tissue from normal tissue.
Discussion
The present study showed that the average
methylation degree of 5 CpG sites in the Gal-3 gene regulatory
region is significantly decreased in thyroid cancer tissues
compared with non-neoplastic thyroid tissues. Although Gal-3 gene
expression is an established marker of thyroid malignancy, to the
authors’ knowledge this is the first report investigating the Gal-3
DNA methylation state in thyroid tumors. Previous studies have
reported that aberrant hypermethylation at various genes is
associated with thyroid malignancy, including genes involved in the
control of cell proliferation and invasion, such as p16INK4A
(21), Rassf1A (22), PTEN (23), Rap1GAP (24), TIMP3, RAR-b2, DAPK (15,16,25),
CDH1 (26,27), TGFb and CITED1 (28), as well as genes specific to thyroid
differentiation, such as Na+/I− symporter
(NIS), TSH receptor, pendrin, SL5A8 (29,30)
and TTF-1 (31), as reviewed by
Catalano et al (32). In the
majority of these studies, which were performed by
methylation-specific PCR, a considerable overlap was observed in
the methylation levels between benign and malignant tumors, with
the exception of hypermethylation at RAR-b2 (15,16),
NIS (33), TSHR (34), ECAD (26) and ATM (35), which was observed to be more
prevalent in patients with papillary thyroid carcinoma than in
non-malignant thyroid diseases. However, none of the observed
tumor-related gene hypermethylation is considered to be a
stand-alone marker for distinguishing malignant from benign tumors.
Galusca et al (36)
evaluated the global DNA methylation status in several types of
thyroid tumor using a monoclonal anti-5-methylcytidine (5-mC)
antibody in an immunohisto-chemical quantitative analysis. The
authors observed global DNA hypomethylation in thyroid carcinomas
compared with benign lesions with an overall accuracy estimated to
be similar to Gal-3 immunostaining. Notably, the combination of
5-mC and Gal-3 led to an accuracy of 96% (36). The presently reported data, obtained
by high resolution methylation analysis at the gene-specific level,
shows that hypomethylation of the Gal-3 gene clearly distinguishes
papillary and anaplastic carcinoma from non-neoplastic thyroid
tissues. Marked differential methylation was observed between the
neoplastic and non-neoplastic groups. This difference was easily
detectable by considering the average methylation state of the 5
CpG sites included in the Gal-3 gene region from +134 to +156,
rather than considering the methylation state of individual sites.
In fact, the identity of the hyper-methylated CpG sites in
non-neoplastic tissues was highly variable among the samples.
Although in the present study the Gal-3 methylation state was not
investigated in thyroid adenomas, the data suggest that the
evaluation of the Gal-3 methylation state at the five identified
CpG sites may greatly aid in thyroid tumor diagnosis. Further
studies on a larger range of samples, including malignant and
benign thyroid tumors, are likely to clarify whether the assessment
of the methylation state of CpG sites +134, +137, +142, +147 and
+156, possibly in combination with Gal-3 immunostaining, may be a
candidate analysis which substantially contributes to increasing
the accuracy of the currently used markers for distinguishing
thyroid cancer from benign thyroid adenomas.
Acknowledgements
The present study was supported by
grants from MIUR and Regione Campania (DGRC 1091/09) to L.C. and
F.L., from AIRC to A.F., and from Progetto Bandiera Epigenomica,
EPIGEN, CNR (to L.C. and A.F.).
References
|
1.
|
Hedinger C, Williams ED and Sobin LH: The
WHO histological classification of thyroid tumours: a commentary on
the second edition. Cancer. 63:908–911. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Kondo T, Ezzat S and Asa SL: Pathogenetic
mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer.
6:292–306. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Xing M: BRAF mutation in thyroid cancer.
Endocr Relat Cancer. 12:245–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Suarez HG, du Villard JA, Severino M, et
al: Presence of mutations in all three ras genes in human thyroid
tumors. Oncogene. 5:565–570. 1990.PubMed/NCBI
|
|
5.
|
Santoro M, Melillo RM, Grieco M,
Berlingieri MT, Vecchio G and Fusco A: The TRK and RET tyrosine
kinase oncogenes cooperate with ras in the neoplastic
transformation of a rat thyroid epithelial cell line. Cell Growth
Differ. 4:77–84. 1993.PubMed/NCBI
|
|
6.
|
de Nigris F, Cerutti J, Morelli C, et al:
Isolation of a SIR-like gene, SIR-T8, that is overexpressed in
thyroid carcinoma cell lines and tissues. Br J Cancer. 86:917–923.
2002.
|
|
7.
|
Melillo RM, Castellone MD, Guarino V, et
al: The RET/PTC-RAS-BRAF linear signaling cascade mediates the
motile and mitogenic phenotype of thyroid cancer cells. J Clin
Invest. 115:1068–1081. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Chiu CG, Strugnell SS, Griffith OL, et al:
Diagnostic utility of galectin-3 in thyroid cancer. Am J Pathol.
176:2067–2081. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Xu XC, el-Naggar AK and Lotan R:
Differential expression of galectin-1 and galectin-3 in thyroid
tumors. Potential diagnostic implications. Am J Pathol.
147:815–822. 1995.PubMed/NCBI
|
|
10.
|
Gharib H: Fine-needle aspirate biopsy of
thyroid nodules: advantage, limitation and effect. Mayo Clin Proc.
69:44–49. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Bartolazzi A, Orlandi F, Saggiorato E, et
al: Galectin-3-expression analysis in the surgical selection of
follicular thyroid nodules with indeterminate fine-needle
aspiration cytology: a prospective multicentre study. Lancet Oncol.
9:543–549. 2008. View Article : Google Scholar
|
|
12.
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2(Suppl 1): S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Baylin S and Bestor TH: Altered
methylation patterns in cancer cell genomes: cause or consequence?
Cancer Cell. 1:299–305. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Czarnecka K, Pastuszak-Lewandoska D,
Migdalska-Sek M, et al: Aberrant methylation as a main mechanism of
TSGs silencing in PTC. Front Biosci (Elite Ed). 3:137–157. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Hoque MO, Rosenbaum E, Westra WH, et al:
Quantitative assessment of promoter methylation profiles in thyroid
neoplasms. J Clin Endocrinol Metab. 90:4011–4018. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Cras A, Darsin-Bettinger D, Balitrand N,
Cassinat B, Soulié A, Toubert ME, Delva L and Chomienne C:
Epigenetic patterns of the retinoic acid receptor
β2promoter in retinoic acid-resistant thyroid cancer
cells. Oncogene. 26:4018–4024. 2007.
|
|
17.
|
Ehrich M, Nelson MR, Stanssens P, et al:
Quantitative high-throughput analysis of DNA methylation patterns
by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci
USA. 102:15785–15790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Keller S, Sarchiapone M, Zarrilli F, et
al: Increased BDNF promoter methylation in the Wernicke area of
suicide subjects. Arch Gen Psychiatry. 67:258–267. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Tost J and Gut IG: DNA methylation
analysis by pyrosequencing. Nat Protoc. 2:2265–2275. 2007.
View Article : Google Scholar
|
|
20.
|
Keller S, Sarchiapone M, Zarrilli F, et
al: TrkB gene expression and DNA methylation state in Wernicke area
does not associate with suicidal behavior. J Affect Disord.
135:400–404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Elisei R, Shiohara M, Koeffler HP and
Fagin JA: Genetic and epigenetic alterations of the
cyclin-dependent kinase inhibitor-sp15INK4b and p16INK4a in human
thyroid carcinoma cell lines and primary thyroid carcinomas.
Cancer. 83:2185–2193. 1998. View Article : Google Scholar
|
|
22.
|
Schagdarsurengin U, Gimm O, Hoang-Vu C,
Dralle H, Pfeifer GP and Dammann R: Frequent epigenetic silencing
of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer
Res. 62:3698–3701. 2002.PubMed/NCBI
|
|
23.
|
Alvarez-Nuñez F, Bussaglia E, Mauricio D,
et al: PTEN promoter methylationin sporadic thyroid carcinomas.
Thyroid. 16:17–23. 2006.PubMed/NCBI
|
|
24.
|
Zuo H, Gandhi M, Edreira MM, et al:
Downregulation of Rap1GAP through epigenetic silencing and loss of
heterozygosity promotes invasion and progression of thyroid tumors.
Cancer Res. 70:1389–1397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Hu S, Liu D, Tufano RP, et al: Association
of aberrant methylation of tumor suppressor genes with tumor
aggressiveness and BRAF mutation in papillary thyroid cancer. Int J
Cancer. 119:2322–2329. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Graff JR, Greenberg VE, Herman JG, et al:
Distinct patterns of E-cadherin CpG island methylation in
papillary, follicular, Hurthle’s cell, and poorly differentiated
human thyroid carcinoma. Cancer Res. 58:2063–2066. 1998.PubMed/NCBI
|
|
27.
|
Wiseman SM, Masoudi H, Niblock P, et al:
Derangement of the E-cadherin/catenin complex is involved in
transformation of differentiated to anaplastic thyroid carcinoma.
Am J Surg. 191:581–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Sassa M, Hayashi Y, Watanabe R, et al:
Aberrant promoter methylation in overexpression of CITED1 in
papillary thyroid cancer. Thyroid. 21:511–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Xing M: BRAF mutation in papillary thyroid
cancer: pathogenic role, molecular bases, and clinical
implications. Endocr Rev. 28:742–762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Xing M: Gene methylation in thyroid
tumorigenesis. Endocrinology. 148:948–953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Kondo T, Nakazawa T, Ma D, et al:
Epigenetic silencing of TTF-1/NKX2-1 through DNA hypermethylation
and histone H3 modulation in thyroid carcinomas. Lab Invest.
89:791–799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Catalano MG, Fortunati N and Boccuzzi G:
Epigenetics modifications and therapeutic prospects in human
thyroid cancer. Front Endocrinol (Lausanne). 3:402012.PubMed/NCBI
|
|
33.
|
Venkataraman GM, Yatin M, Marcinek R and
Ain KB: Restoration of iodide uptake in dedifferentiated thyroid
carcinoma: relationship to human Na+/I−
symporter gene methylation status. J Clin Endocrinol Metab.
84:2449–2457. 1999.PubMed/NCBI
|
|
34.
|
Xing M, Usadel H, Cohen Y, et al:
Methylation of the thyroid-stimulating hormone receptor gene in
epithelial thyroid tumors: a marker of malignancy and a cause of
gene silencing. Cancer Res. 63:2316–2321. 2003.PubMed/NCBI
|
|
35.
|
Smith JA, Fan CY, Zou C, Bodenner D and
Kokoska MS: Methylation status of genes in papillary thyroid
carcinoma. Arch Otolaryngol Head Neck Surg. 133:1006–1011. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Galusca B, Dumollard JM, Lassandre S,
Niveleau A, Prades JM, Estour B and Peoc’h M: Global DNA
methylation evaluation: potential complementary marker in
differential diagnosis of thyroid neoplasia. Virchows Arch.
447:18–23. 2005. View Article : Google Scholar : PubMed/NCBI
|