Introduction
Exposure to ionizing radiation poses a threat to
astronauts on space missions. Of particular concern is the
radiation emitted from high atomic mass (Z) and high-energy (HZE)
particles. Unlike the relatively infrequent energy deposition by
low linear energy transfer (LET) radiation along its track, high
LET radiation from HZE particles exhibits a highly energetic and
dense core and a laterally extending secondary radiation. The
secondary radiation, termed δ-rays, generates an external region
called the penumbra as a consequence of the HZE particle traversing
through a medium. It has been estimated that there are
approximately 32 cells hit by δ-rays for each cell traversed by a
primary HZE particle of 1 GeV/n iron ions (1). The combination of the highly energetic
core and δ-rays suggests a more damaging effect of HZE particle
radiation compared to that observed from the same dose of low LET
radiation. For example, the ratio of double-strand breaks to
single-strand breaks is observed to be higher for HZE particle
radiation when compared to photon radiation (2). There is evidence that differentially
expressed genes respond uniquely to either photons or HZE particles
(3). The number and types of gene
expression changes attributable to δ-rays are thought to be similar
to those expected from exposure to other types of low LET radiation
(4). In addition, a significant
number of genes not associated with DNA damage have been shown to
be responsive to ionizing radiation (5). The response of target genes for
products necessary for cell communication, such as those involved
with the extracellular matrix (ECM) (6) and gap junctions (e.g., connexin 43)
(7), further suggests an important
role of ionizing radiation in altering events other than those
associated with the DNA damage/repair response.
Ionizing radiation is typically viewed as a
genotoxic stress to cells. As such, growth arrest of
radiation-damaged cells through the control of cell cycle
checkpoints allows the cells to recognize and repair DNA and other
damage before DNA synthesis or mitosis occurs (8), whereas apoptosis serves as the means
for removing cells with irreparable damage from the cell
population. Studies of the effects of low doses have been difficult
due to the inherently small changes in gene expression patterns.
However, experiments with X- and γ-rays at doses ranging from 0.02
to 20 Gy have previously been successful in demonstrating the
effects of low LET radiation on gene expression by microarray
analyses (9–12). Low-dose experiments (0.2–2 Gy
γ-rays) on peripheral blood cells revealed the induction of genes
up to 72 h post-irradiation following 0.2-Gy exposure, whereby a
linear dose-response correlation was observed between 0.2 and 2 Gy
over the 24-48 h post-irradiation period for selected genes
associated with growth arrest, such as CDKN1A/WAF1 and GADD45A
(10). Exposures of human
fibroblasts to 0.02 and 4 Gy X-rays over a time course of 1–24 h
revealed the expression of genes exclusive to low or high-dose
exposures, with overlaps (4). While
the alteration of gene expression for genes associated with
cell-cell signaling and DNA damage response was observed following
exposure to a low dose, apoptosis and proliferation genes were
modulated following exposure to a high dose of radiation (11).
Selenium is known to be a cancer chemopreventive
agent (13) with anticarcinogenic
activities against the development of cancers occurring in several
different organ systems, such as the prostate, lung and colon
(14,15). Selenite and L-selenomethionine (SeM)
are the two forms of seleno compounds used in most cancer
prevention studies. Although the mechanism for their mode of action
is not clear, their ability to promote apoptosis (15) and cytotoxicity (16) in various cancer cell lines suggests
a pro-oxidant effect from the inorganic form of selenium. SeM is
the organic form of selenium found in selenized yeast supplements
and used in clinical trials. The ability of SeM to effectively
mitigate oxidative stress in vitro and in vivo
(17–19) supports a role for SeM in antioxidant
activities. Moreover, SeM treatment was shown to suppress iron ion
radiation-induced transformation in human thyroid epithelial cells
(HTori-3) (17).
It was reported previously that a 10 cGy dose to
HTori-3 cells did not affect cell survival levels, and that a 20
cGy dose led to cell survival levels of approximately 85% in cells
exposed to iron ion irradiation (20). In the current study, genomic
profiling was performed to assess the effects of non-toxic (10 cGy)
and slightly toxic (20 cGy) radiation exposure in cultured HTori-3
cells in the presence and absence of SeM.
Materials and methods
Cell culture and radiation exposure
Human thyroid epithelial cells (HTori-3) were
maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F12
supplemented with 1% glutamine and 10% FBS (growth medium).
Twenty-four hours prior to irradiation with iron ions, fresh medium
with or without 5 μM SeM (Sigma-Aldrich, St. Louis, MO, USA)
was added. At the time of radiation exposure, the cells were
approximately 80% confluent. Irradiation was performed at the NASA
Space Radiation Laboratory (NSRL) facility at the Brookhaven
National Laboratory (Upton, NY, USA). Radiation exposure was from 1
GeV/n iron ions delivered as a horizontal beam of approximately
20×20 cm in dimension at a dose rate of approximately 40 cGy/min.
Six or 16 h post-irradiation, the cells were harvested and frozen
in RNAlater solution (Qiagen, Valencia, CA, USA). Three replicates
of two independent experiments were generated for each radiation
dose/SeM supplement combination. For sham-irradiated controls, SeM
treated or untreated cells were maintained in the same manner as
utilized for the irradiated cells at the NSRL facility. For mock
SeM pretreatment, the medium was supplemented with
phosphate-buffered saline (PBS).
RNA preparation, microarray and real-time
RT-PCR
RNA was extracted from frozen cells using the RNeasy
kit (Qiagen) according to the manufacturer’s instructions. Each
microarray probe was prepared and hybridized at the Penn
Bioinformatics Core (University of Pennsylvania) using 1 μg
total RNA. First-strand cDNA was synthesized using Superscript II
First Strand cDNA Synthesis System (Invitrogen, Carlsbad, CA, USA).
Following RNA degradation with RNase H, second-strand cDNA was
synthesized with DNA polymerase I and extracted with 25:24:1 (v/v)
phenol:chloroform:isoamyl alcohol. The double-stranded cDNA was
used as a template to generate biotinylated cRNA using the BioArray
HighYield RNA Transcript Labeling kit (Enzo Life Sciences,
Farmingdale, NY, USA). The resulting cRNA was purified, fragmented
and hybridized to U133Av2 Gene Chips (Affymetrix, Santa Clara, CA,
USA) according to the manufacturer’s instructions, and further
processed at the Penn Bioinformatics Core.
For real-time RT-PCR analysis, cDNA was initially
synthesized with Superscript II using ∼1-2 μg total RNA.
Two-step PCR (initial denaturation, 95°C, 30 sec; 40–50 cycles of
95°C, 5 sec and 65°C, 34 sec) was monitored in real-time by the
SYBR-Green DNA intercalating dye (SYBR Advantage qPCR Premix;
Clontech Laboratories, Inc., Mountain View, CA, USA) according to
the manufacturer’s instructions on an Applied Biosystems 7300
Real-Time PCR System instrument. Primers were designed by
ProbeFinder (Roche Applied Science, Indianapolis, IN, USA) or
Primer Designer (equipped with the LightCycler real-time RT-PCR
instrument, Roche Applied Science) with both β-actin and GAPDH
serving as reference genes. The primer sequences used are shown in
Table I. PCR products were
confirmed by melting curve analysis and/or running on 1% agarose
gels. cDNA microarray analysis was performed with biological
triplicates, whereas n=4–6 for real-time RT-PCR analyses.
 | Table I.Primer sets used for real-time RT-PCR
experiments. |
Table I.
Primer sets used for real-time RT-PCR
experiments.
| Symbol | Forward primer
(5′→3′) | Reverse primer
(5′→3′) | Amplicon size
(nt) |
|---|
| ATF3 |
TTTGCCATCCAGAACAAGC |
CATCTTCTTCAGGGGCTACCT | 121 |
| CDC6 |
CCTGTTCTCCTCGTGTAAAAGC |
GTGTTGCATAGGTTGTCATCG | 73 |
| FAS |
GTGGACCCGCTCAGTACG |
TCTAGCAACAGACGTAAGAACCA | 112 |
| GADD45A |
TTGCAATATGACTTTGGAGGAA |
CATCCCCCACCTTATCCAT | 71 |
| β-actin |
TCGTGCGTGACATTAAGG | ACAGGTCTTTGCGGAT | 258 |
| GAPDH |
AGCCACATCGCTCAGACAC |
GCCCAATACGACCAAATCC | 66 |
Statistical analysis and DAVID
Probe intensities were summarized and normalized
using log scale robust multi-array analysis (RMA). ANOVA was
performed using the GeneSpring GX software (Agilent, Santa Clara,
CA, USA) using a 1.5-fold minimum expression cut-off and a false
discovery rate (FDR) of 5%. Determination of differential gene
expression was performed using dose (sham irradiation, 10 or 20 cGy
exposure), time (6 or 16 h post-irradiation) or SeM treatment as
independent variables. The Database for Annotation, Visualization
and Integrated Discovery (DAVID) (21) was used for functional annotation and
analysis by uploading Affymetrix-based nomenclature of
statistically significant genes (1.5-fold expression cut-off; 5%
FDR; P<0.05; http://david.abcc.ncifcrf.gov). Gene enrichment for
pathway analysis (KEGG pathway) was accomplished by the integrated
Expression Analysis System Explorer (EASE), with a count threshold
of 2 (minimum number of genes for a particular term) and EASE
threshold of 0.1 (a modified Fisher’s exact P-value). The Student’s
t-test was used for statistical comparisons of real-time RT-PCR
results.
Results
Global gene expression change in response
to low-dose iron ion irradiation
To determine the impact of low-dose HZE particle
radiation on gene expression, HTori-3 cells were exposed to 10 or
20 cGy iron ion irradiation (in the absence of SeM) and compared to
the sham-irradiated cells at 6 h post-irradiation. With the
GeneSpring software, global gene expression patterns are
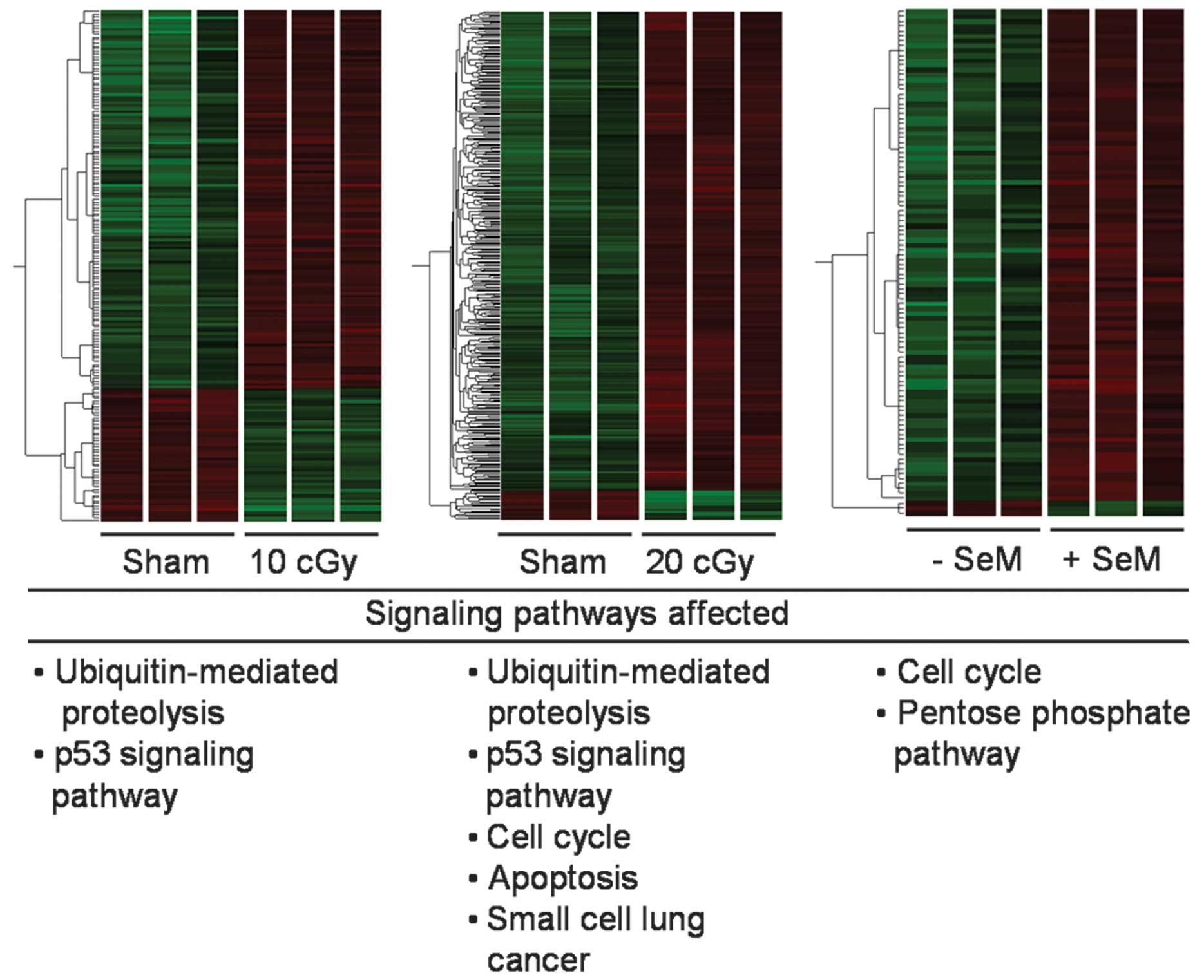
illustrated as heatmaps depicting the clustering of gene expression
upon exposure to radiation (Fig.
1). Pooled averages from three independent experiments were
compared. At a dose of 10 cGy, 121 and 42 unique genes were shown
to be up- and downregulated, respectively, following the removal of
redundant terms by DAVID. Pathway analysis by the KEGG pathway
revealed the upregulation of genes associated with
ubiquitin-mediated proteolysis and the p53 signaling pathway
(Fig. 1 and Table II). Following an increased exposure
to the 20 cGy dose, 285 unique genes were upregulated, while 34
genes were downregulated. Moreover, genes associated with the cell
cycle, apoptosis and small cell lung cancer were observed to be
upregulated, in addition to the pathways elicited at 10 cGy
(Fig. 1 and Table III). No significant pathways were
identified by DAVID for down-regulated genes in either the 10 or 20
cGy dose group. As such, the expression of genes consistent with a
stress response was observed even at non-toxic doses. In addition,
the dose increase (from 10 to 20 cGy) contributed to more stress.
Lists of the total number of differentially expressed genes are
provided in Tables II and III.
 | Table II.Pathway analysis of gene induction at
10 cGy iron ion irradiation. |
Table II.
Pathway analysis of gene induction at
10 cGy iron ion irradiation.
| Symbol | Accession No. | Gene namea | Fold change | Pathway affected |
|---|
| SKP2 | NM_005983 | S-phase
kinase-associated protein 2 | 2.2 | Ubiquitin-mediated
proteolysis |
| CUL2 | NM_003591 | Cullin 2 | 1.8 | Ubiquitin-mediated
proteolysis |
| UBE2G2 | NM_003343 | Ubiquitin-conjugating
enzyme E2 G2 | 1.5 | Ubiquitin-mediated
proteolysis |
| DDB2 | NM_000107 | Damage-specific DNA
binding protein 2 | 1.6 | Ubiquitin-mediated
proteolysis, p53 signaling |
| MDM2 | NM_002392 | Double minute 2 | 1.7 | Ubiquitin-mediated
proteolysis, p53 signaling |
| FAS | NM_000043 | Fas (TNF receptor
superfamily, member 6) | 1.7 | Ubiquitin-mediated
proteolysis, p53 signaling |
 | Table III.Pathway analysis of gene induction at
20 cGy iron ion irradiation. |
Table III.
Pathway analysis of gene induction at
20 cGy iron ion irradiation.
| Symbol | Accession No. | Gene namea | Fold change | Pathway affected |
|---|
| SKP2 | NM_005983 | S-phase
kinase-associated protein 2 | 2.1 | Ubiquitin-mediated
proteolysis |
| CUL5 | AAB70253 | Cullin 5 | 1.5 | Ubiquitin-mediated
proteolysis |
| WWP1 | NM_007013 | WW domain
containing E3 ubiquitin protein ligase 1 | 1.5 | Ubiquitin-mediated
proteolysis |
| CUL3 | AAC28621 | Cullin 3 | 1.5 | Ubiquitin-mediated
proteolysis |
| CUL2 | NM_003591 | Cullin 2 | 1.8 | Ubiquitin-mediated
proteolysis |
| CUL4B | AAB67315 | Cullin 4B | 1.5 | Ubiquitin-mediated
proteolysis |
| SKP1 | NM_006930 | S-phase
kinase-associated protein 1A | 1.6 | Ubiquitin-mediated
proteolysis |
| PIAS2 | AAC36704 | Protein inhibitor
of activated STAT, 2 | 1.5 | Ubiquitin-mediated
proteolysis |
| DDB2 | NM_000107 | Damage-specific DNA
binding protein 2 | 1.7 | p53 signaling |
| FAS | NM_000043 | Fas (TNF receptor
superfamily, member 6) | 2.3 | p53 signaling |
| TNFRSF10B | NM_003842 | TNF receptor
superfamily, member 10B | 1.8 | p53 signaling |
| GADD45A | NM_001924 | Damage-specific DNA
binding protein 2 | 1.6 | p53 signaling, cell
cycle |
| CDK2 | NM_001798 | Cyclin-dependent
kinase 2 | 1.7 | p53 signaling, cell
cycle |
| SKP2 | NM_005983 | S-phase
kinase-associated protein 2 | 2.1 | p53 signaling, cell
cycle |
| BUB3 | AAC06258 | Budding uninhibited
by benzimidazoles 3 homolog (yeast) | 1.7 | Cell cycle |
| YWHAZ | NM_003406 | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
polypeptide | 1.5 | Cell cycle |
| SKP1 | NM_006930 | S-phase
kinase-associated protein 1A | 1.6 | Cell cycle |
| RIPK1 | NM_003804 | Receptor
(TNFRSF)-interacting serine-threonine kinase 1 | 1.7 | Apoptosis |
| FAS | NM_000043 | Fas (TNF receptor
superfamily, member 6) | 2.3 | Apoptosis |
| BCL2L1 | NM_001191 | BCL2-like 1 | 1.5 | Apoptosis |
| TNFRSF10B | NM_003842 | TNF receptor
superfamily, member 10B | 1.8 | Apoptosis |
| CFLAR | NM_003879 | Tumor necrosis
factor receptor superfamily, member 10B | 1.5 | Apoptosis |
| SKP2 | NM_005983 | S-phase
kinase-associated protein 2 | 2.1 | Small cell lung
cancer |
| CDK2 | NM_001798 | Cyclin-dependent
kinase 2 | 1.7 | Small cell lung
cancer |
| BCL2L1 | NM_001191 | BCL2-like 1 | 1.5 | Small cell lung
cancer |
| TRAF3 | AAA56753 | TNF
receptor-associated factor 3 | 1.5 | Small cell lung
cancer |
| PIAS2 | AAC36704 | Protein inhibitor
of activated Stat, 2 | 1.5 | Small cell lung
cancer |
SeM pretreatment alone promotes the
upregulation of genes
HTori-3 cells were pretreated with 5 μM SeM
for 24 h, processed 6 h post-sham irradiation, and compared to the
control cells (mock SeM pretreatment and sham irradiation). In
comparison to the untreated cells, 100 unique transcripts were
differentially modulated, with 97 and 3 genes up- or down-regulated
(Fig. 1 and Table IV). Pathway analysis revealed that
the upregulation of genes was associated with the cell cycle and
pentose phosphate pathway, with no identifiable pathways affected
by the downregulated genes.
 | Table IV.Pathway analysis of gene modulation
after SeM treatment in sham-irradiated cells and in cells exposed
to a 10 cGy dose of radiation. |
Table IV.
Pathway analysis of gene modulation
after SeM treatment in sham-irradiated cells and in cells exposed
to a 10 cGy dose of radiation.
| Symbol | Accession No. | Gene namea | Fold change | Pathway
affected |
|---|
|
Sham-irradiated | | | | |
| CCND3 | NM_001760 | Cyclin D3 | 1.5 | Cell cycle |
| CDC20 | NM_001255 | Cdc20 cell division
cycle 20 homolog (S. cerevisiae) | 1.5 | Cell cycle |
| MAD1L1 | NM_001013836 | Mad1 mitotic arrest
deficient-like 1 (yeast) | 2.0 | Cell cycle |
| CCND1 | NM_053056 | Cyclin D1 | 1.5 | Cell cycle |
| BUB1 | AAB97855 | Budding uninhibited
by benzimidazoles 1 homolog | 1.5 | Cell cycle |
| PGM3 | NM_015599 | Phosphoglucomutase
3 | 1.9 | Pentose
phosphate |
| GPI | NM_000175 | Glucose phosphate
isomerase | 1.7 | Pentose
phosphate |
| PGD | NM_002631 | Phosphogluconate
dehydrogenase | 1.7 | Pentose
phosphate |
| 10 cGyb | | | | |
| PGD | NM_002631 | Phosphogluconate
dehydrogenase | 1.5 | Pentose phosphate,
glycolysis/gluconeogenesis |
| PGM3 | NM_015599 | Phosphoglucomutase
3 | 1.6 | Pentose phosphate
glycolysis/gluconeogenesis |
| GPI | NM_000175 | Glucose phosphate
isomerase | 1.7 | Pentose phosphate
glycolysis/gluconeogenesis |
| ENO1 | NM_001428 | Enolase 1 | 1.9 |
Glycolysis/gluconeogenesis |
| MET | NM_000245 | Met
proto-oncogene | 0.6 | Adherens
junction |
| IQGAP1 | NM_003870 | IQ motif containing
GTPase activating protein 1 | 0.6 | Adherens
junction |
SeM supplementation mitigates gene
expression associated with the cellular stress response from 10 cGy
irradiation
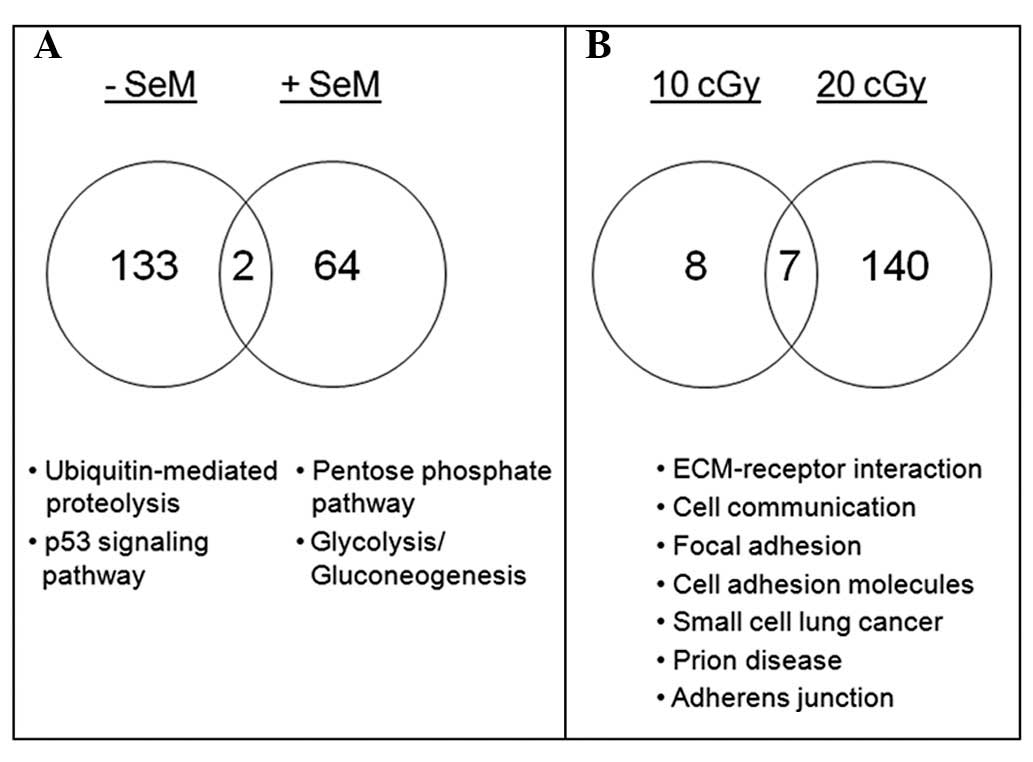
As shown in Fig. 2A,
SeM supplementation in irradiated cell cultures resulted in a
reduction in the number of regulated genes representing less than
half of the total differentially expressed transcripts, in
comparison to the untreated population. Cells were irradiated with
10 cGy iron ions in the absence and presence of SeM pretreatment
and processed 6 h post-irradiation. Datasets from both radiation
treatments were initially compared pairwise to the sham-irradiated
samples and then compared for SeM treatment. Upon SeM pretreatment,
ubiquitin-mediated proteolysis and p53 signaling (Table II) were no longer significant.
Moreover, genes associated with the pentose phosphate pathway and
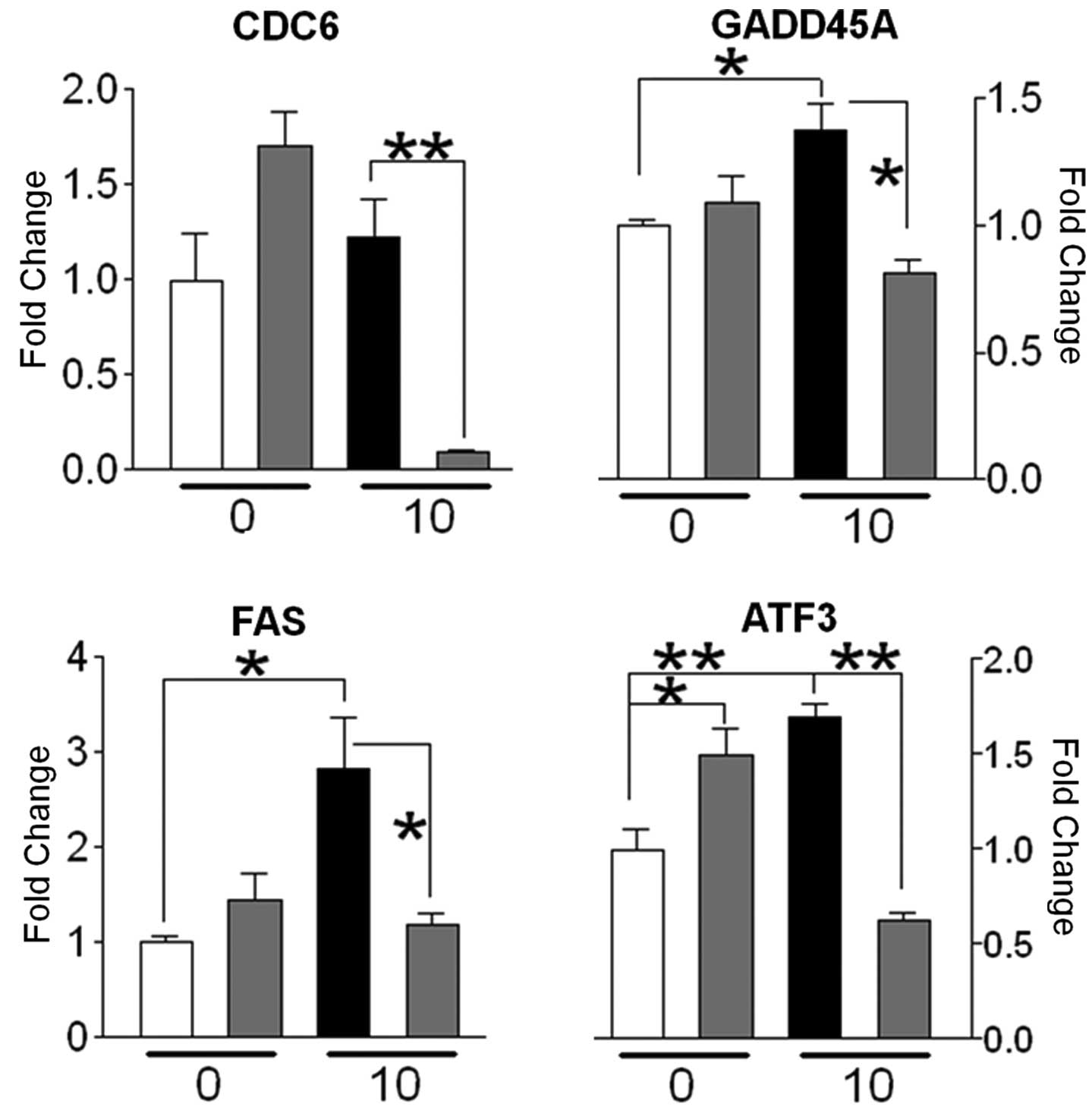
glycolysis/gluconeogenesis were upregulated. Of the stress-related
genes represented in the microarray data, CDC6, GADD45A and FAS
were shown to be downregulated in irradiated cells pretreated with
SeM, as compared to radiation alone (Fig. 3). Additionally, the
well-characterized stress response gene, ATF3 (22), which showed a significant increase
in expression in response to irradiation, was observed to be
downregulated when supplemented with SeM (Fig. 3). Taken together, the downregulation
of these genes associated with the cellular stress response by SeM
in irradiated cells may be indicative of the ability of SeM to
mitigate cellular stress resulting from radiation treatment.
SeM supplementation downregulates cell
communication genes following an extended period
post-irradiation
Having observed the ability of SeM to mitigate
radiation-induced activation of stress-associated pathways at 6 h
post-irradiation, the ability of the cells to recover was further
examined with an extended recovery period. Cells exposed to a 10
cGy dose yielded negligible gene modulation when processed 16 h
post-irradiation, thereby suggesting the ability of the cells to
recover with time (Fig. 2B).
Attempts to observe cells in the absence of SeM supplementation
following 10 cGy exposure at 16 h post-irradiation also yielded
negligible activation of pathways (data not shown). However, upon
exposure to 20 cGy and in the presence of SeM, certain genes
associated with cell communication were observed to be
downregulated, when compared to the gene expression levels in
irradiated cells (Fig. 2B).
Particularly, the suppression of genes encoding for integrin αv,
fibronectin 1 (FN1) and type IV collagen (Table V) may suggest the ability of SeM to
curtail aggressive cell behavior following exposure to a higher
dose of radiation.
 | Table V.Pathway analysis of downregulated
genes in response to SeM treatment with cells harvested at 16 h
post-irradiation.** |
Table V.
Pathway analysis of downregulated
genes in response to SeM treatment with cells harvested at 16 h
post-irradiation.**
| Symbol | Accession No. | Gene namea | Fold change | Pathway
affected |
|---|
| 10 cGy | | | | |
| No identifable
pathways | | | | |
| 20 cGy | | | | |
| MET | NM_000245 | Met
proto-oncogene | 0.7 | Focal adhesion |
| VCL | NM_003373 | Vinculin | 0.6 | Focal adhesion |
| ITGAV | NM_002210 | Integrin, alpha
V | 0.6 | Focal adhesion,
extracellular matrix receptor interaction |
| LAMB1 | NM_002291 | Laminin, beta
1 | 0.5 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| TNC | NM_002160 | Tenascin C | 0.3 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| THBS1 | NM_003246 | Thrombospondin
1 | 0.6 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| COL4A2 | NM_001846 | Collagen, type IV,
alpha 2 | 0.5 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| COL4A1 | NM_001845 | Collagen, type IV,
alpha 1 | 0.5 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| FN1 | NM_002026 | Fibronectin 1 | 0.5 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| LAMC1 | NM_002293 | Laminin, gamma
1 | 0.5 | Focal adhesion,
cell communication, extracellular matrix receptor interaction |
| DSC3 | NM_001941 | Desmocollin 3 | 0.6 | Cell
communication |
| DSG2 | AAH99655 | Desmoglein 2 | 0.6 | Cell
communication |
| CDH2 | NM_001792 | Cadherin 2, type 1,
n-cadherin | 0.6 | Cell adhesion
molecules |
| PTPRF | NM_002840 | Protein tyrosine
phosphatase, receptor type F | 0.7 | Cell adhesion
molecules |
| NEO1 | NM_002499 | Neogenin homolog
1 | 0.7 | Cell adhesion
molecules |
| GLG1 | NM_012201 | Golgi apparatus
protein 1 | 0.6 | Cell adhesion
molecules |
| VCAN | NM_004385 | Chondroitin sulfate
proteoglycan 2 (Versican) | 0.4 | Cell adhesion
molecules |
| NCAM1 | NM_000615 | Neural cell
adhesion molecule 1 | 0.6 | Cell adhesion
molecules |
| ITGAV | NM_002210 | Integrin, alpha
V | 0.6 | Cell adhesion
molecules, small cell lung cancer |
| COL4A1 | NM_001845 | Collagen, type IV,
alpha 1 | 0.5 | Small cell lung
cancer |
| FN1 | NM_001846 | Fibronectin 1 | 0.5 | Small cell lung
cancer |
| COL4A2 | NM_002026 | Collagen, type IV,
alpha 2 | 0.5 | Small cell lung
cancer |
| LAMB1 | NM_002291 | Laminin, beta
1 | 0.5 | Small cell lung
cancer, Prion disease |
| LAMC1 | NM_002293 | Laminin, gamma
1 | 0.6 | Small cell lung
cancer, Prion disease |
| HSPA5 | NM_005347 | Heat shock 70 kDa
protein 5 | 0.5 | Prion disease |
| PTPRF | NM_002840 | Protein tyrosine
phosphatase, receptor type F | 0.6 | Adherens
junction |
| INSR | NM_000208 | Insulin
receptor | 0.6 | Adherens
junction |
| MET | NM_000245 | Met
proto-oncogene | 0.7 | Adherens
junction |
| VCL | NM_003373 | Vinculin | 0.6 | Adherens
junction |
Discussion
Selenium is a promising cancer chemopreventive
agent; its contribution to antioxidant activities is thought to be
a likely mode of action. SeM has also been shown to contribute to
the mitigation of radiation-induced oxidative stress (17–19).
Since SeM alone is redox-inactive, the antioxidant contribution is
expected to be indirect. Specifically, SeM is a nutritional source
of selenium cofactor for selenoproteins and enzymes, such as
glutathione peroxidase and thioredoxin reductase (13), which serve as essential antioxidant
enzymes with activities which protect against oxidative stress.
Indeed, the function of certain transcription factors associated
with oxidative stress, including p53 (23,24),
AP-1 (25) and NF-κB (25,26),
are known to be oxidant-sensitive and maintained in the active
state through reduction. Thus, SeM may contribute to downstream
signaling via the modulation of the oxidation state of these key
transcription factors during a stress response.
The signaling events associated with selenium
treatment have been shown to target malignant cell lines, while
sparing normal cells. For example, SeM has the ability to
selectively induce apoptosis and growth arrest in cancerous, but
not normal, human prostate cells (27). Likewise, gene profiling of
premalignant lesions in rat mammary glands and cultured human
mammary epithelial cells treated with methylseleninic acid (a
potent pro-oxidant form of selenium) revealed the modulation of
regulatory genes linked to cell cycle and apoptosis, properties
consistent with a role in the reduction of lesion development or
the death of the premalignant cells (28,29).
Mechanistically, SeM treatment was shown to promote growth
inhibition in human colon cancer cells through the phosphorylation
of histone H3 via the activation of MAPK extracellular regulated
kinase (ERK) and subsequent phosphorylation of the ribosomal S6
kinase (RSK) (30,31). Recently, the ability of selenium to
alter the ECM and stroma has been suggested as being responsible
for its cancer chemopreventive potential (32). A comparison of both normal and
malignant prostate cell lines adapted to Se-methylselenocysteine (a
source of selenium in alliums) for one month, in order to achieve
steady-state selenium levels, revealed significant modulation of
ECM-associated genes, such as collagens (32). As such, we observed various gene
products associated with cell-cell and cell-ECM communication to be
downregulated when treated with SeM and processed 16 h after 20 cGy
irradiation (Table V). Although the
significance of this observation is unclear, the contributions to
stromal remodeling (from the alteration of ECM gene products) and
cell-cell communication may suggest a possible mechanism for the
chemopreventive property of selenium. As SeM suppresses radiation
transformation when applied to cultures of irradiated HTori-3 cells
at time periods of longer than 16 h post-irradiation (Ware et
al, unpublished data), the effects of SeM at 16 h
post-irradiation may be particularly significant for understanding
the mechanism(s) by which SeM suppresses radiation transformation
in vitro.
Our results demonstrate the ability of low-dose HZE
particle radiation to elicit changes in global gene expression
consistent with a stress response. The activation of genes
associated with cell cycle checkpoint (CDC6), apoptosis (GADD45A
and FAS) and general stress response (ATF3) is consistent with a
scenario of cellular growth arrest and repair. Whereas SeM
mitigates the expression of genes associated with the stress
response brought about following a short period of time (6 h)
post-irradiation, SeM effects on gene expression patterns at a
longer period of time post-irradiation (16 h) indicate that it
potentially alters cell behavior by modulating the expression of
genes associated with cell adhesion.
Acknowledgements
This study was supported by a grant
from the National Aeronautics and Space Administration (NASA; No.
NAG9-1517) and an NIH Training Grant (No. 2T32CA009677).
References
|
1.
|
Brooks A, Bao S, Rithidech K, Couch LA and
Braby LA: Relative effectiveness of HZE iron-56 particles for the
induction of cytogenetic damage in vivo. Radiat Res. 155:353–359.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Nelson GA: Fundamental space radiobiology.
Gravit Space Biol Bull. 16:29–36. 2003.PubMed/NCBI
|
|
3.
|
Nelson GA, Jones TA, Chesnut A and Smith
AL: Radiation-induced gene expression in the nematode
Caenorhabditis elegans. J Radiat Res. 43(Suppl): S199–S203.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ding LH, Shingyoji M, Chen F, et al: Gene
expression profiles of normal human fibroblasts after exposure to
ionizing radiation: a comparative study of low and high doses.
Radiat Res. 164:17–26. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Fornace AJ Jr, Amundson SA, Bittner M,
Myers TG, Meltzer P, Weinsten JN and Trent J: The complexity of
radiation stress responses: analysis by informatics and functional
genomics approaches. Gene Expr. 7:387–400. 1999.PubMed/NCBI
|
|
6.
|
Barcellos-Hoff MH: The potential influence
of radiation-induced microenvironments in neoplastic progression. J
Mammary Gland Biol Neoplasia. 3:165–175. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Azzam EI, de Toledo SM and Little JB:
Expression of CONNEXIN43 is highly sensitive to ionizing radiation
and other environmental stresses. Cancer Res. 63:7128–7135.
2003.PubMed/NCBI
|
|
8.
|
Zhou BB and Elledge SJ: The DNA damage
response: putting checkpoints in perspective. Nature. 408:433–439.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Amundson SA, Do KT and Fornace AJ Jr:
Induction of stress genes by low doses of gamma rays. Radiat Res.
152:225–231. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Amundson SA, Bittner M, Meltzer P, Trent J
and Fornace AJ Jr: Induction of gene expression as a monitor of
exposure to ionizing radiation. Radiat Res. 156:657–661. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ding LH, Shingyoji M, Chen F, Chatterjee
A, Kasai KE and Chen DJ: Gene expression changes in normal human
skin fibroblasts induced by HZE-particle radiation. Radiat Res.
164:523–526. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Franco N, Lamartine J, Frouin V, Le Minter
P, et al: Low-dose exposure to gamma rays induces specific gene
regulations in normal human keratinocytes. Radiat Res. 163:623–635.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Combs GF Jr and Gray WP: Chemopreventive
agents: selenium. Pharmacol Ther. 79:179–192. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Clark LC, Combs GF Jr, Turnbull BW, Slate
EH, et al: Effects of selenium supplementation for cancer
prevention in patients with carcinoma of the skin. A randomized
controlled trial Nutritional Prevention of Cancer Study Group JAMA.
276:1957–1963. 1996.
|
|
15.
|
Lu J, Kaeck M, Jiang C, Wilson AC and
Thompson HJ: Selenite induction of DNA strand breaks and apoptosis
in mouse leukemic L1210 cells. Biochem Pharmacol. 47:1531–1535.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Garberg P, Stahl A, Warholm M and Hogberg
J: Studies of the role of DNA fragmentation in selenium toxicity.
Biochem Pharmacol. 37:3401–3406. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kennedy AR, Ware JH, Guan J, Donahue JJ,
et al: Selenomethionine protects against adverse biological effects
induced by space radiation. Free Radic Biol Med. 36:259–266. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Guan J, Wan XS, Zhou Z, Ware J, Donahue
JJ, Biaglow JE and Kennedy AR: Effects of dietary supplements on
space radiation-induced oxidative stress in Sprague-Dawley rats.
Radiat Res. 162:572–579. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Guan J, Stewart J, Ware JH, Zhou Z,
Donahue JJ and Kennedy AR: Effects of dietary supplements on the
space radiation-induced reduction in total antioxidant status in
CBA mice. Radiat Res. 165:373–378. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wan XS, Ware JH, Zhou Z, Donahue JJ, Guan
J and Kennedy AR: Protection against radiation-induced oxidative
stress in cultured human epithelial cells by treatment with
antioxidant agents. Int J Radiat Oncol Biol Phys. 64:1475–1481.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC, et al: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Hai T, Wolfgang CD, Marsee DK, Allen AE
and Sivaprasad U: ATF3 and stress responses. Gene Expr. 7:321–335.
1999.
|
|
23.
|
Gaiddon C, Moorthy NC and Prives C: Ref-1
regulates the trans-activation and pro-apoptotic functions of p53
in vivo. EMBO J. 18:5609–5621. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Seo YR, Kelley MR and Smith ML:
Selenomethionine regulation of p53 by a ref1-dependent redox
mechanism. Proc Natl Acad Sci USA. 99:14548–14553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Gius D, Botero A, Shah S and Curry HA:
Intracellular oxidation/reduction status in the regulation of
transcription factors NF-kappaB and AP-1. Toxicol Lett. 106:93–106.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Reynaert NL, van der Vliet A, Guala AS,
McGovern T, Hristova M, Pantano C, et al: Dynamic redox control of
NF-kappaB through glutaredoxin-regulated S-glutathionylation of
inhibitory kappaB kinase beta. Proc Natl Acad Sci USA.
103:13086–13091. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Menter DG, Sabichi AL and Lippman SM:
Selenium effects on prostate cell growth. Cancer Epidemiol
Biomarkers Prev. 9:1171–1182. 2000.PubMed/NCBI
|
|
28.
|
Dong Y, Ip C and Ganther H: Evidence of a
field effect associated with mammary cancer chemoprevention by
methylseleninic acid. Anticancer Res. 22:27–32. 2007.PubMed/NCBI
|
|
29.
|
Dong Y, Ganther HE, Stewart C and Ip C:
Identification of molecular targets associated with
selenium-induced growth inhibition in human breast cells using cDNA
microarrays. Cancer Res. 62:708–714. 2002.PubMed/NCBI
|
|
30.
|
Goulet AC, Chigbrow M, Frisk P and Nelson
M: Selenomethionine induces sustained ERK phosphorylation leading
to cell-cycle arrest in human colon cancer cells. Carcinogenesis.
26:109–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Goulet AC, Watts G, Lord JL and Nelson MA:
Profiling of selenomethionine responsive genes in colon cancer by
microarray analysis. Cancer Biol Ther. 6:494–503. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Hurst R, Elliott RM, Goldson AJ and
Fairweather-Tait SJ: Se-methylselenocysteine alters collagen gene
and protein expression in human prostate cells. Cancer Lett.
269:117–126. 2008. View Article : Google Scholar : PubMed/NCBI
|