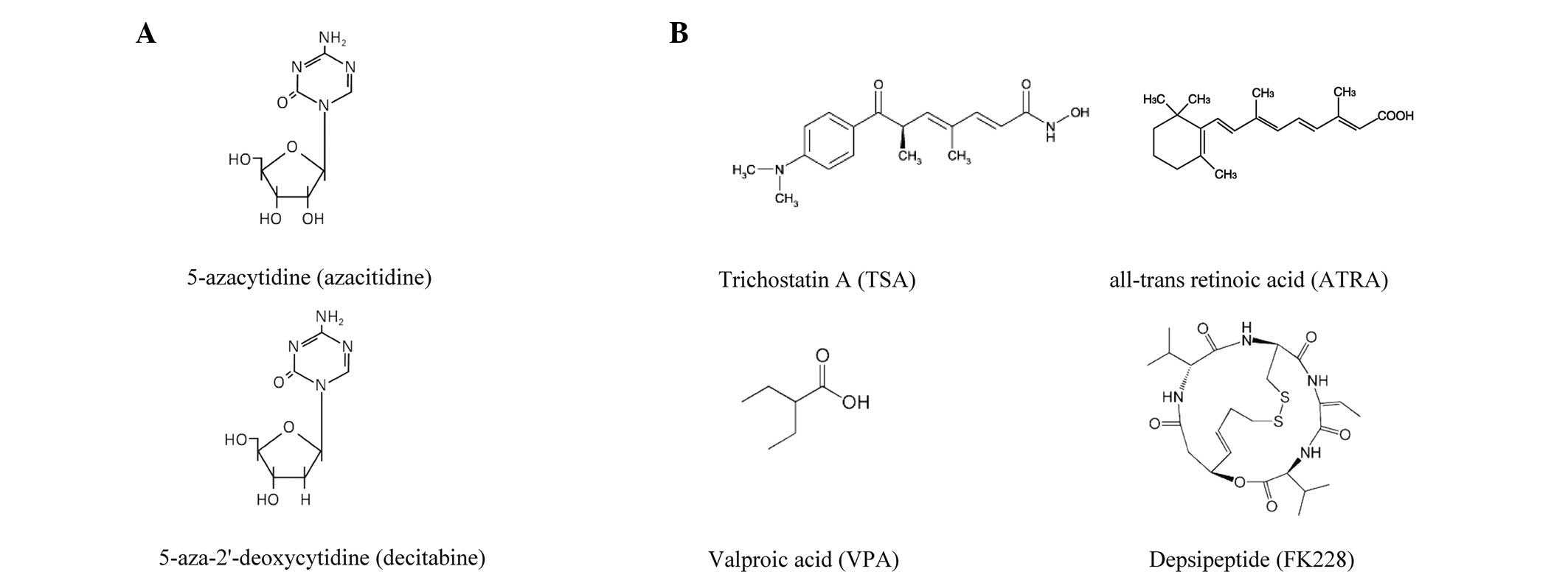
Telomeres serve as essential structures that protect
the ends of linear chromosomes from DNA repair and degradation, and
their maintenance is critical for long-term cell proliferation and
survival (1,2). Mammalian telomeres consist of tandem
TTAGGG repeats that are bound by a specialized six-protein complex
known as shelterin and may be replenished by telomerase (3). Telomerase is composed of two essential
components, a catalytic subunit with reverse transcriptase
activity, telomerase reverse transcriptase (TERT), and a telomerase
RNA component (TERC), that acts as a template for DNA synthesis
(4–6). Telomerase activity is overexpressed in
the majority of cancer cells but is barely detectable in the
predominance of normal somatic cells (7).
Among the various aspects of gene control,
epigenetic alterations have gained attention as critical
determinants for tumor initiation and subsequent cancer progression
(8,9). The forms of epigenetic control of gene
expression include DNA methylation and histone modification. DNA
methylation involves a covalent modification at the fifth carbon
position of cytosine residues within CpG dinucleotides, resulting
in the transcriptional silencing of the affiliated gene (10). Promoter hypermethylation of tumor
suppressor genes has been increasingly considered as a fundamental
mechanism for the silencing of these genes in cancer cells,
resulting in tumor initiation and progression (11,12).
In addition to DNA methylation, another key element in the
epigenetic control of gene expression is histone modification,
including acetylation, methylation, phosphorylation and
ubiquitination. Aberrant patterns of histone modifications have
been associated with a large number of human malignancies (13,14).
DNA methylation and histone modifications have been extensively
recognized as epigenetic mechanisms that regulate gene
transcription in carcinogenesis.
Telomere length, maintained by telomerase, is a
prominent mechanism for long-term cell proliferation and survival,
and is strongly involved in cancer, cell senescence and aging
(21–23). It has been demonstrated that the
epigenetic plasticity of the hTERT gene promoter is a
determinant for the control of telomerase activity. Therefore,
inhibiting the expression of the hTERT gene through
epigenetic mechanisms usually results in telomeric attrition. The
epigenetic changes associated with the inhibition of telomerase
activity include hypermethylation and histone modifications of the
hTERT promoter.
In addition to DNA methylation, another prevalent
epigenetic mechanism that affects hTERT transcription is
histone modification, including histone acetylation, methylation,
phosphorylation and ubiquitinization. Histone tails carry basic
charges and are associated with DNA molecules by electrostatic
attraction. The acetylation of the histone proteins neutralizes the
charge status of the histone tails, which decreases the attraction
force between DNA and the histone tails, thus conferring an opened
chromatin structure, allowing transcription factors, including
c-MYC, MAD1 and CTCF, to bind to the DNA. Conversely, the
deacetylation of histones results in the transcription factors
having less access to the DNA (33,34).
It has been demonstrated that Trichostatin A (TSA), a histone
deacetylase (HDAC) inhibitor, is able to induce hTERT
transcription and telomerase activity in normal cells and
telomerase-negative immortal cell lines through the inhibition of
histone deacetylation (Fig. 1)
(35,36). Furthermore, FR901288, a novel cyclic
peptide inhibitor of HDAC, has also been shown to activate
hTERT mRNA expression in oral cancer cell lines (37). However, there are conflicting
studies with regard to hTERT transcription and telomerase
activity in cancer cells induced by HDAC inhibitors. Zhu et
al reported that HDAC inhibitors prevented cell proliferation
and induced apoptosis, but had no effect on the expression of hTERC
and hTERT mRNA, or on telomerase activity (38). In prostate and brain cancer cells,
the hTERT gene expression and telomerase activity were
inhibited by HDAC inhibitors (30,40).
Therefore, the HDAC inhibitors may exhibit various effects on
hTERT transcription and telomerase activity in cancer cells.
In addition to histone acetylation, hTERT transcription was
also reported to be associated with histone methylation, of which
three varying forms, including mono-, di- and trimethylation, may
emerge in methylation modifications of the histone lysine residues.
It has been demonstrated that mono- and dimethylated
histone3-lysine9 (H3-K9) are localized to distinct domains of
silent chromatin, where they are associated with inactive genes,
whereas trimethylated H3-K9 is enriched in pericentric
heterochromatin (41). Further
studies have shown that a lack of hTERT expression in
telomerase-negative cell lines is associated with histone H3 and H4
hypoacetylation and the methylation of H3-K9. However, hTERT
transcription in telomerase-positive cell lines is associated with
the hyperacetylation of H3 and H4 and the methylation of Lys4-H3
(H3-K4) (42). Histone
methyltransferase (HMTase) is considered to be responsible for
histone methylation at the hTERT promoter. Liu et al
reported that SET and MYND domain-containing protein 3 (SMYD3), a
HMTase, may directly transactivate hTERT transcription and
telomerase activity in normal human fibroblasts and cancer cell
lines through histone H3-K4 trimethylation (43). These results suggest that the
epigenetic regulation of histones may contribute to hTERT
gene expression and telomerase activity (Fig. 2).
Telomerase activity is a hallmark of the immortal
cell phenotype and several mechanisms have been reported to be
involved in its regulation, including transcriptional factors, DNA
methylation and histone deacetylation. Furthermore, it has been
shown that cells in numerous types of leukemia are able to maintain
their telomere length and prevent replicative senescence or
apoptosis by the epigenetic regulation of hTERT (44–46).
Therefore, telomerase suppression using epigenetic modifications
should be a promising target for the treatment of leukemia.
In addition to inducing cell differentiation,
telomerase inhibition through epigenetic mechanisms has been
reported to promote growth arrest, apoptosis and sensitivity to
certain chemotherapeutic reagents in human acute leukemia cells.
Woo et al demonstrated that TSA had an antiproliferative and
apoptosis-inducing effect on the human leukemic cell line U937, and
that these growth-inhibitory effects were associated with the
inhibition of hTERT expression and telomerase activity.
Therefore, a loss of telomerase activity may be a good surrogate
biomarker to assess the antitumor activity of TSA in human leukemic
cells (61). The resistance to
imatinib is a major problem in chronic myelogenous leukemia (CML)
treatment, and recent studies have shown that by targeting
telomerase expression using a dominant-negative form of the
catalytic protein subunit of hTERT, or by the treatment with
HDAC inhibitors, the risk of imatinib resistance may be reduced and
the imatinib-induced apoptosis in leukemia cells may be enhanced,
suggesting that antitelomerase strategies may be able to prevent,
or at least delay the onset of such resistance (62,63).
Epigenetic modifiers that are currently available in
preclinical and early clinical trials of leukemia target DNMTs
through DNMT inhibitors, or alter the status of the histones using
HDAC inhibitors, in order to modulate gene transcription. It has
been noted that epigenetic modifications contribute to hTERT
gene expression and telomerase activity, resulting in a positive
effect in the treatment of leukemia (59,61).
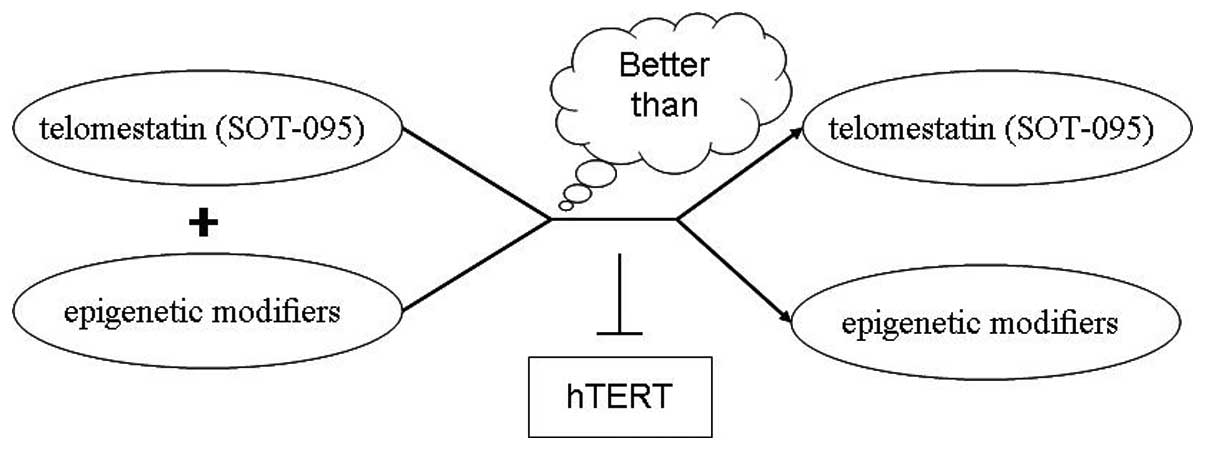
In addition to epigenetic modifiers, the use of several telomerase
hTR subunit small molecule inhibitors has resulted in the specific
inhibition of telomerase activity. Therefore, an antitelomerase
strategy involving a combination of epigenetic modifiers and
telomerase hTR subunit small molecule inhibitors may exert a more
potent effect for the treatment of human leukemia (Fig. 3).
Although epigenetic modifiers have shown promise as
therapies for human leukemia in early clinical trials, certain
limitations prevent their widespread clinical application. Firstly,
the exact molecular mechanisms underlying the epigenetic regulation
and hTERT expression remain to be elucidated, as do numerous
details with regard to telomerase regulation. An improved
understanding of the linkage will facilitate the identification of
more specific and selective epigenetic modifiers for leukemia cells
(66). Secondly, a broad spectrum
of biological and potentially adverse effects have been identified
following treatment using epigenetic modifiers. Further
investigation with regard to these effects is required in
large-scale and multicentric populations of treated patients
(67). Thirdly, further studies
will be required to identify whether the inhibition of hTERT
gene expression is causal or consequential to the anticancer
effects of epigenetic modifiers, and whether the hTERT gene
or telomerase activity may be an appropriate predictive biomarker
for assessing the antitumor activity of these agents in human
leukemia cells (68). Finally, it
should be taken into account whether the antitelomerase approach
using epigenetic modifiers with telomerase hTR subunit small
molecule inhibitors may be a better combinatorial strategy when
compared with methods that are already used in prospective clinical
trials.
Despite the unanswered biological questions, an
increased understanding of the role of epigenetic regulation in
hTERT gene expression and the treatment of leukemia may
provide a prospective anticancer therapeutic approach in the form
of the antitelomerase strategy.
This study is supported by Zhejiang
University and grants from the National Natural Science Foundation
of China (grant no. 81272593) and the Zhejiang Provincial Natural
Science Foundation of China (grant no. LQ13H160008).
|
1.
|
de Lange T: Shelterin: the protein complex
that shapes and safeguards human telomeres. Genes Dev.
19:2100–2110. 2005.PubMed/NCBI
|
|
2.
|
Palm W and de Lange T: How shelterin
protects mammalian telomeres. Annu Rev Genet. 42:301–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Morin GB: The human telomere terminal
transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG
repeats. Cell. 59:521–529. 1989. View Article : Google Scholar
|
|
4.
|
Cohen SB, Graham ME, Lovrecz GO, Bache N,
Robinson PJ and Reddel RR: Protein composition of catalytically
active human telomerase from immortal cells. Science.
315:1850–1853. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Blackburn EH: Switching and signaling at
the telomere. Cell. 106:661–673. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Pinto AR, Li H, Nicholls C and Liu JP:
Telomere protein complexes and interactions with telomerase in
telomere maintenance. Front Biosci. 16:187–207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kim NW, Piatyszek MA, Prowse KR, Harley
CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL and Shay
JW: Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Agrelo R and Wutz A: Cancer progenitors
and epigenetic contexts: an Xisting connection. Epigenetics.
4:568–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Vineis P, Chuang SC, Vaissière T, Cuenin
C, Ricceri F; Genair-EPIC Collaborators; Johansson M, Ueland P,
Brennan P and Herceg Z: DNA methylation changes associated with
cancer risk factors and blood levels of vitamin metabolites in a
prospective study. Epigenetics. 6:195–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer - a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Shao C, Lacey M, Dubeau L and Ehrlich M:
Hemimethylation footprints of DNA demethylation in cancer.
Epigenetics. 4:165–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sawan C and Herceg Z: Histone
modifications and cancer. Adv Genet. 70:57–85. 2010. View Article : Google Scholar
|
|
14.
|
Rivière G, Lienhard D, Andrieu T, Vieau D,
Frey BM and Frey FJ: Epigenetic regulation of somatic
angiotensin-converting enzyme by DNA methylation and histone
acetylation. Epigenetics. 6:478–489. 2011.PubMed/NCBI
|
|
15.
|
Cairney CJ and Keith WN: Telomerase
redefined: integrated regulation of hTR and hTERT for telomere
maintenance and telomerase activity. Biochimie. 90:13–23. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Dessain SK, Yu H, Reddel RR, Beijersbergen
RL and Weinberg RA: Methylation of the human telomerase gene CpG
island. Cancer Res. 60:537–541. 2000.PubMed/NCBI
|
|
17.
|
Zhu J, Zhao Y and Wang S: Chromatin and
epigenetic regulation of the telomerase reverse transcriptase gene.
Protein Cell. 1:22–32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Iliopoulos D, Oikonomou P, Messinis I and
Tsezou A: Correlation of promoter hypermethylation in hTERT, DAPK
and MGMT genes with cervical oncogenesis progression. Oncol Rep.
22:199–204. 2009.PubMed/NCBI
|
|
19.
|
Gigek CO, Leal MF, Silva PN, Lisboa LC,
Lima EM, Calcagno DQ, Assumpção PP, Burbano RR and Smith Mde A:
hTERT methylation and expression in gastric cancer. Biomarkers.
14:630–636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wang S, Hu C and Zhu J: Distinct and
temporal roles of nucleosomal remodeling and histone deacetylation
in the repression of the hTERT gene. Mol Biol Cell. 21:821–832.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Jia W, Wang S, Horner JW, Wang N, Wang H,
Gunther EJ, DePinho RA and Zhu J: A BAC transgenic reporter
recapitulates in vivo regulation of human telomerase reverse
transcriptase in development and tumorigenesis. FASEB J.
25:979–989. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Baird DM, Rowson J, Wynford-Thomas D and
Kipling D: Extensive allelic variation and ultrashort telomeres in
senescent human cells. Nat Genet. 33:203–207. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Harley CB, Futcher AB and Greider CW:
Telomeres shorten during ageing of human fibroblasts. Nature.
345:458–460. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Li Y, Liu L, Andrews LG and Tollefsbol TO:
Genistein depletes telomerase activity through cross-talk between
genetic and epigenetic mechanisms. Int J Cancer. 125:286–296. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Lopatina NG, Poole JC, Saldanha SN, Hansen
NJ, Key JS, Pita MA, Andrews LG and Tollefsbol TO: Control
mechanisms in the regulation of telomerase reverse transcriptase
expression in differentiating human teratocarcinoma cells. Biochem
Biophys Res Commun. 306:650–659. 2003. View Article : Google Scholar
|
|
27.
|
Shin KH, Kang MK, Dicterow E and Park NH:
Hypermethylation of the hTERT promoter inhibits the expression of
telomerase activity in normal oral fibroblasts and senescent normal
oral keratinocytes. Br J Cancer. 89:1473–1478. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Zinn RL, Pruitt K, Eguchi S, Baylin SB and
Herman JG: hTERT is expressed in cancer cell lines despite promoter
DNA methylation by preservation of unmethylated DNA and active
chromatin around the transcription start site. Cancer Res.
67:194–201. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Devereux TR, Horikawa I, Anna CH, Annab
LA, Afshari CA and Barrett JC: DNA methylation analysis of the
promoter region of the human telomerase reverse transcriptase
(hTERT) gene. Cancer Res. 59:6087–6090. 1999.PubMed/NCBI
|
|
30.
|
Kumari A, Srinivasan R, Vasishta RK and
Wig JD: Positive regulation of human telomerase reverse
transcriptase gene expression and telomerase activity by DNA
methylation in pancreatic cancer. Ann Surg Oncol. 16:1051–1059.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Kumari A, Srinivasan R and Wig JD: Effect
of c-MYC and E2F1 gene silencing and of 5-azacytidine treatment on
telomerase activity in pancreatic cancer-derived cell lines.
Pancreatology. 9:360–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Guilleret I, Yan P, Grange F, Braunschweig
R, Bosman FT and Benhattar J: Hypermethylation of the human
telomerase catalytic subunit (hTERT) gene correlates with
telomerase activity. Int J Cancer. 101:335–341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Krajewski WA: Histone acetylation status
and DNA sequence modulate ATP-dependent nucleosome repositioning. J
Biol Chem. 277:14509–14513. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lai SR, Phipps SM, Liu L, Andrews LG and
Tollefsbol TO: Epigenetic control of telomerase and modes of
telomere maintenance in aging and abnormal systems. Front Biosci.
10:1779–1796. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Takakura M, Kyo S, Sowa Y, Wang Z, Yatabe
N, Maida Y, Tanaka M and Inoue M: Telomerase activation by histone
deacetylase inhibitor in normal cells. Nucleic Acids Res.
29:3006–3011. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Hou M, Wang X, Popov N, Zhang A, Zhao X,
Zhou R, Zetterberg A, Björkholm M, Henriksson M, Gruber A and Xu D:
The histone deacetylase inhibitor trichostatin A derepresses the
telomerase reverse transcriptase (hTERT) gene in human cells. Exp
Cell Res. 274:25–34. 2002. View Article : Google Scholar
|
|
37.
|
Murakami J, Asaumi J, Kawai N, Tsujigiwa
H, Yanagi Y, Nagatsuka H, Inoue T, Kokeguchi S, Kawasaki S, Kuroda
M, Tanaka N, Matsubara N and Kishi K: Effects of histone
deacetylase inhibitor FR901228 on the expression level of
telomerase reverse transcriptase in oral cancer. Cancer Chemother
Pharmacol. 56:22–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Zhu K, Qu D, Sakamoto T, Fukasawa I,
Hayashi M and Inaba N: Telomerase expression and cell proliferation
in ovarian cancer cells induced by histone deacetylase inhibitors.
Arch Gynecol Obstet. 277:15–19. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Suenaga M, Soda H, Oka M, Yamaguchi A,
Nakatomi K, Shiozawa K, Kawabata S, Kasai T, Yamada Y, Kamihira S,
Tei C and Kohno S: Histone deacetylase inhibitors suppress
telomerase reverse transcriptase mRNA expression in prostate cancer
cells. Int J Cancer. 97:621–625. 2002. View Article : Google Scholar
|
|
40.
|
Khaw AK, Silasudjana M, Banerjee B, Suzuki
M, Baskar R and Hande MP: Inhibition of telomerase activity and
human telomerase reverse transcriptase gene expression by histone
deacetylase inhibitor in human brain cancer cells. Mutat Res.
625:134–144. 2007. View Article : Google Scholar
|
|
41.
|
Rice JC, Briggs SD, Ueberheide B, Barber
CM, Shabanowitz J, Hunt DF, Shinkai Y and Allis CD: Histone
methyltransferases direct different degrees of methylation to
define distinct chromatin domains. Mol Cell. 12:1591–1598. 2003.
View Article : Google Scholar
|
|
42.
|
Atkinson SP, Hoare SF, Glasspool RM and
Keith WN: Lack of telomerase gene expression in alternative
lengthening of telomere cells is associated with chromatin
remodeling of the hTR and hTERT gene promoters. Cancer Res.
65:7585–7590. 2005.PubMed/NCBI
|
|
43.
|
Liu C, Fang X, Ge Z, Jalink M, Kyo S,
Björkholm M, Gruber A, Sjöberg J and Xu D: The telomerase reverse
transcriptase (hTERT) gene is a direct target of the histone
methyltransferase SMYD3. Cancer Res. 67:2626–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Röth A, Vercauteren S, Sutherland HJ and
Lansdorp PM: Telomerase is limiting the growth of acute myeloid
leukemia cells. Leukemia. 17:2410–2417. 2003.PubMed/NCBI
|
|
45.
|
Annaloro C, Onida F, Saporiti G and
Lambertenghi Deliliers G: Cancer stem cells in hematological
disorders: current and possible new therapeutic approaches. Curr
Pharm Biotechnol. 12:217–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Liu L, Saldanha SN, Pate MS, Andrews LG
and Tollefsbol TO: Epigenetic regulation of human telomerase
reverse transcriptase promoter activity during cellular
differentiation. Genes Chromosomes Cancer. 41:26–37. 2004.
View Article : Google Scholar
|
|
47.
|
Bechter OE, Eisterer W, Dlaska M, Kühr T
and Thaler J: CpG island methylation of the hTERT promoter is
associated with lower telomerase activity in B-cell lymphocytic
leukemia. Exp Hematol. 30:26–33. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Terrin L, Trentin L, Degan M, Corradini I,
Bertorelle R, Carli P, Maschio N, Bo MD, Noventa F, Gattei V,
Semenzato G and De Rossi A: Telomerase expression in B-cell chronic
lymphocytic leukemia predicts survival and delineates subgroups of
patients with the same igVH mutation status and different outcome.
Leukemia. 21:965–972. 2007.
|
|
49.
|
Lübbert M, Suciu S, Baila L, Rüter BH,
Platzbecker U, Giagounidis A, Selleslag D, Labar B, Germing U,
Salih HR, Beeldens F, Muus P, Pflüger KH, Coens C, Hagemeijer A,
Eckart Schaefer H, Ganser A, Aul C, de Witte T and Wijermans PW:
Low-dose decitabine versus best supportive care in elderly patients
with intermediate- or high-risk myelodysplastic syndrome (MDS)
ineligible for intensive chemotherapy: final results of the
randomized phase III study of the European Organisation for
Research and Treatment of Cancer Leukemia Group and the German MDS
Study Group. J Clin Oncol. 29:1987–1996. 2011.
|
|
50.
|
Kantarjian HM, Giles FJ, Greenberg PL,
Paquette RL, Wang ES, Gabrilove JL, Garcia-Manero G, Hu K, Franklin
JL and Berger DP: Phase 2 study of romiplostim in patients with
low- or intermediate-risk myelodysplastic syndrome receiving
azacitidine therapy. Blood. 116:3163–3170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Baer MR and Gojo I: Novel agents for the
treatment of acute myeloid leukemia in the older patient. J Natl
Compr Canc Netw. 9:331–335. 2011.
|
|
52.
|
Garcia-Manero G, Gore SD, Cogle C, Ward R,
Shi T, Macbeth KJ, Laille E, Giordano H, Sakoian S, Jabbour E,
Kantarjian H and Skikne B: Phase I study of oral azacitidine in
myelodysplastic syndromes, chronic myelomonocytic leukemia, and
acute myeloid leukemia. J Clin Oncol. 29:2521–2527. 2011.
View Article : Google Scholar
|
|
53.
|
Claus R and Lübbert M: Epigenetic targets
in hematopoietic malignancies. Oncogene. 22:6489–6496. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Lane AA and Chabner BA: Histone
deacetylase inhibitors in cancer therapy. J Clin Oncol.
27:5459–5468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Pendino F, Sahraoui T, Lanotte M and
Ségal-Bendirdjian E: A novel mechanism of retinoic acid resistance
in acute promyelocytic leukemia cells through a defective pathway
in telomerase regulation. Leukemia. 16:826–832. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Pendino F, Flexor M, Delhommeau F, Buet D,
Lanotte M and Segal-Bendirdjian E: Retinoids down-regulate
telomerase and telomere length in a pathway distinct from leukemia
cell differentiation. Proc Natl Acad Sci USA. 98:6662–6667. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Kretzner L, Scuto A, Dino PM, Kowolik CM,
Wu J, Ventura P, Jove R, Forman SJ, Yen Y and Kirschbaum MH:
Combining histone deacetylase inhibitor vorinostat with aurora
kinase inhibitors enhances lymphoma cell killing with repression of
c-Myc, hTERT, and microRNA levels. Cancer Res. 71:3912–3920. 2011.
View Article : Google Scholar
|
|
58.
|
Quintás-Cardama A, Santos FP and
Garcia-Manero G: Histone deacetylase inhibitors for the treatment
of myelodysplastic syndrome and acute myeloid leukemia. Leukemia.
25:226–235. 2011.PubMed/NCBI
|
|
59.
|
Azouz A, Wu YL, Hillion J, Tarkanyi I,
Karniguian A, Aradi J, Lanotte M, Chen GQ, Chehna M and
Ségal-Bendirdjian E: Epigenetic plasticity of hTERT gene promoter
determines retinoid capacity to repress telomerase in
maturation-resistant acute promyelocytic leukemia cells. Leukemia.
24:613–622. 2010. View Article : Google Scholar
|
|
60.
|
Love WK, Berletch JB, Andrews LG and
Tollefsbol TO: Epigenetic regulation of telomerase in
retinoid-induced differentiation of human leukemia cells. Int J
Oncol. 32:625–631. 2008.PubMed/NCBI
|
|
61.
|
Woo HJ, Lee SJ, Choi BT, Park YM and Choi
YH: Induction of apoptosis and inhibition of telomerase activity by
trichostatin A, a histone deacetylase inhibitor, in human leukemic
U937 cells. Exp Mol Pathol. 82:77–84. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Okabe S, Tauchi T, Nakajima A, Sashida G,
Gotoh A, Broxmeyer HE, Ohyashiki JH and Ohyashiki K: Depsipeptide
(FK228) preferentially induces apoptosis in BCR/ABL-expressing cell
lines and cells from patients with chronic myelogenous leukemia in
blast crisis. Stem Cells Dev. 16:503–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Deville L, Hillion J, Pendino F, Samy M,
Nguyen E and Ségal-Bendirdjian E: hTERT promotes imatinib
resistance in chronic myeloid leukemia cells: therapeutic
implications. Mol Cancer Ther. 10:711–719. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Nakajima A, Tauchi T, Sashida G, Sumi M,
Abe K, Yamamoto K, Ohyashiki JH and Ohyashiki K: Telomerase
inhibition enhances apoptosis in human acute leukemia cells:
possibility of anti-telomerase therapy. Leukemia. 17:560–567. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Tauchi T, Shin-ya K, Sashida G, Sumi M,
Okabe S, Ohyashiki JH and Ohyashiki K: Telomerase inhibition with a
novel G-quadruplex-interactive agent, telomestatin: in vitro and in
vivo studies in acute leukemia. Oncogene. 25:5719–5725. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Altucci L, Clarke N, Nebbioso A,
Scognamiglio A and Gronemeyer H: Acute myeloid leukemia:
therapeutic impact of epigenetic drugs. Int J Biochem Cell Biol.
37:1752–1762. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67.
|
Bhalla KN: Epigenetic and chromatin
modifiers as targeted therapy of hematologic malignancies. J Clin
Oncol. 23:3971–3993. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Piekarz RL and Bates SE: Epigenetic
modifiers: basic understanding and clinical development. Clin
Cancer Res. 15:3918–3926. 2009. View Article : Google Scholar : PubMed/NCBI
|