Introduction
Epithelial ovarian cancer (EOC) is the leading cause
of reproductive system cancer mortality in females (1). When epithelial ovarian carcinoma is
diagnosed at early stages, the survival rate is high (90%).
However, the majority of cases of ovarian carcinoma are not
identified until the late stage and the five-year relative survival
rates for the late stage of EOC are <10% (2). Despite advances in surgery and the
wide use of platinum-based chemotherapy, the survival rate of
patients with late stage EOC has changed little since
platinum-based treatment was introduced >30 years ago.
Consequently, the identification of the molecular changes that
occur during the development and progression of ovarian cancer is
urgently required.
MicroRNAs (miRNAs), a class of small, non-coding
RNAs, have been identified as gene expression regulators that
induce mRNA degradation or a translation blockade through pairing
to the 3′ untranslated region (3′-UTR) of the target mRNAs
(3). There is significant evidence
that the dysregulation of the miRNAs is a hallmark of cancer
(4). Emerging evidence shows that
miRNAs are abnormally expressed in various types of cancer and are
involved in various cell functions, including tumor proliferation,
drug resistance, apoptosis and metastasis. miR-222 is overexpressed
in various types of tumors (5–8).
miR-222 expression has been shown to induce cell growth,
oncogenesis, invasion, migration and drug resistance in tumor cells
(9–11), and was also reported to be a
significant marker of a poor prognosis (12). However, for miR-222, the possible
roles and associated target genes in ovarian cancer remain poorly
elucidated. In the present study, the role of miR-222 on the
carcinogenesis of ovarian cancer and the underlying mechanisms were
examined.
Materials and methods
Human EOC tissue collection
EOC tissues were obtained from patients who had
undergone surgery at the Department of Gynecological Cancer of
Tongji Hospital (Huazhong University of Science and Technology,
Wuhan, China), between 2009 and 2010. All patients underwent
debulking and subsequently received first-line
platinum/taxane-based chemotherapy. All the patients were diagnosed
with EOC (stages III and IV) based on a histopathological
evaluation. Informed consent was obtained from all patients. All
the tissue samples were collected, immediately snap-frozen in
liquid nitrogen and stored at −80°C. The tumor content of the
specimens was assessed by hematoxylin and eosin staining at the
Department of Pathology, Tongji Hospital. Only specimens containing
>60% tumor tissue were used. This study was approved by the
ethics committee of Tongji Hospital, Wuhan, China.
Cell culture and transfection
The OV2008 and C13* cells were gifts from
Professor Rakesh of the Ottawa Regional Cancer Center, Ottawa,
Canada. The A2780 ovarian cancer cell line was obtained from The
European Collection of Cell Cultures (ECACC, Salisbury, UK). These
cells were maintained in RPMI-1640 supplemented with 2 mmol/l
L-glutamine and 10% fetal bovine serum (FBS). ES2, SKOV-3 and
CAOV-3 were purchased from the American Type Culture Collection
(ATCC) and maintained in McCoy’s 5A or Dulbecco’s modified Eagle’s
medium (DMEM) containing 10% FBS. All cells were used within six
months of thawing and were cultured in a humidified 5%
CO2 incubator at 37°C. The cells were plated without
antibiotics ∼24 h prior to the transfections. Transient
transfections of the miRNA mimics/inhibitor (RiboBio, Guangzhou,
China) were performed using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
All transfections were performed for 48 h.
RNA extraction and qPCR
Total RNAs, including miRNAs were extracted from
cultured cells or fresh ovarian cancer tissues using the GeneJET
RNA Purification kit (Fermentas, Vilnius, Lithuania) according to
the manufacturer’s instructions. The expression of mature miR-222
was determined with the Bulge-Loop™ miRNA qPCR Primer Set (RiboBio)
with SYBR-Green qPCR; U6 snRNA was used as an internal control.
P27Kip1 mRNA expression was analyzed with qPCR using the
SYBR-Green method. All protocols were performed according to the
manufacturer’s instructions and the results were normalized to the
expression of GAPDH. The primer sequences were as follows:
P27Kip1 forward, 5′-TCCGGCTAACTCTGAGGACA-3′ and reverse,
5′-AGAAGAATCGTCGGTTGCAGG-3′; GAPDH forward,
5′-AGAGGCAGGGATGATGTTCTG-3′ and reverse, 5′-GACTCATGACCA
CAGTCCATGC-3′.
Cell cycle analysis
For the cell cycle experiments, the cells were
trypsinized, harvested and processed with standard methods using
propidium iodide (PI) to stain cellular DNA. The cell samples were
analyzed using a FACSCalibur system (BD Biosciences, San Jose, CA,
USA). Histograms were analyzed for the cell cycle distribution
using ModFit version 2.0 (Verity Software House, Topsham, ME,
USA).
Proliferation assays
The cells (5,000 per well) were plated in 96-well
plates and grown for 96 h following transfection (final miRNA
concentration of 100 nmol/l) in normal culture conditions. Cell
proliferation was documented every 24 h for four days using a CCK8
assay.
Plasmid construction and luciferase
assay
The full-length 3′-UTR of P27Kip1 was
amplified by PCR from genomic DNA of normal patients using specific
primers: sense, 5′-TAAGAATATGTTTCCTTGTTTATCAGAT-3′ and anti-sense,
5′-AATAGCTATGGAAGTTTTCTTTATTGAT-3′. The amplified DNA was then
cloned into psi-Check2 to generate a luciferase reporter vector.
The 3′-UTR of the mutant vector of P27Kip1 was also
constructed via the overlap extension PCR method using the
following primers: sense, 5′-CTCTAAAAGCGTTGGAGCATTATGCAATTAGG-3′
and anti-sense, 5′-CCTAATTGCATAATGCTCCAACGCTTTTAGAG-3′. For the
reporter assays, the cells were cultured in 24-well plates and
transfected with psi-Check2-P27-3′-UTR (mutant) and miR-222 mimics
(miR control). Luciferase activity was measured at 48 h
post-transfection using the Dual-Luciferase® reporter
assay system (Promega, Madison, WI, USA) and an LB 960 Centro XS3
luminometer (Molecular Devices, Sunnyvale, CA, USA). All
experiments were performed in triplicate.
Western blotting
The cells were lysed using mammalian protein
extraction reagent RIPA (Beyotime, Haimen, China) supplemented with
a protease inhibitor cocktail (Roche, Mannheim, Germany). The
proteins (50 μg) were electrophoresed by SDS-PAGE and
transferred onto PVDF membranes. Immunoblotting was performed using
human anti-P27Kip1 antibody (1:1,000; Cell Signaling,
Danvers, MA, USA), and GAPDH antibody (1:5,000) was used as a
control (Cell Signaling).
EdU incorporation and immunofluorescence
microscopy
The incorporation of 5-ethynyl-2’-deoxyuridine (EdU)
into actively proliferating SKOV3 cells was evaluated using a
Cell-Light™ EdU Cell Proliferation Detection kit; RiboBio)
following the manufacturer’s instructions. Cell immunostaining was
observed with an epifluorescence microscope (Axioplan II; Carl
Zeiss AG, Oberkochen, Germany) equipped with a charge-coupled
device camera. Digital images were acquired and analyzed with
ImageJ software (NIH, Bethesda, MD, USA).
Statistical analysis
All values are expressed as the mean ± SD.
Statistical analysis was performed with the two-tailed Student’s
t-test. The Wilcoxon signed rank test was used to analyze the
statistically significant upregulation of miR-222 expression in
ovarian cancers. The Pearson correlation analysis was performed
between the expression of miR-222 and P27Kip1. P<0.05
was considered to indicate a statistically significant difference
in all results.
Results
miR-222 expression is upregulated in
ovarian cancer
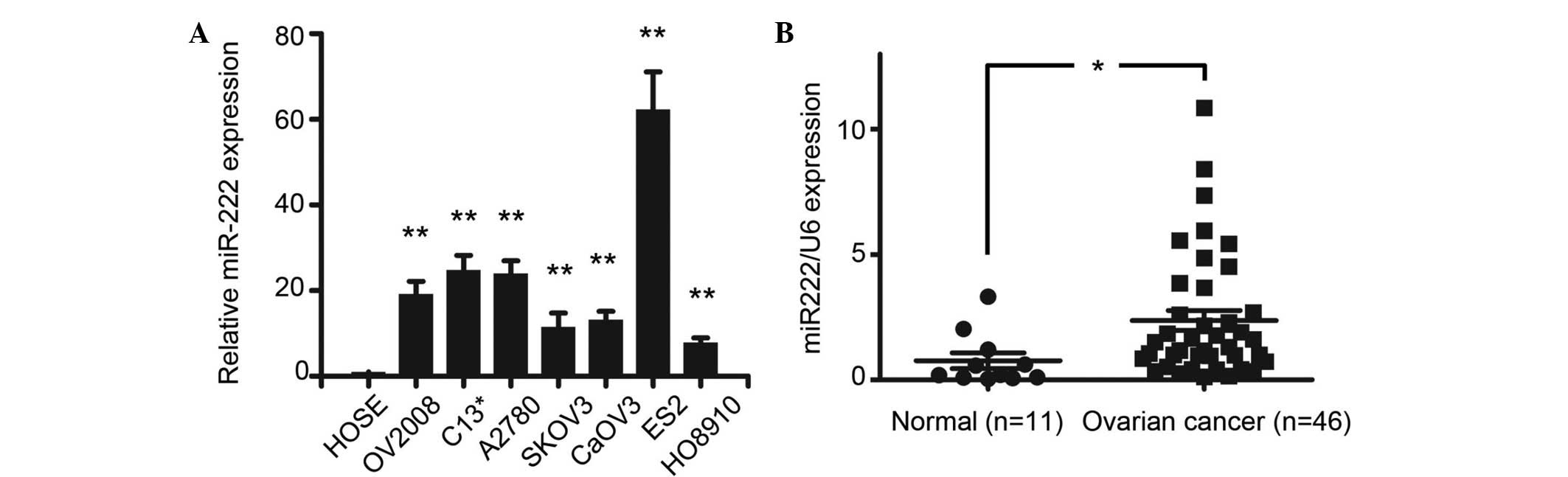
To analyze the expression levels of miR-222, a qPCR
analysis of miR-222 expression was conducted in several ovarian
cancer cell lines (OV2008, C13*, A2780, SKOV3, CaOV3,
ES2 and HO8910) and a human ovarian surface epithelial (HOSE) cell.
The results showed that miR-222 was overexpressed in all ovarian
cancer cell lines compared with the HOSE cells (P<0.01).
Moreover, when miR-222 expression was measured in 11 normal ovarian
tissues and 40 epithelial ovarian cancer tissues, qPCR showed that
miR-222 was also overexpressed in tumor samples associate with the
normal ovaries (P<0.05; Fig. 1A and
B).
Effect of miR-222 on ovarian cancer cell
proliferation in vitro
To demonstrate the function of miR-222 in ovarian
cancer cells, miR-222 mimics or miR-222 inhibitors were transiently
transfected into the SKOV3 cells to upregulate or downregulate the
expression of miR-222, respectively. qPCR confirmed the increased
or decreased expression of miR-222 in the miR-222 mimics or
inhibitor transfectants, respectively (data not shown). The cell
proliferation assays showed that the numbers of SKOV3 cells
transfected with miR-222 mimics were increased compared with the
cells transfected with the miR-NC controls or blanks (Fig. 2A). However, the SKOV3 cells
transfected with the miR-222 inhibitors exhibited significantly
reduced growth (Fig. 2B). It is
well known that enhanced cell proliferation is associated with
altered cell cycles. Consequently, a cell cycle analysis was
conducted in the SKOV3 cells transiently transfected with the
miR-222 mimics. The results showed that the cells transfected with
the miR-222 mimics exhibited an increased S phase with a decreased
G0-G1 phase (Fig.
2C). The proportion of cells corresponding to the S phase was
higher in the miR-222 mimic-transfected cells (38.4±4.2%) compared
with the control cells (21.5±3.8%; P<0.05; Fig. 2D), while the proportion of cells
corresponding to the G0-G1 phase was lower in
the miR-222 mimic-transfected cells (45.9±6.2%) compared with the
control cells (68.8±5.8%; P<0.05; Fig. 2D). EdU incorporation was monitored
by immunofluorescence microscopy following the transfection of the
SKOV3 cells with miR-222 for 48 h. A quantitative analysis of the
EdU-positive cells showed that the miR-222 overexpressing cells
were 49% EdU-positive compared with 24% in the miR control (>100
cells were counted). All these results indicated that miR-222 acts
to promote ovarian cancer cell proliferation by increasing the
population of cells in the S phase.
miR-222 directly targets
P27Kip1 by interacting with its 3′-UTR
To further investigate the molecular mechanism by
which miR-222 promotes ovarian cancer proliferation, a TargetScan
algorithm analysis was performed to identify the putative
downstream target genes of miR-222, particularly those with tumor
suppressive effects and the ability to regulate the cell cycle to
promote tumor proliferation. Based on this rationale, CDNK1B
(encoding protein P27Kip1), a tumor suppressive gene
critical for cell cycle regulation, was selected. As shown in
Fig. 3A, the 3′-UTR of the
P27Kip1 mRNA has a single predicted binding site that is
highly conserved in numerous species. To investigate whether
miR-222 is able to directly target P27Kip1 by
interacting with its 3′-UTR in vitro, the wild-type 3′-UTRs
of P27Kip1 were cloned and inserted downstream of a
luciferase reporter gene (Fig. 3B).
Subsequently, the miR-222 mimics or miR controls were cotransfected
with wild-type 3′-UTR reporter vectors into the SKOV3 cells. It was
observed that the miR-222 mimics decreased the relative luciferase
activity of the wild-type 3′-UTR reporter vector by 37% (Fig. 3C). Furthermore, the predicted
binding site of miR-222 in the 3′-UTR was mutated using overlapping
PCR (Fig. 3B). Luciferase reporter
assays showed that the mutant 3′-UTRs of the P27Kip1
vector abrogated the repression of luciferase activity caused by
miR-222 overexpression, indicating that such regulation was
dependent on a specific sequence (Fig.
3D). Next, the present study investigated whether miR-222 was
able to regulate the expression of the P27Kip1 protein
levels. A western blot analysis showed that the enforced expression
of miR-222 significantly repressed endogenous P27Kip1
expression compared with the cells transfected with the negative
control and blank (Fig. 3E). By
contrast, following transfection with the miR-222 inhibitor in the
SKOV3 cells, the expression of P27Kip1 was clearly
increased compared with the cells transfected with the negative
control or blank (Fig. 3F).
Together, these data indicate that P27Kip1 protein
expression is regulated by miR-222 in ovarian cancer.
Expression levels of miR-222 and
P27Kip1 are inversely correlated in patients with
ovarian cancer
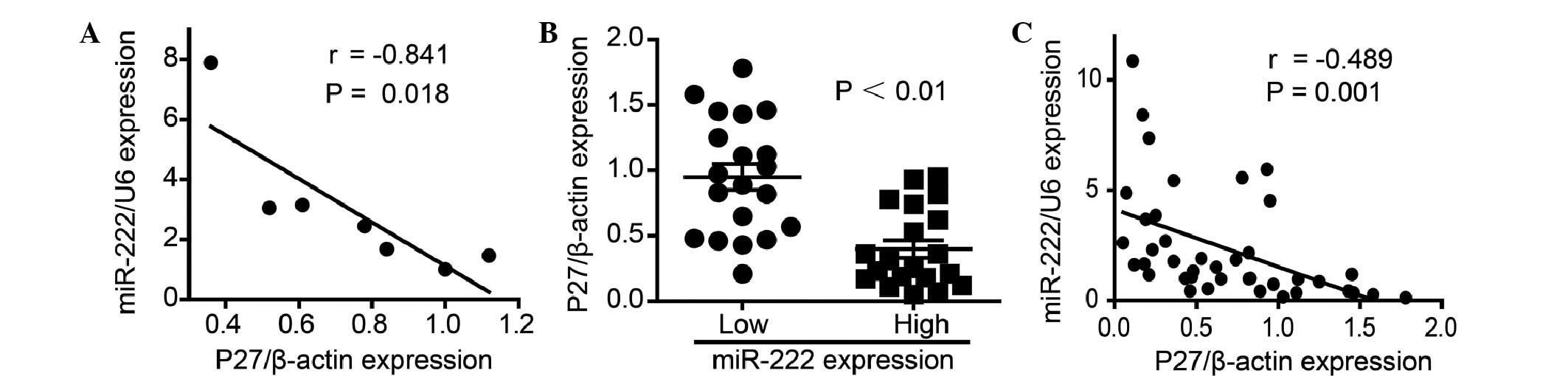
P27Kip1 mRNA expression was first
evaluated using qPCR in the ovarian cancer cells to determine the
association between the expression of P27Kip1 and
miR-222. Just as P27Kip1 was demonstrated to be a target
gene of miR-222, an inverse correlation was observed between
P27Kip1 and miR-222 in the ovarian cancer cells
(r=−0.841, P=0.018; Fig. 4A).
Furthermore, qPCR was performed in 40 ovarian cancer samples. The
data showed that P27Kip1 expression varied between
carcinomas, and that when patients were divided into two groups
according to miR-222 expression, the patients with high miR-222
expression levels presented with significantly lower
P27Kip1 mRNA expression compared with patients with low
miR-222 expression levels (P<0.01; Fig. 4B). Moreover, a Pearson correlation
analysis of these patients revealed a significant inverse
correlation between P27Kip1 and miR-222 expression
(r=−0.489, P=0.001; Fig. 4C).
Therefore, P27Kip1 mRNA expression was associated with
the expression of miR-222, suggesting that miR-222 was able to
negatively regulate P27Kip1 expression in human ovarian
cancer.
Discussion
P27Kip1, encoded by the CDKN1B gene, is a
member of the Cip/Kip family of cyclin-dependent kinase inhibitors
that function to negatively control cell cycle progression.
P27Kip1 binds to CDK2 and cyclin E complexes to prevent
cell-cycle progression from the G1 to S phase.
P27Kip1 also acts as a tumor suppressor, and its
expression is often disrupted in human cancers (13), indicating that its levels of
expression have prognostic and potentially therapeutic
implications. In ovarian cancer, decreased P27Kip1
levels have been correlated with tumor grade, chemotherapy
resistance and poor patient survival (14–16).
P27Kip1 dysfunction is generally regarded as being due
to gene mutation or loss of heterozygosity of P27, but it would be
of great interest to study other potential post-transcriptional
mechanisms such as miRNA-dependent translation suppression.
miR-222 has been shown to be overexpressed in
various types of cancer, such as pancreatic cancer, papillary
thyroid carcinoma, glioblastoma and prostate carcinoma (5,17–19),
and has been reported to an important marker of poor prognosis in
ovarian cancers (14–15). In the present study, it was observed
that miR-222 was frequently upregulated in ovarian cancer.
Furthermore, the results also provided functional evidence
concerning the possible role of miR-222 in ovarian cancer, as it
was demonstrated that miR-222 upregulation is able to induce an
enhancement of ovarian cancer cell proliferation potential,
possibly by downregulating its target, P27Kip1. A
bioinformatic analysis showed that the 3′-UTR of the
P27Kip1 mRNA contained the highly-conserved putative
miR-222 binding site. A luciferase reporter assay demonstrated that
P27Kip1 was a direct target of miR-222. Consistently,
there was an inverse correlation between the P27Kip1 and
miR-222 expression levels in the ovarian cancer cell lines and
tissues. Overall, the present results suggest that miR-222
upregulation in human ovarian cancer may promote ovarian cancer
cell proliferation during ovarian carcinogenesis. We propose that
this fine-tuning regulatory action exerted by miR-222 on the levels
of P27Kip1 protein present in the ovarian cancer cells
may be considered as a sophisticated mechanism, which ensures a
rapid response in P27Kip1 levels to any environmental
and intracellular variations.
In conclusion, the present study suggests that the
over-expression of miR-222 may contribute to the growth and
progression of ovarian cancer, at least in part by repressing
P27Kip1 expression. Additional functional studies are
now required to aid in our comprehension of the molecular basis of
the formation of this carcinoma, and to provide new evidence for
developing innovative therapies targeting the specific molecular
mechanisms of ovarian cancer.
Acknowledgements
The present study was supported by the
National Basic Research Program of China (973 Program,
2009CB521808), the National Nature and Science Foundation of China
(81272859, 81000979, 81230038, 81071663, 81025011 and 81272422),
the Nature and Science Foundation of Hubei Province (2011CBD542)
and Fundamental Research Funds for the Central Universities (HUST:
2012TS058).
References
|
1.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2.
|
No authors listed. Ovarian cancer,
five-year stage-specific relative survival rates (2004–2008). J
Natl Cancer Inst. 103:12872011.PubMed/NCBI
|
|
3.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Galardi S, Mercatelli N, Giorda E, et al:
miR-221 and miR-222 expression affects the proliferation potential
of human prostate carcinoma cell lines by targeting p27Kip1. J Biol
Chem. 282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Miller TE, Ghoshal K, Ramaswamy B, et al:
MicroRNA-221/222 confers tamoxifen resistance in breast cancer by
targeting p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Zhang CZ, Zhang JX, Zhang AL, et al:
MiR-221 and miR-222 target PUMA to induce cell survival in
glioblastoma. Mol Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Lu Y, Roy S, Nuovo G, et al:
Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of
tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein
and modulating mitogenic signal. J Biol Chem. 286:42292–42302.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Stinson S, Lackner MR, Adai AT, et al:
miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes
epithelial-to-mesenchymal transition in breast cancer. Sci Signal.
4:pt52011.PubMed/NCBI
|
|
10.
|
Stinson S, Lackner MR, Adai AT, et al:
TRPS1 targeting by miR-221/222 promotes the
epithelial-to-mesenchymal transition in breast cancer. Sci Signal.
4:ra412011.PubMed/NCBI
|
|
11.
|
Yang CJ, Shen WG, Liu CJ, et al: miR-221
and miR-222 expression increased the growth and tumorigenesis of
oral carcinoma cells. J Oral Pathol Med. 40:560–566. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wurz K, Garcia RL, Goff BA, et al: MiR-221
and MiR-222 alterations in sporadic ovarian carcinoma: Relationship
to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer.
49:577–584. 2010.PubMed/NCBI
|
|
13.
|
Koff A: How to decrease p27Kip1 levels
during tumor development. Cancer Cell. 9:75–76. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Duncan TJ, Al-Attar A, Rolland P, Harper
S, Spendlove I and Durrant LG: Cytoplasmic p27 expression is an
independent prognostic factor in ovarian cancer. Int J Gynecol
Pathol. 29:8–18. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Xing H, Wang S, Hu K, et al: Effect of the
cyclin-dependent kinases inhibitor p27 on resistance of ovarian
cancer multicellular spheroids to anticancer chemotherapy. J Cancer
Res Clin Oncol. 131:511–519. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Masciullo V, Sgambato A, Pacilio C, et al:
Frequent loss of expression of the cyclin-dependent kinase
inhibitor p27 in epithelial ovarian cancer. Cancer Res.
59:3790–3794. 1999.PubMed/NCBI
|
|
17.
|
Greither T, Grochola LF, Udelnow A,
Lautenschlager C, Würl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Visone R, Russo L, Pallante P, et al:
MicroRNAs (miR)-221 and miR-222, both overexpressed in human
thyroid papillary carcinomas, regulate p27Kip1 protein levels and
cell cycle. Endocr Relat Cancer. 14:791–798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Quintavalle C, Garofalo M, Zanca C, et al:
miR-221/222 over-expression in human glioblastoma increases
invasiveness by targeting the protein phosphate PTPμ.
Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|