Introduction
Oral cancer is the sixth most common type of cancer
worldwide. The annual estimated incidence is ~275,000 (1) and has recently been reported to be
increasing in frequency (2).
Despite advances in diagnostic techniques and improvement in
treatment modalities, the prognosis of oral cancer remains poor,
mainly owing to the high rate of local and regional recurrence and
to the development of new malignant changes within the original
field of precancerization (3–5).
Numerous pathways have been reported to be activated during oral
cancer progression (6). In order to
develop therapeutic approaches that hit multiple targets, the
identification of molecular targets for oral cancer is urgently
required.
MicroRNAs (miRNAs) are short single-stranded
nucleotide RNA molecules, which function as master regulators of
gene expression by post-transcriptional modifications of target
mRNAs (7). These noncoding RNAs are
emerging as important modulators in cellular pathways and appear to
play a key role in tumorigenesis (8–10).
With increasing understanding of the cellular behaviors affected by
them, modulation of miRNA activity may provide exciting
opportunities for cancer therapy.
miRNAs have been reported to serve as tumor
suppressors (11). Specifically, it
has been indicated that miR-145 is a tumor suppressor capable of
inhibiting tumor cell growth (12)
and expression levels of miR-145 have been found to be decreased in
human lung adenocarcinoma (13).
However, the role of miR-145 in human oral cancer remains largely
unknown. In the present study, the expression levels of miR-145
were investigated in the human TCA8113 oral cancer line and the
effect of miR-145 transfection on the proliferation, migration and
invasion abilities of TCA8113 cells was determined.
Materials and methods
Cell lines
Human oral TCA8113 cancer and human normal oral
keratinocytes (hNOK) cell lines were obtained from the Center of
Biomedical Experimental Research at the Xi’an Jiaotong University
School of Medicine (Xi’an, China).
Cell culture
TCA8113 and hNOK cells were separately cultured in
Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies,
Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin (Invitrogen Life Technologies), and
incubated at 37°C in an atmosphere containing 5%
CO2.
Quantitative PCR (qPCR) of miR-145
qPCR was used to measure miR-145 expression levels
in hNOK and TCA8113 cells. miR-145 was amplified using the
Bulge-Loop™ miRNA qRT-PCR Primer Set (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) (14). The
thermal profile for the qPCR was 95°C for 1 min, followed by 40
cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 5 sec on a
Bio-Rad CFX96 RT-qPCR system (Bio-Rad, Hercules, CA, USA). All
qPCR, including no-template controls, were performed in triplicate.
Expression levels of miR-145 were evaluated using the comparative
threshold cycle method and normalized against U6.
TCA8113 cell transfection with
miR-145
The miR-145 expression vector (miRNASelect
pEP-miR-145) and miRNA negative control vector (miRNASelect
pEP-miR-Null) were obtained from Cell Biolabs Inc., (San Diego, CA,
USA). TCA8113 cells were transfected with pEPmiR-145 or
pEP-miR-Null using Effectene transfection reagent (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. Vector DNA
(1 μg) was diluted in 100 μl extracellular buffer mixed with 8 μl
enhancer and incubated for 2 min. Effectene transfection reagent (5
μl) was added to the DNA-enhancer mixture and incubated for 10 min
to allow transfection complex formation. The normal growth medium
was replaced by 2.5 ml antibiotic free medium containing 10% (v/v)
FBS during complex formation. The transfection complex was then
applied to the cells and incubated for 48 h. Transfection
efficiency was analyzed by qPCR at 48 h following transfection, as
aforementioned.
The following four groups were established in this
study: blank control, reagent (cells treated with transfection
reagent), negative control (cells treated with transfection reagent
plus pEP-miR-Null) and miR-145 group (cells treated with
transfection reagent plus pEPmiR-145).
Cell proliferation assay
The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich, St. Louis, MO, USA) colorimetric assay was used to
screen for cell proliferation. In brief, cells were seeded into
eight 96-well plates at a density of 2×103 cells/well
and maintained in normal growth medium in a 5% CO2
humidified incubator at 37°C for 48, 72 and 96 h following
treatment. Subsequently, 20 μl MTT (5 mg/ml) was added into each
well and cell culture was continued for a further 4 h. Following
aspiration of the medium, the cells were lysed with
dimethylsulfoxide (Sigma-Aldrich). The absorbance was measured
using a microplate reader at 490 nm. The cell growth curve was
plotted with optical density values as ordinate against time as
abscissa. The experiment was repeated three times.
Cell migration assay
The effect of miR-145 transfection on TCA8113 cell
migration was measured as the ability of cells to migrate through
Transwell filters (6.5-mm diameter; 5-mm pore size). Transwell
filters were coated with reconstituted basement membrane substance
(Matrigel; BD Biosciences, San Diego, CA, USA) for 1.5 h prior to
adding the cells. At 24 h following miR-145 transfection, cells
were detached by trypsinization and 1×105 cells were
seeded into Transwell filters in 100 ml starvation medium.
Subsequently, 500 ml growth medium was placed in the lower
compartment and the cells were left to migrate for 24 h.
Nonmigrated cells were removed using a cotton swab and the
transmigrated cells at the rear side of the filter were stained
with Giemsa. TCA8113 cells on each filter were counted at
magnification, ×400 to quantitate TCA8113 cell migration. Images of
three random fields from three replicate wells were captured.
Migration was determined as the mean of cells that had migrated per
×400 field and expressed as a percentage of the blank control.
In vitro invasion assay
TCA81135 cell invasion was evaluated using 24-well
Transwell units with 8-μm porosity polycarbonate filters. The
filters were coated with 50 μl Matrigel (8 mg/ml). The coated
filters were air-dried at 4°C prior to the addition of the cells.
The basement membrane was hydrated with 50 μl serum-free RPMI-1640
medium 30 min prior to use. At 24 h following miR-145 transfection,
cells were digested with trypsin and the cell density was adjusted
to 1×106 cells/ml using serum-free RPMI-1640 medium. A
total of 200 μl cell suspension was added into each upper Transwell
chamber and 600 μl RPMI-1640 medium containing 5% fetal bovine
serum was added into the lower chamber. There were three duplicates
for each cell group. Following this, the cells were incubated for
24 h in a humidified atmosphere of 5% CO2 at 37°C. Cells
were fixed with methanol and stained with Giemsa. Cells on the
upper surface of the filter were removed by wiping with a cotton
swab and invasion was determined by counting the cells that
migrated to the lower side of the filter with optical microscopy at
magnification, ×400. A total of five visual fields at the center
and in the surrounding areas were counted, and the average was
calculated (15). The experiment
was repeated three times.
Statistical analysis
All data are presented as the means ± SEM.
Statistical analysis was performed using SPSS 16.0 software (SPSS,
Inc., Chicago, IL, USA). Differences among groups were tested by
one-way analysis of variance. P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of miR-145 in hNOK and TCA8113
cells
As demonstrated by qPCR, the expression levels of
miR-145 in human oral cancer TCA8113 cells were ~50% of those of
the hNOK cells (0.342±0.029 vs. 0.695±0.033). There was a
significant difference between the two groups (P<0.05).
Validation of miR-145 transfection in
TCA8113 cells
qPCR was performed to validate the transfection of
miR-145 in TCA8113 cells. As shown in Fig. 1, compared with the blank control,
reagent and negative control group cells, the expression of miR-145
in the miR-145 group cells was significantly increased at 48 h
following transfection (P<0.05), which continued for at least 96
h (data not shown).
Transfection with miR-145 decreases cell
proliferation in TCA8113 cells
To examine whether miR-145 transfection had an
effect on TCA8113 cell growth, an MTT cell proliferation assay was
performed. Compared with the blank control, reagent and negative
control group cells, the miR-145 group cells showed decreased cell
proliferation at 48, 72 and 96 h post-transfection, which is
consistent with the role of miR-145 in cell growth in TCA8113 cells
(P<0.05; Fig. 2).
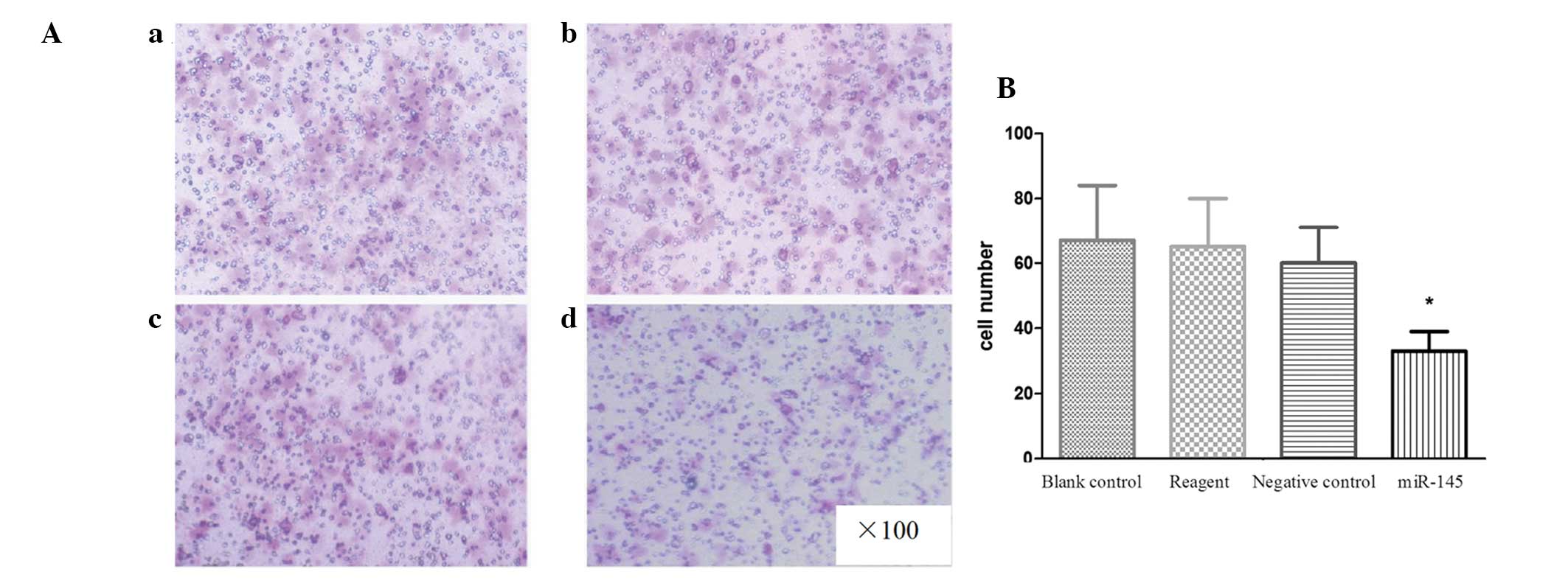
Transfection with miR-145 reduces cell
migration in TCA8113 cells
The effect of miR-145 transfection on the cell
migration ability of TCA8113 cells was investigated by Transwell
invasion assay (Fig. 3A). The
results indicated that miR-145-transfected cells had a
significantly reduced ability to pass through the basement membrane
when compared with the cells in the other three groups (all
P<0.05; Fig. 3B). These data are
consistent with the hypothesis that miR-145 may serve as a tumor
suppressor.
Transfection with miR-145 reduces the
invasive ability of TCA8113 cells
The invasive ability of TCA8113 cells was
investigated using a Matrigel invasion assay. The invasion assay
results indicated that miR-145-transfected TCA8113 cells had a
significantly lower ability to pass through the basement membrane
compared with the cells in the other three groups (all P<0.05;
Fig. 4A and B). These result
indicate that miR-145 is involved in the suppression of TCA8113
cell invasion.
Discussion
At present, changes in the miRNA profile in various
cancer cells and their roles in carcinogenesis have been
increasingly analyzed (16).
Studies have shown that miR-145 is closely associated with
tumorigenesis and the expression levels of miR-145 have been found
to be significantly downregulated in bladder (17), breast (18), colorectal (19), esophageal (20) and gastric cancers (21). Downregulated expression levels of
miR-145 in oral cancer have been shown previously in a hamster oral
squamous cell cancer model (22).
However, the role of miR-145 in human oral cancer tumorigenesis
remains poorly understood. In the present study, the expression of
miR-145 was investigated in the human TCA8113 oral cancer line, and
the effect of miR-145 transfection on the proliferation, migration
and invasion abilities of TCA8113 cells was also observed.
qPCR analysis indicated that miR-145 expression
levels were significantly lower in TCA8113 cells compared with hNOK
cells, indicating that miR-145 may be associated with the genesis
of human oral cancer. These data are consistent with previous
studies showing decreased miR-145 expression in a variety of
malignant tumors. Sachdeva et al showed that the expression
levels of miR-145 decreased gradually during the transition from
normal breast tissue to cancer tissue (23). Chen et al also reported that
the expression levels of miR-145 declined gradually from the
tumorigenesis to the progression stage in prostate cancer (24). In addition, a previous study by
Drebber et al showed that, in patients with advanced colon
cancer undergoing neoadjuvant chemoradiotherapy, a significant
upregulation of miR-145 in post-therapeutic tumor tissue was noted
compared with that in pre-therapeutic tumor tissue. Patients with
low intratumoral post-therapeutic expression had a significantly
poorer response to neoadjuvant therapy compared with patients with
a high expression of miR-145 (25).
These results indicate that miR-145 may be an important molecular
biomarker in early diagnosis and prediction of treatment response
and prognosis of tumors.
At present, the diagnosis and clinical staging of
tumors are mainly based on conventional histology and radiological
imaging. Due to its convenience and non-invasiveness, blood
sampling for the detection of miRNAs as tumor markers has been
increasingly applied in recent years. Serum miR-132, miR-26a,
let-7b and miR-145 have been reported as potential candidates for
novel biomarkers in serous ovarian cancer (26). Due to its decreased expression in
TCA8113 cells, we hypothesize that miR-145 may be used as a
potential biomarker in the early diagnosis of oral cancer.
We further investigated the effect of miR-145
transfection on in vitro proliferation, migration and
invasion of TCA8113 cells. Upregulation of miR-145 resulted in a
suppression of tumor cell proliferation, migration and invasion.
Our observations are consistent with previous studies on the
functional roles of miR-145. Shi et al(27) reported that, in colon cancer cells,
miR-145 is directly bound to the insulin receptor substrate-1
(IRS-1) 3′-untranslated region and downregulates IRS-1 protein,
inhibiting the growth of colon cancer cells. Gregersen et
al(19) employed a
microarray-based approach to identify miR-145 targets in colon
cancer cells, and YES and STAT1 were verified as direct miR-145
targets. In the PC3 prostate cancer cell line, Zaman et
al(28) found that
overexpression of miR-145 by transfection resulted in increased
apoptosis and an increase in cells in the G2/M phase. Microarray
analysis of miR-145-overexpressing PC3 cells showed upregulation of
the pro-apoptotic gene, TNFSF10. In HCT-116 and MCF-7 cells,
Sachdeva et al(23) showed
that c-Myc is a direct target of miR-145. In addition, the blockade
of miR-145 by anti-miR-145 was able to reverse p53-mediated c-Myc
repression, defining a role for miR-145 in the post-transcriptional
regulation of c-Myc by p53 and indicating that miR-145 provides a
direct link between p53 and c-Myc in this gene regulatory network.
miR-145 was also found to play a negative regulatory role in cell
growth through RTKN (29), OCT,
SOX-2 and KLF4 pathways (30). A
further study (12) showed that
miR-145 significantly suppresses cell invasion in MCF-7 and HCT-116
cells, and that miR-145 is also able to suppress lung metastasis in
an experimental metastasis animal model. This miR-145-mediated
suppression of cell invasion is, in part, due to the silencing of
the metastasis gene, mucin 1 (MUC1). Furthermore, suppression of
MUC1 by miR-145 causes a reduction of β-catenin as well as
oncogenic cadherin 11. Overall, these results indicate that, as a
tumor suppressor, miR-145 inhibits not only tumor growth but also
cell invasion and metastasis, and miR-145 is a promising new
therapeutic target for the treatment of various types of cancer,
including oral cancer. In the future, studies of the regulation of
target genes by miR-145 in oral cancer TCA8113 cells are likely to
continue to identify the action mechanisms of miR-145, which may be
significant for the diagnosis and treatment of oral cancer through
miR-145.
In conclusion, our data indicate that the expression
levels of miR-145 in TCA8113 cells are significantly lower than
those of hNOK cells. miR-145 may be a valuable molecular biomarker
for the early diagnosis of oral cancer. Cellular proliferation,
migration and invasion abilities in miR-145-transfected TCA8113
cells were significantly decreased, indicating that miR-145 may
participate in oral cancer genesis and progression. Thus, miR-145
is a potential therapeutic target for the treatment of oral
cancer.
References
|
1
|
Warnalulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar
|
|
2
|
Hernández-Guerrero JC, Jacinto-Alemán LF,
Jiménez-Farfán MD, Macario-Hernández A, Hernández-Flores F and
Alcántara-Vázquez A: Prevalence trends of oral squamous cell
carcinoma. Mexico City’s General Hospital experience. Med Oral
Patol Oral Cir Bucal. 18:e306–e311. 2013.PubMed/NCBI
|
|
3
|
Bagan J, Sarrion G and Jimenez Y: Oral
cancer: clinical features. Oral Oncol. 46:414–417. 2010. View Article : Google Scholar
|
|
4
|
Shah JP and Gil Z: Current concepts in
management of oral cancer-surgery. Oral Oncol. 45:394–401. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurita H, Nakanishi Y, Nishizawa R, Xiao
T, Kamata T, et al: Impact of different surgical margin conditions
on local recurrence of oral squamous cell carcinoma. Oral Oncol.
46:814–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pérez I, Varona A, Blanco L, Gil J,
Santaolalla F, et al: Increased APN/CD13 and acid aminopeptidase
activities in head and neck squamous cell carcinoma. Head Neck.
31:1335–1340. 2009.PubMed/NCBI
|
|
7
|
Yi R and Fuchs E: MicroRNAs and their
roles in mammalian stem cells. J Cell Sci. 124:1775–1783. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valastyan S, Chang A, Benaich N, Reinhardt
F and Weinberg RA: Activation of miR-31 function in
already-established metastases elicits metastatic regression. Genes
Dev. 25:646–659. 2011. View Article : Google Scholar
|
|
9
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, et
al: Identification of miRNomes in human liver and hepatocellular
carcinoma reveals miR-199a/b-3p as therapeutic target for
hepatocellular carcinoma. Cancer Cell. 19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu J, Guo H, Li H, Liu Y, Liu J, et al:
MiR-145 regulates epithelial to mesenchymal transition of breast
cancer cells by targeting Oct4. PLoS One. 7:e459652012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hammond SM: MicroRNAs as tumor
suppressors. Nat Genet. 39:582–583. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo L, Liu Y, Bai Y, Sun Y, Xiao F and Guo
Y: Gene expression profiling of drug-resistant small cell lung
cancer cells by combining microRNA and cDNA expression analysis.
Eur J Cancer. 46:1692–1702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Y, Chen W, Zhang Y, Hamburger AW, Pan H
and Zhang Z: Suppression of salivary adenoid cystic carcinoma
growth and metastasis by ErbB3 binding protein Ebp1 gene transfer.
Int J Cancer. 120:1909–1913. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho WC: MicroRNAs: potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chiyomaru T, Enokida H, Tatarano S,
Kawahara K, Uchida Y, et al: miR-145 and miR-133a function as
tumour suppressors and directly regulate FSCN1 expression in
bladder cancer. Br J Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Spizzo R, Nicoloso MS, Lupini L, Lu Y,
Fogarty J, et al: miR-145 participates with Tp53 in a
death-promoting regulatory loop and targets estrogen receptor-alpha
in human breast cancer cells. Cell Death Differ. 17:246–254. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gregersen LH, Jacobsen AB, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-145 targets YES and STAT1 in colon
cancer cells. PLoS One. 5:e88362010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, et al: MiR-145, miR-133a and miR-133b: Tumor suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu T, Wang XY, Gong RG, Li A, Yang S, et
al: The expression profile of microRNAs in a model of
7,12-dimethyl-benz[a]anthrance-induced oral carcinogenesis in
Syrian hamster. J Exp Clin Cancer Res. 28:642009.PubMed/NCBI
|
|
23
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V, et
al: p53 represses c-Myc through induction of the tumor suppressor
miR-145. Proc Natl Acad Sci USA. 106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Gong J, Zeng H, Chen N, Huang R,
et al: MicroRNA-145 targets BNIP3 and suppresses prostate cancer
progression. Cancer Res. 70:2728–2738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Drebber U, Lay M, Wedemeyer I, Vallböhmer
D, Bollschweiler E, et al: Altered levels of the onco-microRNA 21
and the tumor-supressor microRNAs 143 and 145 in advanced rectal
cancer indicate successful neoadjuvant chemoradiotherapy. Int J
Oncol. 39:409–415. 2011.
|
|
26
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
et al: Detection of microRNA as novel biomarkers of epithelial
ovarian cancer from the serum of ovarian cancer patient. Int J
Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA-145 targets in insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zaman MS, Chen Y, Deng G, Shahryari V, Suh
SO, et al: The functional significance of microRNA-145 in prostate
cancer. Br J Cancer. 103:256–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang S, Bian C, Yang Z, Bo Y, Li J, et al:
miR-145 inhibits breast cancer cell growth through RTKN. Int J
Oncol. 34:1461–1466. 2009.PubMed/NCBI
|
|
30
|
Xu N, Papagiannakopoulos T, Pan G, Thomson
JA and Kosik KS: MicroRNA-145 regulates OCT4, SOX2, and KLF4 and
represses pluripotency in human embryonic stem cells. Cell.
137:647–658. 2009. View Article : Google Scholar : PubMed/NCBI
|