Introduction
Gastric cancer (GC) is one of the most common types
of gastrointestinal malignancy worldwide and is the second leading
cause of cancer-related mortality (1–3). The
majority of patients are diagnosed at an advanced clinical stage,
however, the molecular mechanisms are not fully understood.
microRNAs (miRNAs) are a class of small endogenous
non-coding RNAs consisting of 19–24 nt, which have high
evolutionary conservation. miRNA are important in organism growth,
development and the incidence of disease by base pairing to
complementary sites in the target mRNA 3′-untranslated region.
Thus, miRNA may inhibit the translation or degradation of target
mRNA (4–7). Previous studies have indicated that
aberrant expression of miRNAs contributes to the initiation and
progression of human malignancies, including colon cancer, GC and
breast cancer (8,9). However, to date, no specific studies
have been conducted to investigate the correlation between miR-32
and GC. The present study investigated the effect of miR-32 on the
biological behaviors of the human gastric carcinoma cell line,
SGC-7901.
Materials and methods
Cell culture and transfection
The human GC cell line, SGC-7901, was obtained from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China)
and cultured in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich,
St. Louis, MO, USA) containing 10% fetal bovine serum, at 37°C in a
humidified 5% CO2 incubator. Throughout the experiment,
cells were used in the logarithmic phase of growth. Transfection
was performed using Lipofectamine™ 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. On the day prior to transfection, cells were seeded
onto 6-well plates (5×105 cells/well) and inoculated in
complete medium without antibiotics (2 ml/well). At 85% confluence,
the plasmid DNA (4.0 μg/well) and transfection reagent (10 μl/well)
were diluted with RPMI-1640 (250 μl) and stood at room temperature
for 5 min. The transfection reagent and dilution of the plasmid DNA
were then mixed and stood at room temperature for 30 min. Next, the
transfection complexes were added onto 6-well plates (500 μl/well)
and cultured at 37°C in a humidified 5% CO2 incubator
for 48 h. This study was approved by the ethics committee of the
Affiliated Hospital of Nantong University (Nantong, China).
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted according to the
manufacturer’s instructions and dissolved in DEPC water (40 μg).
The RNA concentration and purity were detected by
ultraviolet-visible spectrophotometer. The specimens, which had
A260/A280 values fluctuating between 1.8–2.0, were used for reverse
transcription. The process of reverse transcription cDNA synthesis
requires the use of RNAse-free centrifuge tubes and must be
performed on ice according to the following reaction system: 4 μl
5X reaction buffer, 2 μl dNTP MIX (10 mmol/l), 1 μl RiboLock™ RNase
inhibitor, 1 μl RevertAid M-MuLV reverse transcriptase (all from
Fermentas Canada Co., Ltd., Burlington, ON, Canada) and RT primer
(Sangon Biotech China Co., Ltd., Shanghai, China) (primer
concentrations were adjusted to 5–50 nmol/l). The final volume was
adjusted to 20 μl with DEPC water. Following transfection, the PCR
reaction solution used to detect the relative expression levels of
miR-32 in GC cells (SGC-7901) included the following: 12.5 μl
SYBR-Green/ROX qPCR master mix, 1.5 μl forward and reverse primers
(including primers for miR-32 and U6) and 1 μl template DNA. The
final volume was adjusted to 25 μl with DEPC water (each sample
experiment was repeated four times). The following reaction
conditions were used: uracil-DNA glycosylase pretreatment at 50°C
for 2 min, initial denaturation at 95°C for 10 min, denaturation at
95°C for 15 sec and extension for 60 sec at 60°C for 40 cycles.
Scratch-wound assay
Transiently transfected cells in logarithmic growth
phase were seeded onto 6-well plates (5×105/well)
following culture for 24 h. At 24 h after seeding, the confluent
cell monolayers were wounded with a pipette. Exfoliated cells were
washed off using phosphate-buffered saline (PBS) and culture was
continued in fresh medium without fetal bovine serum (FBS). Wound
closure was monitored by microscopy at various times (0, 6 and 24
h). Visual fields (n=4) of each insert were randomly frozen under
an inverted fluorescence microscope (IX71, Olympus, Tokyo, Japan).
Migration activity was calculated as the mean distance between the
edges of three points. Healing rate = (mean original distance -
mean distance at a time point)/mean original distance × 100. Each
test group was assayed in triplicate.
Migration assay
Transiently transfected cells were trypsinized and
suspended without serum-free RPMI-1640 culture medium. The
suspended cells were seeded in the upper chamber of the
Transwell® insert and RPMI-1640 medium containing 20%
FBS (600 μl) was added to the lower chamber of the Transwell
insert. Following culturing at 37°C in a humidified 5%
CO2 incubator for 24 h, the inserts were washed with
PBS. A cotton swab was used to remove adherent cells on the inner
side of the upper chamber membrane, then the chamber membranes were
fixed in paraformaldehyde (4%) for 10 min. Coomassie Blue (600 μl)
was added into each well and incubated at room temperature for 15
min. The inserts were washed again and the upper chamber was left
to dry naturally. Visual fields (n=15) of each insert were randomly
counted under an upright light microscope (BX51, Olympus) and the
average value was calculated.
Cell proliferation assay by cell counting
kit-8 analysis
The cells were digested, resuspended and the cell
concentration was adjusted (5,000/well). Next, the cells were
seeded onto a 96-well plate and cultured at 37°C in a humidified 5%
CO2 incubator until the cells had adhered. The cells
were transiently transfected using Lipofectamine 2000, changing the
medium after 4 h. CCK-8 solution (10 μl) was added to each well of
the plate at 3 time points (24, 48 and 72 h after medium
replacement) and the cells continued to be incubated on the plate.
The absorbance was measured at 450 nm using a multifunctional
microplate reader at 3 time points (1, 2 and 4 h after incubation).
Cell growth curves were determined using the value of absorbance
(at 450 nm) at various time points. Finally, the cell growth
inhibition rate (IR) was calculated using the following formula: IR
= (1 − Aexperimental/Acontrol) × 100, where A
represents the absorbance value.
Statistical analysis
All experiments were repeated at least three times.
Statistical analysis was performed using SPSS 19.0 (SPSS, Inc.,
Chicago, IL, USA). Data are presented as mean ± standard deviation
and groups were compared using one-way analysis of variance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of miR-32 in SGC-7901 cells
transfected with miR-32-mimic and -inhibitor
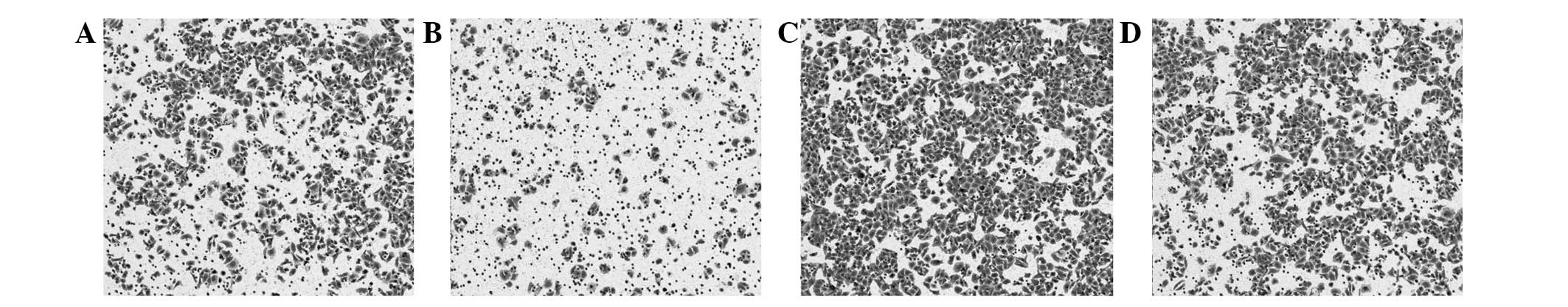
The transfection efficiency was confirmed under the
inverted microscope. The results showed that >80% cells were
labeled with GFP and SGC-7901 cells had been successfully
transfected with miR-32-mimic and -inhibitor (Fig. 1A and B). Compared with the control
group, the expression of miR-32 in the miR-32-mimic group was
significantly downregulated and in the miR-32-inhibitor group
significantly upregulated (P<0.05; Fig. 1C).
miR-32 inhibits the migration and
invasion of SGC-7901 cells in vitro
The scratch-wound assay was used to measure and
compare the scratch breadth at 24 h following transfection. The
results showed that the migration ability of the miR-32-mimic group
(61.39±2.21 vs. 64.42±2.15%; healing rate, 4.71±1.66%) was
significantly lower than the the untreated group (27.49±2.15 vs.
60.4±0.73%; healing rate, 55.97±2.95%) and inhibitor (29.97±0.66
vs. 64.86±0.36%; healing rate, 53.79±0.76%) groups (Fig. 2). Furthermore, the Transwell assay
was used to detect the cell invasion ability. The number of cells
that had traversed the membrane were counted following transfection
for 48 h. Compared with the control group, the miR-32-mimic group
significantly reduced the invasion ability of GC cells. By
contrast, the miR-32-inhibitor group significantly increased the
invasion ability (P<0.05) (Table
I; Fig. 3).
 | Table ICell invasion ability for GC cells by
Transwell® assay. |
Table I
Cell invasion ability for GC cells by
Transwell® assay.
| Variable | Visions, n | Invading cells,
n |
|---|
| miR-32-mimic
group | 15 | 45.93±4.63a |
| miR-32-inhibitor
group | 15 | 76.12±3.62 |
| Empty vector
group | 15 | 82.19±3.32 |
| Untreated group | 15 | 93.93±7.09 |
miR-32 inhibits cell proliferation
To investigate the effect of miR-32 on the growth of
human GC, the SGC-7901 cell line was transfected with miR-32-mimic
and -inhibitor and a control vector for 48 and 72 h. Cell viability
was performed by the CCK-8 assay. The results showed that high
expression of miR-32 in the miR-32-mimic group significantly
inhibited cell proliferation compared with the miR-32-inhibitor,
control and untransfected groups (P<0.05; Table II).
 | Table IICell viability measured by cell
counting kit-8 assay at various times. |
Table II
Cell viability measured by cell
counting kit-8 assay at various times.
| 24 h | 48 h | 72 h |
|---|
|
|
|
|
|---|
| Variable | A450 value | IR, % | A450 value | IR, % | A450 value | IR, % |
|---|
| miR-32-mimic
group | 0.41±0.16 | 36.46±14.33 | 0.58±0.12a | 43.474±18.63 | 0.65±0.10a | 45.05±23.76 |
| miR-32-inhibitor
group | 0.36±0.06 | 38.54±13.45 | 0.78±0.08 | 36.300±14.10 | 1.13±0.09 | 24.81±2.85 |
| Empty vector
group | 0.37±0.07 | 31.50±11.08 | 0.83±0.15 | 33.580±86.69 | 1.22±0.29 | 35.42±23.82 |
| Untreated group | 0.43±0.19 | 0 | 0.97±0.19 | 0 | 1.13±0.67 | 0 |
Discussion
According to the Lauren classification, there are
two major types of GC: Intestinal and diffuse. The diffuse type is
associated with the mutation, deletion and promoter methylation of
E-cadherin (10). While, based on
the Correa hypothesis of gastric carcinogenesis, the intestinal
type of gastric carcinogenesis is a multistage process with the
following order of gastric carcinogenesis: Normal gastric mucosa,
superficial gastritis, atrophic gastritis, intestinal metaplasia,
intraepithelial neoplasia and carcinoma. A number of molecules and
complex regulatory networks are involved in gastric carcinogenesis,
including infection with Helicobacter pylori, activation of
oncogenes, inactivation of cancer suppressor genes and changes in
epigenetic modification (11).
Gastric carcinogenesis is an important issue which remains to be
solved by clinical scientists. Critical molecular mechanisms and
biomarkers for GC must be found to improve the identification of
high risk warning signs, early diagnosis, prognosis and effective
therapeutic options.
Caudal type homeobox transcription factor 2 (CDX2),
is comprised of 311 residues and binds to corresponding DNA
sequences through a helix-loop-helix domain. CDX2 is expressed in
intestinal epithelial, pancreas ductal and acinar epithelial cells
which originate from the endoderm, but is absent in the esophagus
and normal gastric mucosa epithelial cells. CDX2 is an
intestine-specific nuclear transcription factor and is important in
regulating the proliferation and differentiation of normal
intestinal epithelial cells (12).
It has been previously shown that ectopic expression
of CDX2 causes changes associated with gastric intestinal
metaplasia and GC (13–17). Etopic expression of CDX2 in gastric
epithelial cells leads to the genesis of intestinal metaplasia,
followed by intestinal GC. Helicobacter pylori infection
causes the chronic inflammation of gastric mucosa which may
progress to intestinal metaplasia. Intestinal metaplasia epithelial
cells are well recognized as precancerous lesions and increase the
risk of intestinal GC. CDX2 is found in almost 100% of gastric
intestinal metaplasia tissue and the majority of early GC,
particularly intestinal GC (14,15).
Our previous study (17,18) also showed that CDX2 mRNA is absent
in normal gastric tissue, but ectopic expression presents in
intestinal metaplasia, gastric epithelial dysplasia and GC. In
addition, the positive rate of CDX2 in intestinal GC was
significantly higher than in the diffuse type. A previous study
(15) reported that CDX2 locates,
not only in intestinal metaplasia, but also in incomplete
intestinal metaplasia. However, the expression levels in incomplete
intestinal metaplasia were much lower. Consistently, CDX2
expression in the goblet and columnar cells of the incomplete
intestinal metaplasia tissue was much lower than in intestinal
metaplasia. Although the structure of incomplete intestinal
metaplasia cells is similar to colonic epithelial cells, they
express much lower levels of CDX2. From intestinal to dysplasia
metaplasia and then GC, the expression of CDX2 decreases. The
aforementioned studies illustrate that CDX2 functions as a cancer
suppressor gene in gastric carcinogenesis, as well as colon cancer.
The low expression of CDX2 in intestinal and dysplasia metaplasia
is a critical marker of high risk gastric carcinogenesis.
Simultaneously with the progression of malignant lesions, the
expression of CDX2 in GC tissue declines; the expression of CDX2 in
early GC is significantly higher than in advanced GC and low status
lymphatic and distant metastasis GC. Following surgical treatment,
patients with CDX2-positive expression demonstrated a higher
survival rate compared with those with CDX2-negative expression
(19). In addition, the in
vitro experiments showed that GC cells transfected with CDX2
exhibited typical apoptotic morphological changes (20). Overexpression of CDX2 arrests GC
cells in the G0/G1 stage of the cell cycle and induces cell
apoptosis. All the results showed that CDX2 is involved in the
regulation of GC proliferation and metastasis (21). In conclusion, CDX2 is a cancer
suppressor gene of GC.
miRNAs are important for post-transcriptional gene
regulation via binding to target mRNA to mediate destabilization
and translational inhibition. It has been previously confirmed that
miRNAs are involved in cell proliferation, differentiation and
apoptosis. Therefore, miRNAs have been found to closely correlate
with the genesis and progression of tumors. Previous studies have
identified that specific miRNA have the function to suppress
cancer, while others, by contrast, promote cancer. miRNA, as a
newly identified gene regulation factor, is involved in the complex
control network of the cell and mediates a dispensable regulation
model of gene expression (4).
It remains unclear whether CDX2, as a nuclear
transcription regulator, directly upregulates or downregulates
miRNA to mediate GC cell proliferation and metastasis. Our previous
studies showed that SGC-7901 GC cells transfected with CDX2 exhibit
changes in biological behavior. The miRCURY LNA™ array was used to
screen for the altered expression of miRNA in SGC-7901 cells
transfected with CDX2. It was found that the expression of 59
miRNAs was changed significantly in the transfected group
(transfected with pEGFP-N1-CDX2). Of the 59 miRNAs, 25 were
upregulated and 34 were downregulated by more than double. miR-32
and miR-374a, which were found to be upregulated significantly,
were randomly selected to confirm the result of the miRNA
expression chip by qPCR. The qPCR results were consistent with the
results from the chip (unpublished data). Following the
overexpression of miR-32 at 48 and 72 h, cell proliferation and
invasion ability were examined and found to be markedly decreased.
By contrast, the suppressed expression of miR-32 led to a marked
increase in cell proliferation and invasion ability. The target
prediction software miRanda, TargetScan and miRtarget were used to
predict miR-32 targets and SMAD7, GATA6, MYH9 and SOX4 were
identified. SMAD7 is a suppressive member of the SMAD family and is
involved in the transforming growth factor-β/SMAD signaling pathway
and GATA6 and SOX4 are key molecules in the Wnt signaling pathway.
Therefore, CDX2 may mediate miR-32 to achieve its anticancer
effect. As a result, miR-32 was selected in the present study as
the target gene to further study its effect on the biological
behavior of GC cells.
miR-32 eukaryotic expression vectors (mimic and
inhibitor) were successfully constructed and SGC-7901 GC cells were
transfected with these vectors. miR-32 expression was upregulated
and downregulated by various methods and dynamic biological changes
were observed in the GC cells. The results showed that, compared
with the inhibitor and control groups (blank and empty vector
controls), the miR-32-mimic group inhibited cell proliferation and
migration at 48 and 72 h following transfection (P<0.05).
Therefore, miR-32 markedly inhibits the malignant behavior of
SGC-7901 cells. However, the mechanisms by which CDX2 mediates the
miR-32-altered phenotype of GC cells in vivo and in
vitro, and which molecules miR-32 interacts with to regulate
the phenotype of GC cells, remain to be solved in future
studies.
In conclusion, the present study confirmed that
miR-32 notably impacts the biological behavior of GC cells and the
upregulation of miR-32 markedly inhibits the proliferation and
migration of GC cells. These results are likely to contribute to
the identification of the molecular mechanisms of CDX2 antigastric
growth and metastasis and the development of targeted therapeutics
for GC.
References
|
1
|
Compare D, Rocco A and Nardone G: Risk
factors in gastric cancer. Eur Rev Med Pharmacol Sci. 14:302–308.
2010.
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
3
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-regulated microRNAs impair TGFbeta-dependent
cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell.
13:272–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bueno MJ, Pérez de Castro I and Malumbres
M: Control of cell proliferation pathways by microRNAs. Cell Cycle.
7:3143–3148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lauren P: The two histological main types
of gastric carcinoma: diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965.
|
|
11
|
Tamura G, Yin J, Wang S, Fleisher AS, Zou
T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP, et al:
E-Cadherin gene promoter hypermethylation in primary human gastric
carcinomas. J Natl Cancer Inst. 92:569–573. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuasa Y: Control of gut differentiation
and intestinal-type gastric carcinogenesis. Nat Rev Cancer.
3:592–600. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bonhomme C, Duluc I, Martin E,
Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN and
Domon-Dell C: The Cdx2 homeobox gene has a tumour suppressor
function in the distal colon in addition to a homeotic role during
gut development. Gut. 52:1465–1471. 2003. View Article : Google Scholar
|
|
14
|
Bai YQ, Yamamoto H, Akiyama Y, Tanaka H,
Takizawa T, Koike M, Kenji Yagi O, Saitoh K, Takeshita K, Iwai T
and Yuasa Y: Ectopic expression of homeodomain protein CDX2 in
intestinal metaplasia and carcinomas of the stomach. Cancer Lett.
176:47–55. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eda A, Osawa H, Yanaka I, Satoh K, Mutoh
H, Kihira K and Sugano K: Expression of homeobox gene CDX2 precedes
that of CDX1 during the progression of intestinal metaplasia. J
Gastroenterol. 37:94–100. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mutoh H, Sakurai S, Satoh K, Tamada K,
Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N, et al:
Development of gastric carcinoma from intestinal metaplasia in
Cdx2-transgenic mice. Cancer Res. 64:7740–7747. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao ZB, Zhang JF, Xu Z, Zhu HJ, Zhang JG,
Pan ZP, Xiao F and Yang JL: Ectopic expression of guanylyl cyclase
C in gastric cancer as a potential biomarker and therapeutic
target. J Dig Dis. 10:272–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang JF, Zhang JG, Kuai XL, Zhang H,
Jiang W, Ding WF, Li ZL, Zhu HJ and Mao ZB: Reactivation of the
homeotic tumor suppressor gene CDX2 by
5-aza-2′-deoxycytidine-induced demethylation inhibits cell
proliferation and induces caspase-independent apoptosis in gastric
cancer cells. Exp Ther Med. 5:735–741. 2013.PubMed/NCBI
|
|
19
|
Qin R, Wang NN, Chu J and Wang X:
Expression and significance of homeodomain protein Cdx2 in gastric
carcinoma and precancerous lesions. World J Gastroenterol.
18:3296–3302. 2012.PubMed/NCBI
|
|
20
|
Xie Y, Li L, Wang X, Qin Y, Qian Q, Yuan X
and Xiao Q: Overexpression of Cdx2 inhibits progression of gastric
cancer in vitro. Int J Oncol. 36:509–516. 2010.PubMed/NCBI
|
|
21
|
Barros R, Camilo V, Pereira B, Freund JN,
David L and Almeida R: Pathophysiology of intestinal metaplasia of
the stomach: emphasis on CDX2 regulation. Biochem Soc Trans.
38:358–363. 2010. View Article : Google Scholar : PubMed/NCBI
|