Introduction
Stromal cell-derived factor-1 (SDF-1 or CXCL12) is a
cytokine belonging to the CXC chemokine family (1,2). For a
number of years it was thought that CXCR4 was the sole receptor for
this chemokine. However, in 2005, a new SDF-1 receptor was
identified and named CXCR7 (3). The
SDF-1 and CXCR4/CXCR7 axis exerts pleiotropic activity and is
involved in normal development, organogenesis, regeneration and
tumorigenesis (4–6).
The chemokine SDF-1 and its receptors, CXCR4 and
CXCR7, have been implicated in cancer progression and metastasis.
At the mRNA and protein levels, enhanced expression of SDF-1
and CXCR4 has been demonstrated in different malignancies,
including epithelial ovarian cancer and ovarian cancer cell lines
(7–12). These earlier observations on the
chemokine expression profile in ovarian cancer were confirmed more
recently by other groups (13,14).
With regard to the expression of the SDF-1 and CXCR4/CXCR7 axis in
ovarian cancer, the majority of data originates from
immunohistochemical studies. However, commonly used antibodies are
only able to recognize subpopulations of studied molecules
(15–18).
Numerous human genes are known to have transcript
variants and among them SDF-1 (Gene ID: 6387) has four
variants; CXCR4 (Gene ID: 7852) has two variants and
CXCR7 (Gene ID: 57007) has no known alternative splicing
variants. Limited data are available on the expression levels of
SDF-1 and CXCR4 variants and CXCR7 in human
epithelial ovarian cancer. Therefore, the present study aimed to
characterize the expression pattern and levels of SDF-1 and
CXCR4 transcript variants and CXCR7 in epithelial
ovarian cancer and healthy human ovaries. We also aimed to
correlate the obtained data to plasma SDF-1α levels and to specific
clinicopathological variables of studied patients.
Materials and methods
This study included 113 females subjected to surgery
at the Department of Oncology, Poznań University of Medical
Sciences (PUMS; Poznań, Poland). Histopathological examinations
were conducted at the Department of Tumor Pathology, Wielkopolska
Centre for Oncology (Poznań, Poland). The studies were conducted as
accepted by the Bioethical Commission (PUMS; consent no.164/11 of
17th February, 2011). Written informed consent was obtained from
all patients.
Study groups and material sampling
Expression of SDF-1 and CXCR4
transcript variants and CXCR7 in epithelial ovarian cancer and
control ovaries
This study was conducted in 27 patients subjected to
surgery in the Department of Oncology (PUMS) due to ovarian cancer.
The mean age of the patients in this group was 62 years (range,
47–85 years). The most frequent histopathological diagnosis in the
group involved serous carcinoma (18 patients), followed by
endometriod carcinoma (four patients), mucinous carcinoma (three
patients) and macrocellular carcinoma (two patients). Cancers at
stage III of clinical advancement, according to the International
Federation of Gynecology and Obstetrics (FIGO), were exhibited in
18 patients, followed by cancers at stage II (four patients), stage
IV (three patients) and stage I (two patients). The most frequent
were cancers of a low degree of differentiation (grade 3, 13
patients; grade 2, 11 patients; grade 1, three patients). The
control group consisted of 13 females subjected to surgery due to
uterine myomas or uterine prolapse after menopause. The mean age of
the control group patients was 63 years (range, 51–77 years).
During surgery, a section of ovarian tumor or normal
ovary tissue was taken (~0.5 cm3). The fragments were
immersed in RNAlater fluid (Life Technologies, Austin, TX, USA) and
frozen at a temperature of −70°C.
Plasma concentrations of SDF-1α and
cancer antigen 125 (CA 125)
This study was conducted in 43 patients subjected to
surgery at the Department of Oncology (PUMS). In this group, the
mean age of the patients was 57 years (range, 41–84 years). The
most frequent histopathological diagnosis involved serous carcinoma
(29 patients), followed by mucinous carcinoma (five patients),
endometriod carcinoma (three patients), solid carcinoma (two
patients), macrocellular carcinoma (one patient) and other tumors
(three patients). The majority of patients were diagnosed with
stage III cancer according to FIGO, followed by patients with stage
I (6 patients), stage II (2 patients) and stage IV (2 patients).
Poorly differentiated cancers consttuted the majority exhibited by
the patients (grade 3, 27 patients; grade 2, 13 patients; grade 1,
three patients). The control group consisted of 30 females
subjected to surgery at the Department of Oncology (PUMS) for
gynecological reasons distinct from ovarian cancer and with no
neoplastic diseases in anamnesis. The mean age of the control
patients was 45 years (range, 31–72 years). In patients with
ovarian cancer, venous blood was sampled from the antecubital vein
on three occasions: One day prior to surgery, on the 6th day
following surgery and after six cycles of chemotherapy. In the
control group, blood was sampled once, prior to surgery. The blood
was centrifuged and plasma was subsequently frozen at a temperature
of −70°C.
Methods
Quantitative polymerase chain reaction
(qPCR) analysis
Total RNA was extracted from samples using TRIzol
reagent and the RNeasy Mini kit (Qiagen, Hilden, Germany) using the
standard procedure (19–21). RNA concentration and purity was
determined spectrophotometrically (NanoDrop; ThermoScientific,
Waltham, USA). For each sample, 1 μg of total RNA was reversely
transcribed using MMLV reverse transcription kit (Novazym, Poznań,
Poland) using Oligo dT (PE Biosystems, Warrington, UK) as primers.
The reaction was performed at 42.8°C for 60 min (UNO II
thermocycler; Biometra, Goettingen, Germany). All primer sets were
designed to span, with the exception of CXCR4 variant 1, at least
one intron (Table I). The variants
studied were those listed in GenBank (SDF-1: Gene ID, 6387;
CXCR4: Gene ID, 7852; CXCR7: Gene ID, 57007). Primers
were purchased from the Laboratory of DNA Sequencing and
Oligonucleotide Synthesis (Institute of Biochemistry and
Biophysics, Polish Academy of Sciences, Warsaw, Poland).
 | Table IConventional qPCR analyses of
CXCL12, transcript variants 1–4; CXCR4, transcript
variants 1–2 and CXCR7. |
Table I
Conventional qPCR analyses of
CXCL12, transcript variants 1–4; CXCR4, transcript
variants 1–2 and CXCR7.
| cDNA | GenBank accession
number | Primer | Primer sequence
(5′-3′) | Position | PCR product size
(bp) |
|---|
| CXCL12, transcript
variant 1 | NM_199168 | S |
TACAGATGCCCATGCCGATT | 174–193 | 262 |
| A |
GCCCTTTCATCTCTCACAAGGT | 414–435 | |
| CXCL12, transcript
variant 2 | NM_000609 | S |
TGTGCATTGACCCGAAGCTA | 301–320 | 144 |
| A |
CAGGCCCTTCCCTAACACT | 426–444 | |
| CXCL12, transcript
variant 3 | NM_001033886 | S |
ACTGTGCCCTTCAGATTGTAGCC | 253–275 | 260 |
| A |
AGCAAATTTACAAAGCGCCGAGA | 490–512 | |
| CXCL12, transcript
variant 4 | NM_001178134 | S |
TACAGATGCCCATGCCGATT | 174–193 | 196 |
| A |
CGCTGATCAGGTTGTTTAAAG | 349–369 | |
| CXCR4, transcript
variant 1 | NM_001008540 | S |
CTACATTAATTCTCTTGTGCC | 175–195 | 241 |
| A |
ATTTTCTTCACGGAAACAGG | 396–415 | |
| CXCR4, transcript
variant 2 | NM_003467 | S |
CTGAGTGCTCCAGTAGCC | 61–68 | 281 |
| A |
TGCAGCCTGTACTTGTCC | 314–331 | |
| CXCR7 | NM_020311.2 | S |
CAAAGCTGCCATCTAGAGG | 16–34 | 252 |
| A |
CTGATGTCCGAGAAGTTCC | 249–267 | |
| MRLP19 (reference
gene) | NM_014763 | S |
TCCTCGGGTCCAGGAGATT | 529–547 | 58 |
| A |
CAAGCTATCATCCAGCCGTTT | 566–586 | |
qPCR was performed in a Roche Light Cycler 2.0
(Roche, Mannheim, Germany) with software version 4.05. The SYBR
Green detection system was used with the above listed primers. qPCR
reactions were conducted in 20 μl mixtures, containing 4 μl
template cDNA, 0.5 mM of each gene-specific primer and 3.5 mM of
Mg2+ ions. LightCycler FastStart DNA Master SYBR Green I
mix (Roche) was used. The qPCR program included a 10 min
denaturation step to activate the Taq DNA polymerase, followed by a
three-step amplification program as follows: Denaturation at 95.0°C
for 10 sec, annealing at 58.0°C for 5 sec and extension at 72.0°C
for 5 sec. Specificity of the reaction products was routinely
checked by determination of melting points (0.1°C/sec transition
rate) and random sample separation in a 2.0% ethidium
bromide/agarose gel. All qPCR reactions were performed in
triplicate and the mitochondrial ribosomal protein L19 gene was
used as a reference to normalize the data. Templates not submitted
to the reverse transcription reaction served as negative
controls.
PCR efficiency was assessed by a serial dilution
method. Briefly, the products of the qPCR reactions were separated
in a 2.0% agarose gel and specific bands were extracted using a DNA
gel extraction kit (Millipore, Billerica, MA, USA). The quantity of
extracted DNA was estimated spectrophotometrically. Extracted DNA
was diluted (10-fold serial dilutions) in order to generate a
standard curve for efficiency calculation. The LightCycler software
employed (version 4.05) allowed the amplification efficiency to be
evaluated from the plots (19–21).
Plasma concentrations of SDF-1α and CA
125
Blood samples were obtained from the patients from
the antecubital vein between 7 and 8 am following an overnight
fast. They were centrifuged at 4°C and subsequently frozen and
stored at −70°C.
SDF-1 was quantified using a commercially available
EIA kit (Human CXCL12/SDF-1 Immunoassay; R&D Systems Europe
Ltd., Abingdon, UK; catalog no. DSA00) according to the
manufacturer’s instructions. This kit is specific for the protein
encoded by SDF-1 transcript variant 1, SDF-1α. Absorbance
was read at 450 nm (BioTek Synergy 2; BioTek Instruments, Inc.,
Winooski, VT, USA). All steps were performed at room
temperature.
CA 125 was quantified by the automated microparticle
enzyme immunosorbent assay method using the AXSYM system of Abbott
Laboratories (Chicago, IL, USA).
Statistics
The data are expressed as the mean ± SE. Statistical
comparison of obtained data was performed by means of the
Mann-Whitney U or Wilcoxon tests. All calculations were performed
by Statistica 7.0. software (Statsoft, Tulsa, OK, USA).
Results
Expression of SDF-1 and CXCR4 transcript
variants and CXCR7 in epithelial ovarian cancer and control
ovaries
In the epithelial ovarian cancer and control ovary
group, agarose gel electrophoresis of classic PCR products revealed
expression of SDF-1, transcript variants 1–4 (Fig. 1). In all cases, the agarose gel
showed bands of the expected size. By contrast, in the control and
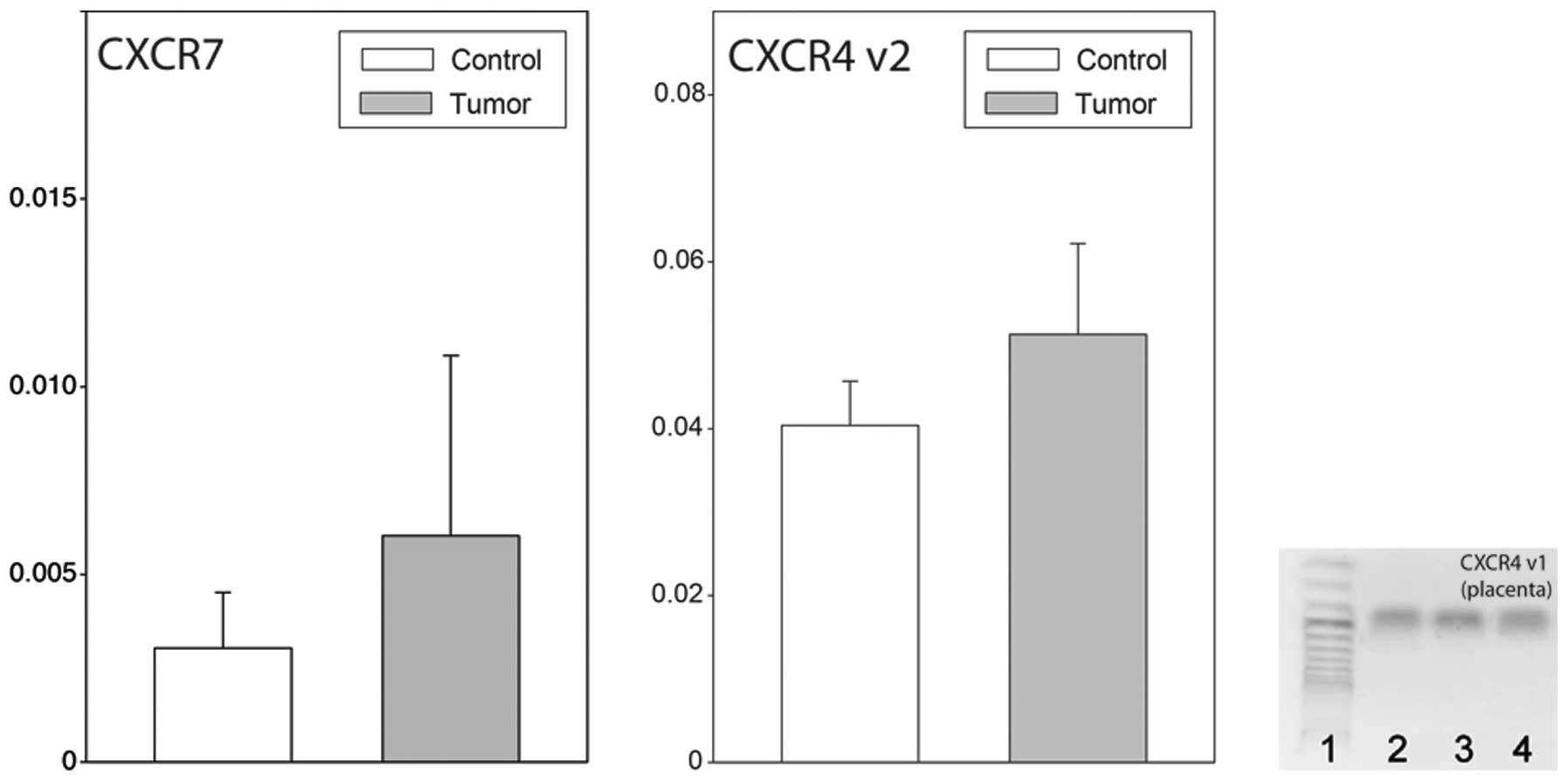
neoplastic ovaries, no reaction product for CXCR4 transcript
variant 1 was found, while that of variant 2 was highly expressed.
It should be emphasized that in human placenta tissue, which was
used as a positive control, the reaction products for CXCR4
transcript variant 1 were abundant (Fig. 2). Furthermore, in normal and
neoplastic ovaries, the expression of CXCR7 was identified.
Following this, we studied SDF-1 transcript variants 1–4
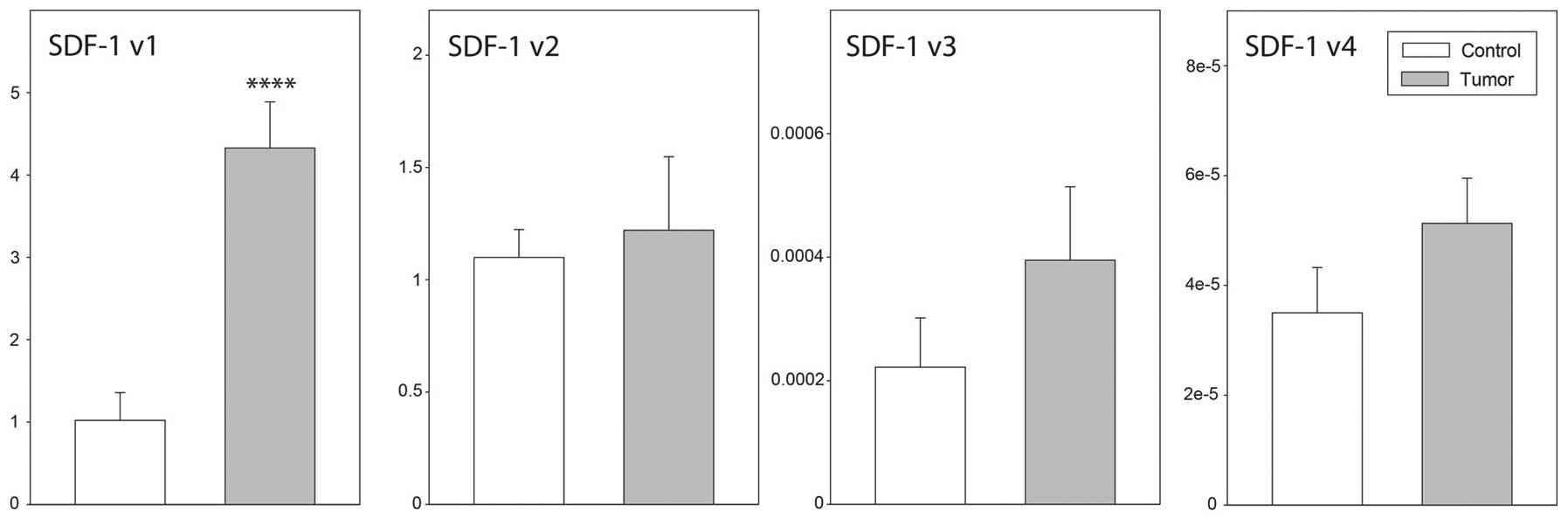
mRNA expression levels by qPCR. As demonstrated in Fig. 3, high SDF-1 transcript
variant 1 expression levels were identified in ovarian cancer and
control ovaries. By contrast, in the two groups studied, the
expression levels of SDF-1 transcript variants 3 and 4 were
extremely low. Furthermore, the expression levels of SDF-1
transcript variant 1 were notably higher in epithelial ovarian
cancer than in control ovaries (P<0.001), while data for the
remaining transcripts were similar in both groups. With regard to
CXCR4 transcript variant 2 and CXCR7, their
expression levels in normal and neoplastic ovaries were similar
(Fig. 3).
 | Figure 1Ethidium bromide-stained 2% agarose
gel demonstrating cDNA amplified with specific primer from RNA of
(A) three ovarian cancers and (B) three control ovaries. Lane 1,
size marker (O’Range Ruler 50-bp DNA Ladder; MBI Fermentas,
Lithuania); lanes 2–4, SDF-1, transcript variant 1; lanes
5–7, SDF-1, transcript variant 2; lanes 8–10, SDF-1,
transcript variant 3; lanes 11–13, SDF-1, transcript variant
4; lanes 14–16, CXCR4, transcript variant 1; lanes 17–19,
CXCR4, transcript variant 2; lanes 20–22, CXCR7. In
the control and neoplastic ovaries, no reaction product for
CXCR4, transcript variant 1 was found. For the remaining
genes, reaction products with expected size were observed. SDF-1,
stromal cell-derived factor-1. |
Plasma concentrations of SDF-1α and CA
125 in epithelial ovarian cancer and control patients
The levels of SDF-1, a circulating chemokine, were
quantified in patients using a kit that is specific for SDF-1α, a
protein encoded by SDF-1 transcript variant 1. As
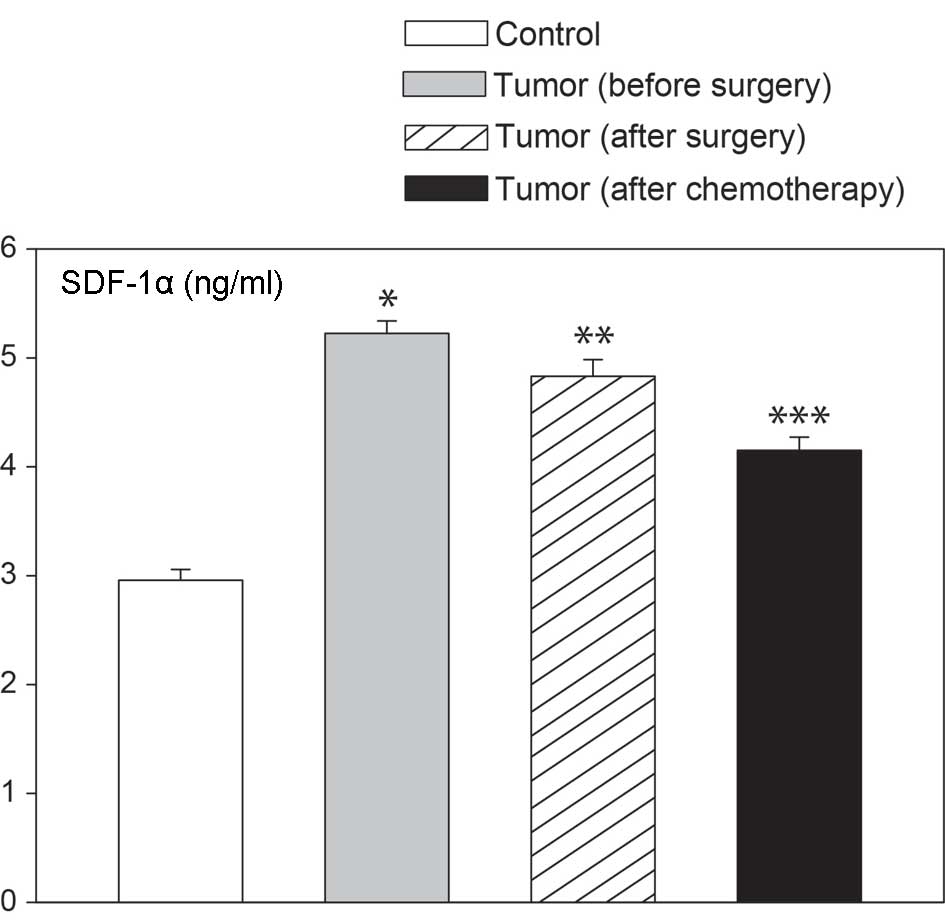
demonstrated in Fig. 4, plasma
SDF-1α levels were notably higher in females with epithelial
ovarian cancer than in control ovaries. Elevated levels of blood
SDF-1α were found prior to surgery, 6 days after surgery and
following the completion of the first chemotherapy course. These
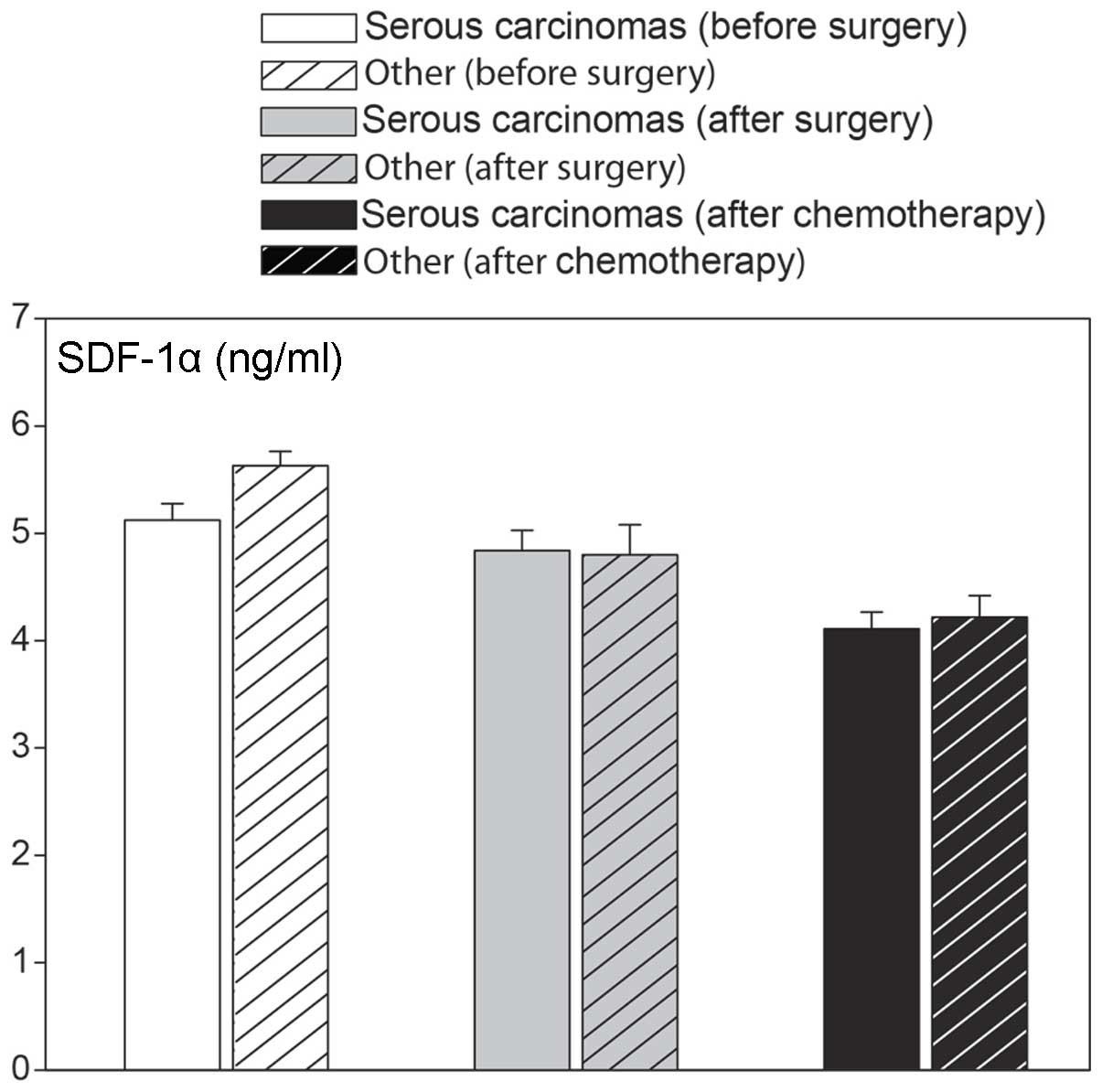
changes were independent of the type of epithelial ovarian cancer,
as similar values were observed in serous cancers, as well as other
types (Fig. 5). We also analyzed
plasma SDF-1α levels in relation to FIGO classifications for
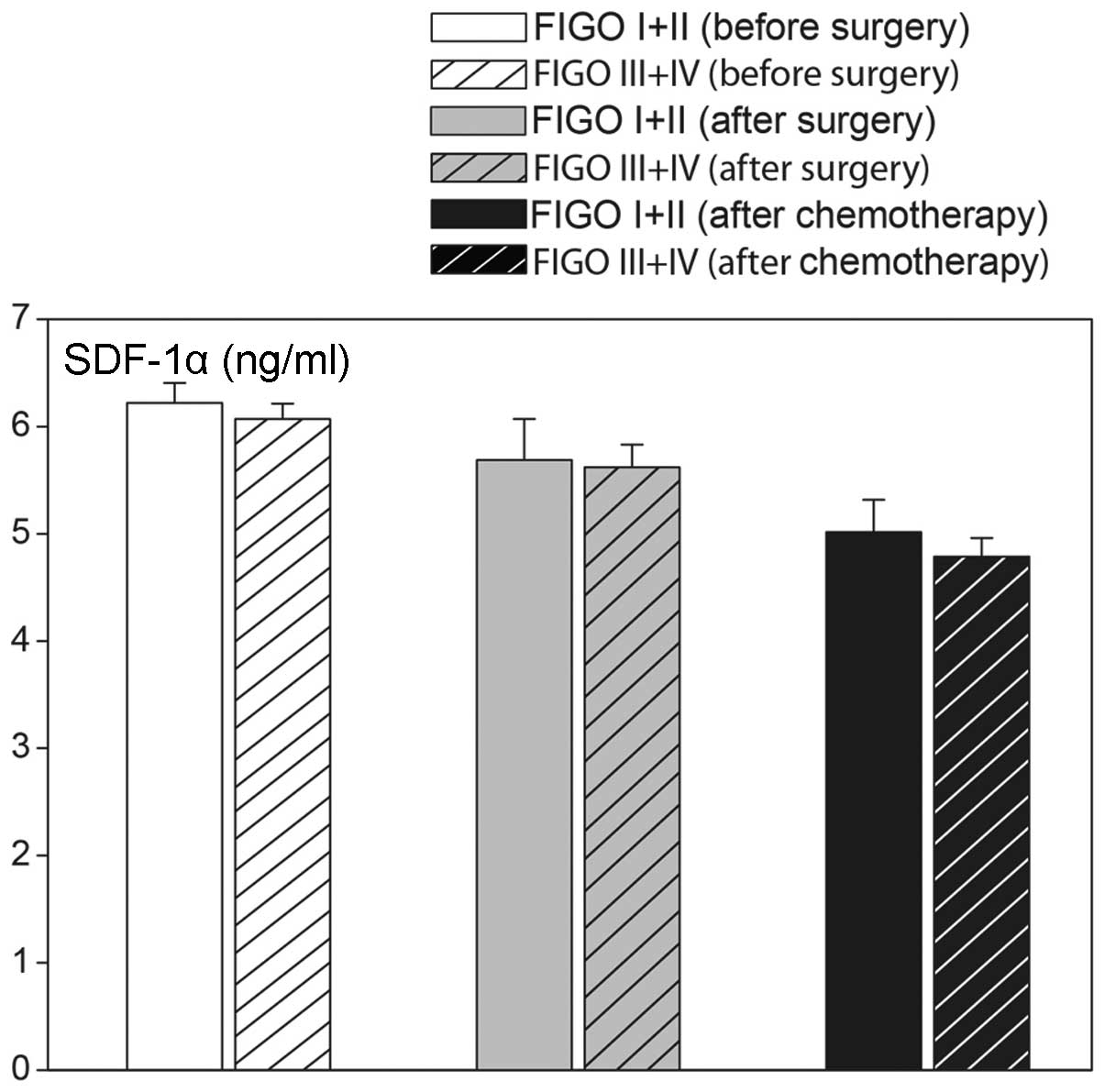
ovarian cancer staging. As shown in Fig. 6, elevated levels of SDF-1α were
found in less (I and II) and more (III and IV) advanced ovarian
cancers (Fig. 7). Plasma CA 125
concentrations were lower in less advanced compared with more
advanced cases of cancer. Furthermore, elevated plasma SDF-1α
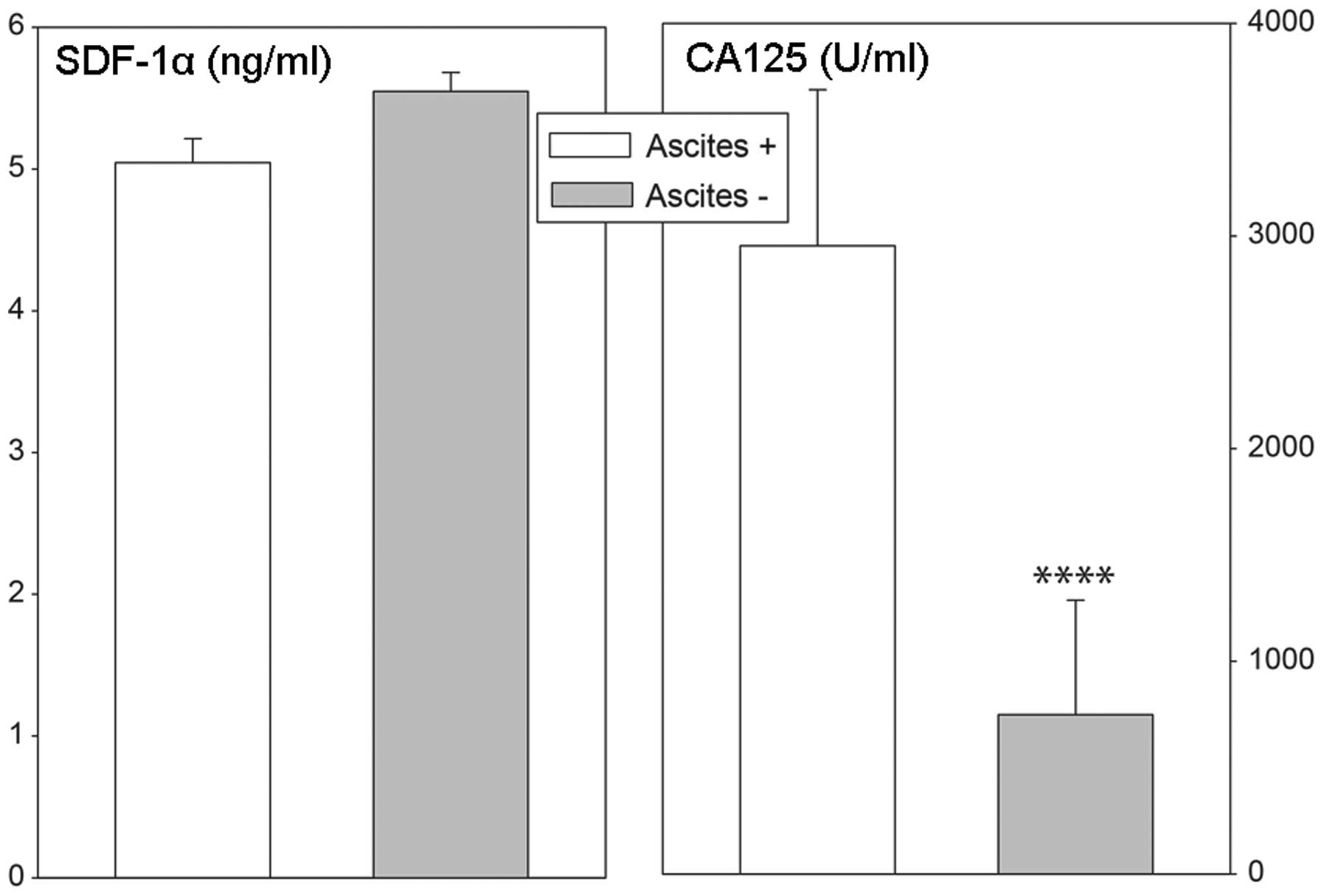
levels in ovarian cancer patients were independent of the presence
of ascites, while in cases with ascites, plasma CA 125
concentrations were notably elevated (Fig. 8).
Discussion
Chemokines are important in the pathogenesis of
several tumors. Early studies demonstrated an evident correlation
between the expression of chemokine receptors and the prognosis or
metastases in various human malignant tumors. This suggests that
chemokines and their corresponding receptors are important in
controlling key biological properties of the microenvironment in
which neoplastic tumors develop (4,15).
SDF-1 is a small cytokine belonging to the CXC
chemokine family. Among its multiple functions, SDF-1 is strongly
chemotactic for lymphocytes, is able to directly activate
leukocytes and can recruit macrophages to malignant tumors. During
embryonic development, SDF-1 is important in the migrational
behavior of hematopoietic cells, angiogenesis and vasculogenesis.
It has also been suggested that this chemokine is involved in
cancer metastasis as tumor cells frequently express specific
receptors for this chemotactic compound (4,15).
The synthesis of SDF-1 is mainly controlled at the
stage of splicing, where alternative splicing events produce a
number of different SDF-1 isoforms, all of which are secreted from
the cell as functional proteins (22–24).
Furthermore, SDF-1 transcription variants appear to be
differentially expressed in various tissues, where they may play
different roles.
SDF-1 is synthesized by bone marrow stromal cells
and the gene has also been identified to be expressed in numerous
other cell types (4,15,24,25).
Its expression at the mRNA and protein levels has been demonstrated
in tumors, including epithelial ovarian carcinoma. The original
data documented SDF-1 expression in >90% of ovarian cancers, in
ovarian cancer cell lines and at the protein level, as well as in
ascites of ovarian cancer patients. Notably, the expression of
SDF-1 was not originally identified in normal ovaries or it was
present at extremely low levels (7,8,16,18,26).
However, it should be noted, that the expression of SDF-1 in tumors
was estimated from immunohistochemsitry, with no attempt made to
identify its isoforms or protein levels.
In this study, using qPCR, we have identified the
expression of four SDF-1 transcriptional variants in
epithelial ovarian cancer. There are numerous human SDF-1 isoforms
(α-ϕ) (24), the present study was
based on current data included in GenBank (SDF-1: Gene ID,
6387; CXCR4: Gene ID, 7852; CXCR7: Gene ID, 57007).
The results of our studies have demonstrated that in the control
and neoplastically altered ovaries, the highest expression level
was exhibited by SDF-1 variant 1, a lower expression by
variant 2 and trace expression by variants 3 and 4. Such a pattern
of transcript expression corroborates the earlier observation that
SDF-1 variant 1 is the most widespread splicing variant
(27). It is known that
SDF-1 is constitutively expressed in tissues (10,28).
While the expression levels of other variants were not altered, a
marked elevation in the expression of SDF-1 transcript
variants in epithelial ovarian cancer was observed. The results may
suggest that in epithelial ovarian cancer, the expression of
SDF-1 and of genes controlling alternative splicing become
elevated, leading to increased formation of SDF-1 variant 1.
As revealed in earlier immunocytochemical studies on epithelial
ovarian cancer, the expression of SDF-1 mainly takes place in
epithelial cells (16,18,26).
While our studies have not permitted us to identify specifically
which cells demonstrate enhanced expression of SDF-1 variant
1 in epithelial ovarian cancer types, it cannot be excluded that
the expression may be linked to cells recruited for tumor
formation. The mechanism(s) that can predict the upregulation of
SDF-1 variant 1 expression in epithelial ovarian cancer is
not yet recognized and remains to be elucidated. However, it should
be mentioned that earlier studies have implicated tissue hypoxia
(which may be present in epithelial ovarian tumors) as a potent
factor in upregulating the expression of SDF-1 and
CXCR4 in various cells and organs (4,28).
CXCR4 is a cognate receptor of SDF-1 (29–31).
SDF-1-induced CXCR4 activation results in the influx of calcium
ions, as well as the activation of other intracellular signaling
pathways, for example MAPK, p42/44-ELK-1, PI3K-AKT-NF-κB and JAK2
and JAC3 (4,15,32).
The SDF-1/CXCR4-CXCR7 system plays a pivotal role in organogenesis,
regeneration and tumorigenesis. Experimental data indicate that
CXCR4-expressing cells follow an SDF-1 gradient (15,32–34).
It is frequently emphasized that CXCR4 is the most widely expressed
chemokine receptor in malignancy. Numerous, if not all neoplastic
cells express this receptor and, therefore, the SDF-1/CXCR4-CXCR7
system may be of particular importance in tumor metastasis
(7,8,32).
Prevailing data on CXCR4 expression in several types of cancer were
obtained by immunohistochemistry; however, the commonly used
anti-CXCR4 antibody is only able to recognize a subpopulation of
CXCR4 molecules (15–18).
Limited data are available with regard to CXCR4 mRNA
expression in epithelial ovarian cancer. Elevated CXCR4 mRNA
expression has recently been reported in ovarian cancer; however,
this particular study lacked important information regarding the
oligonucleotide sequences applied and failed to include
measurements of the resultant CXCR4 protein expression (35). Furthermore, the authors did not
estimate the known CXCR4 transcription variants. In this
regard, our study has demonstrated that in the control and
neoplastic ovaries, CXCR4 transcript variant 2 was highly
expressed while its transcript variant 1 was absent. Furthermore,
the expression levels of CXCR4 transcript variant 2 in
normal ovaries and epithelial ovarian cancer were similar. The
latter finding was unexpected and the explanation for this result
remains to be elucidated. In scope of numerous immunohistochemistry
derived data, it seems legitimate to suggest that in normal ovaries
and epithelial ovarian cancer, CXCR4 transcript variant 2
may be translated into a functional protein.
For a number of years, CXCR4 was considered to be
the only receptor binding to SDF-1 and it was not until 2005 that
CXCR7 was identified as another SDF-1-binding receptor (3,5,36).
Experimental data suggest that due to its expression profile within
tumor cells, CXCR7 may also be important in tumor growth and
metastasis, but investigations of the role of this receptor system
in ovarian cancer have been lacking until now. As demonstrated in
the present study, the expression levels of CXCR7 in normal
and neoplastic ovaries are similar. Thus, we demonstrated that in
epithelial ovarian cancer, CXCR4 and CXCR7 mRNA levels remained
unchanged.
Experimental data suggest that all proteins encoded
by particular SDF-1 transcription variants are secretory
proteins (24). Of these
transcripts, SDF-1 transcript variant 1 encodes circulating
chemokine SDF-1α, which is the main SDF isoform in the blood.
Plasma levels of this cytokine were found to be elevated in breast
cancer (37), pelvic inflammatory
disease (38) and in various
systemic diseases (39–41). In ovarian cancer patients, using
human cytokine microarray technology, plasma SDF-1 levels were
found to be elevated 6.6-fold compared with the control group
(42). We have extended these
earlier findings by demonstrating in the present study notably
higher plasma SDF-1α levels in females with epithelial ovarian
cancer. Elevated plasma SDF-1α levels were independent of the type
of epithelial ovarian cancer or the stage of the cancer. In
addition, these levels remained unaffected by surgery or by
subsequent chemotherapy, and were similar in patients with and
without ascites. These results indicate no direct correlation
between epithelial ovarian cancer and plasma SDF-1α levels. With
regard to these observations, it seems legitimate to suggest that
elevated plasma SDF-1α levels in epithelial ovarian cancer patients
are not correlated to the presence of tumor and/or metastases;
however, they rather reflect a general response to the disease.
These findings are in contrast to data obtained in breast cancer
patients, where plasma SDF-1α levels had a significant correlation
with tumor grade and epithelial subtype (37). Thus, in our opinion, plasma SDF-1α
levels cannot be used as a marker of epithelial ovarian cancer
advancement or progression. Furthermore, we would like to highlight
the fact that, in epithelial ovarian cancer, plasma SDF-1α levels
demonstrate no correlation with blood CA 125 levels.
The available literature contains numerous studies
on the prognostic value of the SDF-1/CXCR4-CXCR7 system in numerous
tumor types, including epithelial ovarian cancer. In this regard,
the majority of data derives from immunohistochemical studies
(including semiquantitative and microarray techniques) and they are
frequently correlated with comprehensive databases of
clinicopathological variables (18,43).
In general, these studies indicate that a high level of SDF-1
expression in ovarian cancer is an independent prognostic factor
for tumor progression and a predictor of poor survival (16,43,44).
Acknowledgements
This study was funded by the grant N N407 347533
from the Poznań University of Medical Sciences (Poznań,
Poland).
References
|
1
|
Tashiro K, Tada H, Heilker R, et al:
Signal sequence trap: a cloning strategy for secreted proteins and
type I membrane proteins. Science. 261:600–603. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murphy PM, Baggiolini M, Charo IF, et al:
International union of pharmacology. XXII Nomenclature for
chemokine receptors. Pharmacol Rev. 52:145–176. 2000.PubMed/NCBI
|
|
3
|
Balabanian K, Lagane B, Infantino S, et
al: The chemokine SDF-1/CXCL12 binds to and signals through the
orphan receptor RDC1 in T lymphocytes. J Biol Chem.
280:35760–35766. 2005. View Article : Google Scholar
|
|
4
|
Ratajczak MZ, Zuba-Surma E, Kucia M, et
al: The pleiotropic effects of the SDF-1-CXCR4 axis in
organogenesis, regeneration and tumorigenesis. Leukemia.
20:1915–1924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burns JM, Summers BC, Wang Y, et al: A
novel chemokine receptor for SDF-1 and I-TAC involved in cell
survival, cell adhesion, and tumor development. J Exp Med.
203:2201–2213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kryczek I, Wei S, Keller E, Liu R and Zou
W: Stroma-derived factor (SDF-1/CXCL12) and human tumor
pathogenesis. Am J Physiol Cell Physiol. 292:C987–C995. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scotton CJ, Wilson JL, Milliken D, Stamp G
and Balkwill FR: Epithelial cancer cell migration: a role for
chemokine receptors? Cancer Res. 61:4961–4965. 2001.PubMed/NCBI
|
|
8
|
Scotton CJ, Wilson JL, Scott K, et al:
Multiple actions of the chemokine CXCL12 on epithelial tumor cells
in human ovarian cancer. Cancer Res. 62:5930–5938. 2002.PubMed/NCBI
|
|
9
|
Feuerer M, Beckhove P, Bai L, et al:
Therapy of human tumors in NOD/SCID mice with patient-derived
reactivated memory T cells from bone marrow. Nat Med. 7:452–458.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zou W, Machelon V, Coulomb-L’Hermin A, et
al: Stromal-derived factor-1 in human tumors recruits and alters
the function of plasmacytoid precursor dendritic cells. Nat Med.
7:1339–1346. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kryczek I, Gryboś M, Dlubek D, Klimczak A,
Rabczyński J and Lange A: Accumulation of CD45RO(+) cells in
peritoneal carcinomatous fluid favours survival of ovarian
carcinoma patients. Cancer Immunol Immunother. 51:513–519.
2002.
|
|
12
|
Jaszczyñska-Nowinka K and Markowska A: New
cytokine: stromal derived factor-1. Eur J Gynaecol Oncol.
30:124–127. 2009.PubMed/NCBI
|
|
13
|
Broxmeyer HE, Orschell CM, Clapp DW, et
al: Rapid mobilization of murine and human hematopoietic stem and
progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med.
201:1307–1318. 2005. View Article : Google Scholar
|
|
14
|
Zhang T, Somasundaram R, Berencsi K, et
al: CXC chemokine ligand 12 (stromal cell-derived factor 1 alpha)
and CXCR4-dependent migration of CTLs toward melanoma cells in
organotypic culture. J Immunol. 174:5856–5863. 2005. View Article : Google Scholar
|
|
15
|
Balkwill F: The significance of cancer
cell expression of the chemokine receptor CXCR4. Semin Cancer Biol.
14:171–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang YP, Wu XH, Shi B, Wu WX and Yin GR:
Expression of chemokine CXCL12 and its receptor CXCR4 in human
epithelial ovarian cancer: an independent prognostic factor for
tumor progression. Gynecol Oncol. 103:226–233. 2006. View Article : Google Scholar
|
|
17
|
Furuya M, Suyama T, Usui H, et al:
Up-regulation of CXC chemokines and their receptors: implications
for proinflammatory microenvironments of ovarian carcinomas and
endometriosis. Hum Pathol. 38:1676–1687. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pils D, Pinter A, Reibenwein J, et al: In
ovarian cancer the prognostic influence of HER2/neu is not
dependent on the CXCR4/SDF-1 signalling pathway. Br J Cancer.
96:485–491. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rucinski M, Andreis PG, Ziolkowska A,
Nussdorfer GG and Malendowicz LK: Differential expression and
function of beacon in the rat adrenal cortex and medulla. Int J Mol
Med. 16:35–40. 2005.PubMed/NCBI
|
|
20
|
Rucinski M, Zok A, Guidolin D, De Caro R
and Malendowicz LK: Expression of precerebellins in cultured rat
calvaria osteoblast-like cells. Int J Mol Med. 22:553–558.
2008.
|
|
21
|
Rucinski M, Ziolkowska A, Tyczewska M and
Malendowicz LK: Expression of prepro-ghrelin and related receptor
genes in the rat adrenal gland and evidences that ghrelin exerts a
potent stimulating effect on corticosterone secretion by cultured
rat adrenocortical cells. Peptides. 30:1448–1455. 2009. View Article : Google Scholar
|
|
22
|
Gleichmann M, Gillen C, Czardybon M, et
al: Cloning and characterization of SDF-1gamma, a novel SDF-1
chemokine transcript with developmentally regulated expression in
the nervous system. Eur J Neurosci. 12:1857–1866. 2000. View Article : Google Scholar
|
|
23
|
De La Luz Sierra M, Yang F, Narazaki M, et
al: Differential processing of stromal-derived factor-1alpha and
stromal-derived factor-1beta explains functional diversity. Blood.
103:2452–2459. 2004.PubMed/NCBI
|
|
24
|
Yu L, Cecil J, Peng SB, Schrementi J, et
al: Identification and expression of novel isoforms of human
stromal cell-derived factor 1. Gene. 374:174–179. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janowski M: Functional diversity of SDF-1
splicing variants. Cell Adh Migr. 3:243–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nowak-Markwitz E, Puła B, Szajnik M,
Dziegiel P, Piotrowska A, Zabel M and Spaczyński M: Expression of
survivin, SDF-1 and CXCR4 on tumor cells in ovarian cancer. Ginekol
Pol. 81:674–677. 2010.(In Polish).
|
|
27
|
Davis DA, Singer KE, De La Luz Sierra M,
et al: Identification of carboxypeptidase N as an enzyme
responsible for C-terminal cleavage of stromal cell-derived
factor-1alpha in the circulation. Blood. 105:4561–4568. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kryczek I, Lange A, Mottram P, et al:
CXCL12 and vascular endothelial growth factor synergistically
induce neoangiogenesis in human ovarian cancers. Cancer Res.
65:465–472. 2005.
|
|
29
|
Oberlin E, Amara A, Bachelerie F, et al:
The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents
infection by T-cell-line-adapted HIV-1. Nature. 382:833–835. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bleul CC, Farzan M, Choe H, Parolin C,
Clark-Lewis I, Sodroski J and Springer TA: The lymphocyte
chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1
entry. Nature. 382:829–833. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
D’Apuzzo M, Rolink A, Loetscher M, et al:
The chemokine SDF-1, stromal cell-derived factor 1, attracts early
stage B cell precursors via the chemokine receptor CXCR4. Eur J
Immunol. 27:1788–1793. 1997.PubMed/NCBI
|
|
32
|
Fulton AM: The chemokine receptors CXCR4
and CXCR3 in cancer. Curr Oncol Rep. 11:125–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kucia M, Reca R, Miekus K, et al:
Trafficking of normal stem cells and metastasis of cancer stem
cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4
axis. Stem Cells. 23:879–894. 2005. View Article : Google Scholar
|
|
34
|
Sun X, Cheng G, Hao M, et al:
CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer
Metastasis Rev. 29:709–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, Cai J, Han F, et al: Silencing of
CXCR4 blocks progression of ovarian cancer and depresses canonical
Wnt signaling pathway. Int J Gynecol Cancer. 21:981–987. 2011.
View Article : Google Scholar
|
|
36
|
Maksym RB, Tarnowski M, Grymula K, et al:
The role of stromal-derived factor-1 - CXCR7 axis in development
and cancer. Eur J Pharmacol. 625:31–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Potter SM, Dwyer RM, Curran CE, Hennessy
E, Harrington KA, Griffin DG and Kerin MJ: Systemic chemokine
levels in breast cancer patients and their relationship with
circulating menstrual hormones. Breast Cancer Res Treat.
115:279–287. 2009. View Article : Google Scholar
|
|
38
|
Tsai HT, Tee YT, Hsieh YH, et al: Elevated
plasma stromal cell-derived factor 1 protein and its gene
polymorphism in patients with pelvic inflammatory disease. Reprod
Sci. 16:610–617. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hansen IB, Ellingsen T, Hornung N, Poulsen
JH, Lottenburger T and Stengaard-Pedersen K: Plasma level of
CXC-chemokine CXCL12 is increased in rheumatoid arthritis and is
independent of disease activity and methotrexate treatment. J
Rheumatol. 33:1754–1759. 2006.
|
|
40
|
Kim HK, Kim JE, Chung J, Lee DS, Han KS,
Park S and Cho HI: Plasma level of stromal derived factor-1 (SDF-1)
is increased in disseminated intravascular coagulation patients who
have poor outcomes: in vitro effect of SDF-1 on coagulopathy.
Thromb Res. 120:559–566. 2007. View Article : Google Scholar
|
|
41
|
Robak E, Kulczycka L, Sysa-Jedrzejowska A,
Wierzbowska A and Robak T: Circulating proangiogenic molecules
PIGF, SDF-1 and sVCAM-1 in patients with systemic lupus
erythematosus. Eur Cytokine Netw. 18:181–187. 2007.
|
|
42
|
Huang R, Lin Y, Flowers L, et al:
Molecular profiling of circulating cytokine levels in human ovarian
cancer patients. Cancer Genomics Proteomics. 1:23–32. 2004.
|
|
43
|
Popple A, Durrant LG, Spendlove I, Rolland
P, Scott IV, Deen S and Ramage JM: The chemokine, CXCL12, is an
independent predictor of poor survival in ovarian cancer. Br J
Cancer. 106:1306–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Machelon V, Gaudin F, Camilleri-Broët S,
et al: CXCL12 expression by healthy and malignant ovarian
epithelial cells. BMC Cancer. 11:972011. View Article : Google Scholar : PubMed/NCBI
|