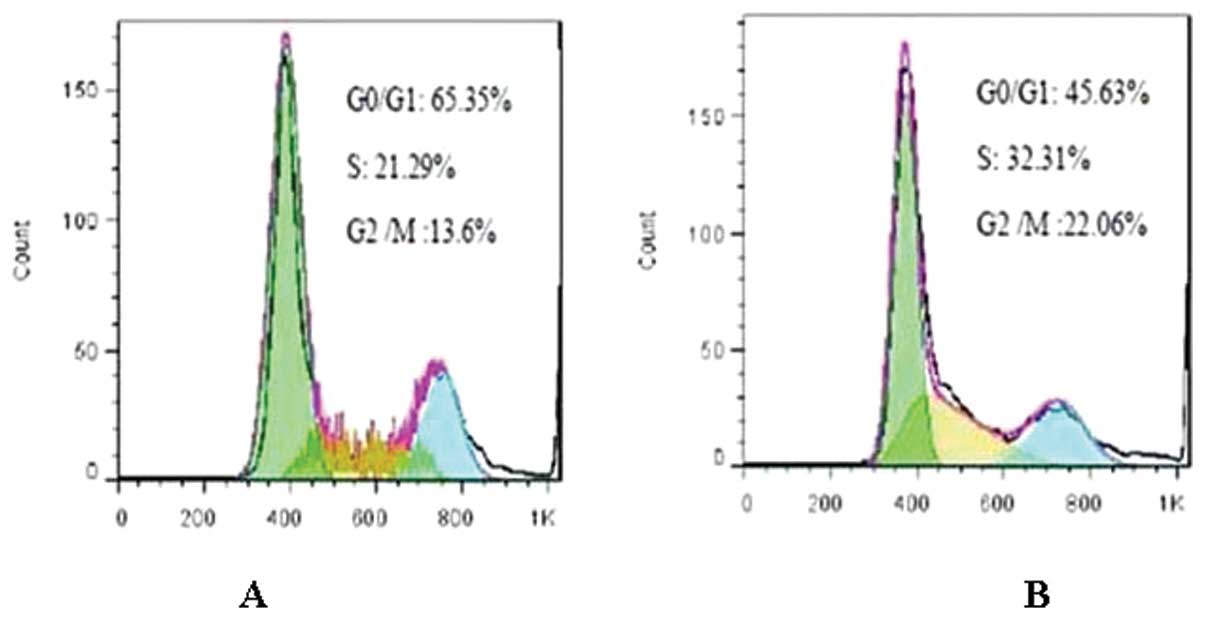
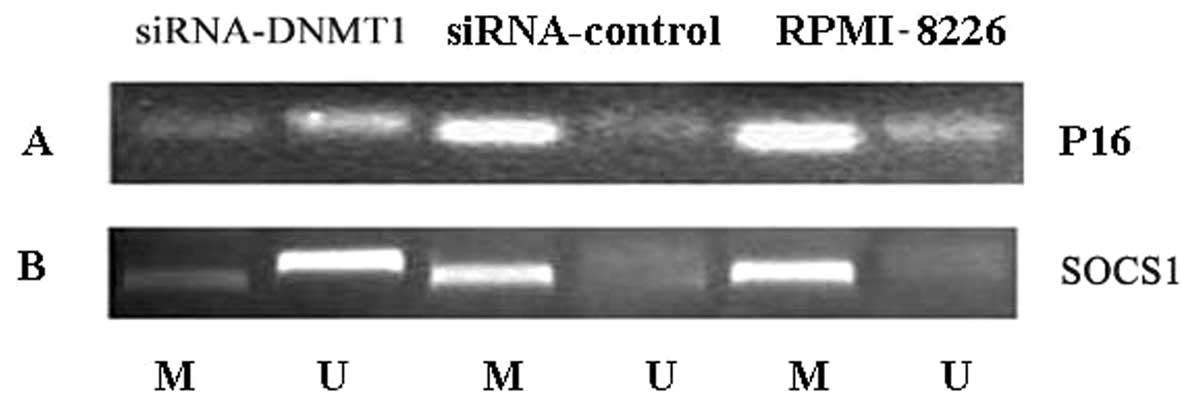
|
1
|
Benjamin M, Reddy S and Brawley OW:
Myeloma and race: a review of the literature. Cancer Metastasis
Rev. 22:87–93. 2003.
|
|
2
|
Siegel R, Ward E, Brawley O, et al: Cancer
statistics, 2011: the impact of eliminating socioeconomic and
racial disparities on premature cancer deaths. CA Cancer J Clin.
61:212–236. 2011.
|
|
3
|
Richardson PG, Barlogie B, Berenson J, et
al: Clinical factors predictive of outcome with bortezomib in
patients with relapsed, refractory multiple myeloma. Blood.
106:2977–2981. 2005.
|
|
4
|
Richardson PG, Weller E, Lonial S, et al:
Lenalidomide, bortezomib, and dexamethasone combination therapy in
patients with newly diagnosed multiple myeloma. Blood. 116:679–686.
2010.
|
|
5
|
Chen T and Li E: Structure and function of
eukaryotic DNA methyltransferases. Curr Top Dev Biol. 60:55–89.
2004.
|
|
6
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005.
|
|
7
|
Belinsky SA, Nikula KJ, Baylin SB, et al:
A microassay for measuring cytosine DNA methyltransferase activity
during tumor progression. Toxicol Lett. 82–83:335–340. 1995.
|
|
8
|
Vertino PM, Yen RW, Gao J and Baylin SB:
De novo methylation of CpG island sequences in human fibroblasts
overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol.
16:4555–4565. 1996.
|
|
9
|
Ng MH, Chung YF, Lo KW, et al: Frequent
hypermethylation of p16 and p15 genes in multiple myeloma. Blood.
89:2500–2506. 1997.
|
|
10
|
Mateos MV, García-Sanz R, Lopez-Perez R,
et al: Methylation is an inactivating mechanism of the p16 gene in
multiple myeloma associated with high plasma cell proliferation and
short survival. Br J Haematol. 118:1034–1040. 2002.
|
|
11
|
Chim CS, Liang R, Fung TK, et al:
Epigenetic dysregulation of the death-associated protein
kinase/p14/HDM2/p53/Apaf-1 apoptosis pathway in multiple myeloma. J
Clin Pathol. 60:664–669. 2007.
|
|
12
|
Ng MH, Lau KM, Wong WS, et al: Alterations
of RAS signalling in Chinese multiple myeloma patients: absent BRAF
and rare RAS mutations, but frequent inactivation of RASSF1A by
transcriptional silencing or expression of a non-functional variant
transcript. Br J Haematol. 123:637–645. 2003.
|
|
13
|
Galm O, Yoshikawa H, Esteller M, et al:
SOCS-1, a negative regulator of cytokine signaling, is frequently
silenced by methylation in multiple myeloma. Blood. 101:2784–2788.
2003.
|
|
14
|
Galm O, Wilop S, Reichelt J, et al: DNA
methylation changes in multiple myeloma. Leukemia. 18:1687–1692.
2004.
|
|
15
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer - a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006.
|
|
16
|
Bestor TH: The DNA methyltransferases of
mammals. Hum Mol Genet. 9:2395–2402. 2000.
|
|
17
|
Szyf M, Pakneshan P and Rabbani SA: DNA
methylation and breast cancer. Biochem Pharmacol. 68:1187–1197.
2004.
|
|
18
|
Xu M, Gao J, Du YQ, et al: Reduction of
pancreatic cancer cell viability and induction of apoptosis
mediated by siRNA targeting DNMT1 through suppression of total DNA
methyltransferase activity. Mol Med Rep. 3:699–704. 2010.
|
|
19
|
Milutinovic S, Knox JD and Szyf M: DNA
methyltransferase inhibition induces the transcription of the tumor
suppressor p21 (WAF1/CIP1/sdi1). J Biol Chem. 275:6353–6359.
2000.
|
|
20
|
Ghoshal K and Bai S: DNA
methyltransferases as targets for cancer therapy. Drugs Today
(Barc). 43:395–422. 2007.
|
|
21
|
Auerkari EI: Methylation of tumor
suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in
carcinogenesis. Oral Oncol. 42:5–13. 2006.
|
|
22
|
Chim CS, Fung TK and Liang R: Disruption
of INK4/CDK/Rb cell cycle pathway by gene hypermethylation in
multiple myeloma and MGUS. Leukemia. 17:2533–2535. 2003.
|
|
23
|
Komazaki T, Nagai H, Emi M, et al:
Hypermethylation-associated inactivation of the SOCS-1 gene, a
JAK/STAT inhibitor, in human pancreatic cancers. Jpn J Clin Oncol.
34:191–194. 2004.
|
|
24
|
Jost E, Gezer D, Wilop S, et al:
Epigenetic dysregulation of secreted Frizzled-related proteins in
multiple myeloma. Cancer Lett. 281:24–31. 2009.
|
|
25
|
Tshuikina M, Jernberg-Wiklund H, Nilsson
K, et al: Epigenetic silencing of the interferon regulatory factor
ICSBP/IRF8 in human multiple myeloma. Exp Hematol. 36:24–31.
2008.
|
|
26
|
Lin RK, Hsu HS, Chang JW, et al:
Alteration of DNA methyltransferases contributes to 5′CpG
methylation and poor prognosis in lung cancer. Lung Cancer.
55:205–213. 2007.
|
|
27
|
Ahluwalia A, Hurteau JA, Bigsby RM, et al:
DNA methylation in ovarian cancer. II. Expression of DNA
methyltransferases in ovarian cancer cell lines and normal ovarian
epithelial cells. Gynecol Oncol. 82:299–304. 2001.
|
|
28
|
Robert MF, Morin S, Beaulieu N, et al:
DNMT1 is required to maintain CpG methylation and aberrant gene
silencing in human cancer cells. Nat Genet. 33:61–65. 2003.
|
|
29
|
Ting AH, Schuebel KE, Herman JG, et al:
Short double-stranded RNA induces transcriptional gene silencing in
human cancer cells in the absence of DNA methylation. Nat Genet.
37:906–910. 2005.
|
|
30
|
Giuliani N, Colla S, Sala R, et al: Human
myeloma cells stimulate the receptor activator of nuclear
factor-kappa B ligand (RANKL) in T lymphocytes: a potential role in
multiple myeloma bone disease. Blood. 100:4615–4621. 2002.
|
|
31
|
Xiao W, Hodge DR, Wang L, et al: NF-kappaB
activates IL-6 expression through cooperation with c-Jun and
IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory
site in autocrine human multiple myeloma cells. Cancer Biol Ther.
3:1007–1017. 2004.
|
|
32
|
Conticello C, Giuffrida R, Adamo L, et al:
NF-κB localization in multiple myeloma plasma cells and mesenchymal
cells. Leuk Res. 35:52–60. 2004.
|
|
33
|
Kannaiyan R, Hay HS, Rajendran P, et al:
Celastrol inhibits proliferation and induces chemosensitization
through down-regulation of NF-κB and STAT3 regulated gene products
in multiple myeloma cells. Br J Pharmacol. 164:1506–1521. 2011.
|
|
34
|
Baliga BC and Kumar S: Role of Bcl-2
family of proteins in malignancy. Hematol Oncol. 20:63–74.
2002.
|
|
35
|
Chen Q, Ray S, Hussein MA, et al: Role of
Apo2L/TRAIL and Bcl-2-family proteins in apoptosis of multiple
myeloma. Leuk Lymphoma. 44:1209–1214. 2003.
|