Introduction
MicroRNAs (miRNAs), which are small, non-coding RNAs
that are 18–25 nucleotides in length, are involved in gene
regulation. miRNAs bind to the 3′untranslated region (3′UTR) of
target mRNAs to inhibit translation or induce the degradation of
mRNA (1–3). A previous study revealed that ~50% of
human miRNAs are located at fragile sites of the genome, which are
associated with cancer (4). This
indicates that miRNAs may be crucial for cancer progression
(1,5). Full-scale analysis of miRNomes has
indicated that only 0.9% of miRNAs are expressed abundantly in the
normal human liver; however, these miRNAs account for 88.2% of all
miRNAs in the liver, and four of the first nine miRNAs belong to
the let-7 family (6). Furthermore,
let-7 family members have been found to be downregulated in a
number of human cancers, including lung, colon, ovarian, uterine
leiomyoma and breast cancer, as well as hepatocellular carcinoma
(HCC) (7–13), indicating that let-7 miRNAs may
present potential tumor suppressors. It has been reported that
let-7c induces apoptosis and inhibits HCC cell proliferation in
vitro (11). In humans, 12
genomic loci have been identified, which encode the let-7 family
members, including let-7a-1, -2, -3, let-7b, let-7c, let-7d,
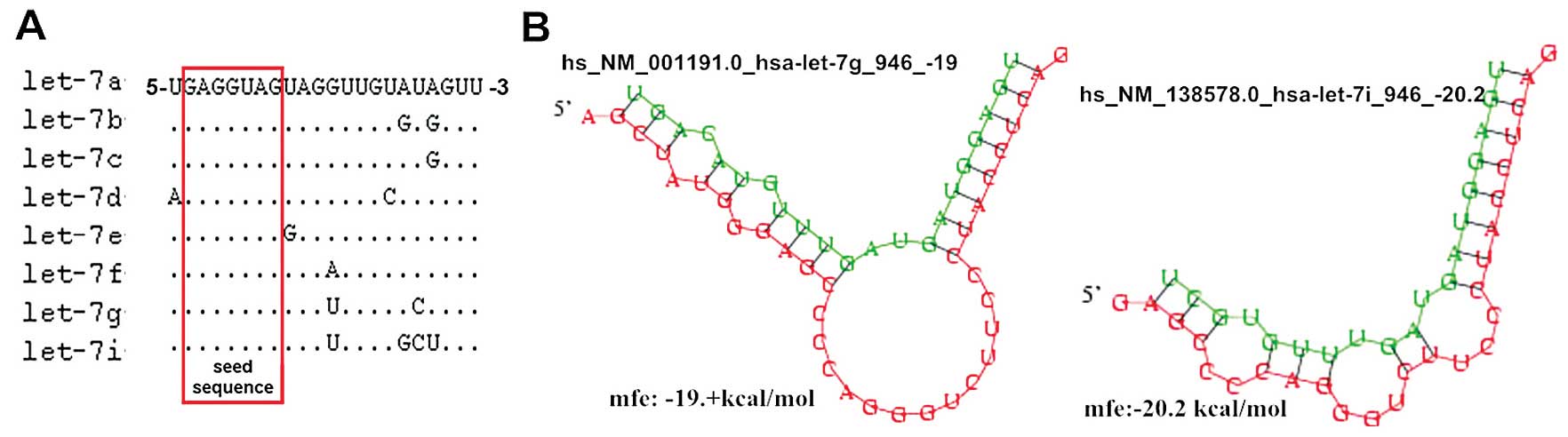
let-7e, let-7f-1, -2, let-7g, let-7i and miR-98 (3). These members share the same nucleotide
sequence between the second and eight consecutive nucleotides at
the 5′end, which are termed ‘seed sequences’, that determine their
target genes (Fig. 1A; miRBase
v.18.0; http://www.mirbase.org/search.shtml) (14); therefore, it has been hypothesized
that each member exhibits the same role. However, the difference in
nucleotides at the 3′end indicates that let-7 families are not
functionally equivalent with regard to combination strength and
efficiency of interactions with target genes (15,16).
It has been reported that members of the let-7 family, mir-48,
mir-84 and mir-241, which have the same seed sequences but
different nucleotide sequences at the 3′end, exhibit specific and
redundant roles in the regulation of developmental timing in
Caenorhabditis elegans (16). An additional study has also
indicated that the degree of complementarity between the 3′end of
the miRNA and target gene is the structural basis of the same
family miRNAs exerting different functions (15).
Bioinformatics analysis (DIANA Lab and PICTAR)
(11) indicates that the
anti-apoptotic protein B-cell lymphoma-extra large (Bcl-xL) is a
target gene of let-7g and let-7i. The aim of the present study was
to investigate whether the let-7 family members exhibit specific
and/or overlapping roles in human HCC as observed in
Caenorhabditis elegans. Thus, the antitumor effect of
let-7g/i on HCC was investigated, and whether let-7g and let-7i
exhibit a concurrent effect on HCC was determined. Furthermore, the
effect of let-7g/i on the Bcl-xL protein in HCC cells was
analyzed.
Materials and methods
Cell culture
The human L-02 liver cell line and hepatoma cell
lines, SMMC-7721 and Bel-7402, were purchased from Shanghai
Institutes for Biological Sciences of Chinese Academy of Sciences
(Shanghai, China). The L-02 and SMMC-7721 cell lines were cultured
in endotoxin-free Dulbecco’s modified Eagle’s medium with 10% fetal
bovine serum (Gibco Life Technologies, Carlsbad, CA, USA). The
BEL-7402 cell line was cultured in RPMI 1640 medium with 10%
(vol/vol) fetal bovine serum (Gibco Life Technologies). The cell
lines were incubated at 37°C in an atmosphere of 5%
CO2.
Transfection
The miR-let-7g/i (let-7g/i) agomir (an engineered
miRNA mimic) and a negative control (similar to agomir-let-7g/i but
with a scramble seeding sequence) were obtained from Guangzhou
RiboBio Co., Ltd., (Guangzhou, China). Cells were plated at 45–50%
confluence. The let-7g/i agomir and/or let-7 agomir negative
control were transfected into the human hepatoma BEL-7402 cell line
using transfection reagent Lipofectamine™ 2000 in Opti-MEM (Gibco
Life Technologies), according to the manufacturer’s instructions.
One negative control group, which was transfected with 100 nm
negative control agomir, and three experimental groups, which were
transfected with 100 nm let-7g agomir, 100 nm let-7i agomir or
co-transfected with 50 nm let-7g and 50 nm let-7i agomir (let-7g +
let-7i) were used. The expression levels of let-7g and let-7i were
quantified using the SYBR Premix Ex TaqTM II (Perfect Real Time)
kit (Takara Bio, Inc., Otsu, Japan), 24 h after transfection.
Briefly, 20 μl PCR reaction mixture was pre-heated at 95°C for 30
sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 34
sec.
5-ethynyl-2′-deoxyuridine (EdU) retention
assay
An EdU assay was performed using the Cell Light EdU
DNA imaging kit (Guangzhou RiboBio Co., Ltd.)to measure the effects
of let-7g/i on cellular proliferation. The EdU assay was performed
48 h after cells were transfected with let-7g/i agomir. Cells were
seeded in 96-well plates and exposed to 25 mm EdU for 2 h at 37°C,
and were then fixed in 4% paraformaldehyde. Following
permeabilization with 0.5% Triton X-100 (Amresco LLC, Solon, OH,
USA), the 16 Apollo reaction cocktail (Guangzhou RiboBio Co., Ltd.)
was added and the cells were incubated for 30 min. Subsequently,
the DNA of the cells was stained with Hoechst 33342 (Sigma-Aldrich,
St. Louis, MO, USA) for 30 min and visualized under a fluorescent
microscope (IX81; Olympus Corporation, Tokyo, Japan). The cell
count was analyzed by Image-Pro Plus 6.0 software (Media
Cybernetics, Inc., Rockville, MD, USA).
Cell apoptosis assay
Apoptosis assay was performed with Annexin
V-fluorescein isothiocyanate Apoptosis Detection Kit I (BD
Pharmingen, San Diego, CA, USA) 48 h following transfection,
according to the manufacturer’s instructions. The cell suspension
(100 μl) was incubated with 5 μl Annexin V and 5 μl propidium
iodide (BD Pharmingen) at room temperature for 10 min. Finally, 400
μl binding buffer was added to each tube and the cells were
suspended. The treated cells were analyzed by
fluorescence-activated cell sorting using a BD LSR II flow
cytometry kit (BD Pharmingen).
Real-time PCR
Total RNA was isolated from cells using the mirVana
miRNA isolation kit (Ambion Life Technologies, Carlsbad, CA, USA)
according to the manufacturer’s instructions. A total of 10 ng
total RNA was reversely transcribed using the TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems, Foster City, CA,
USA). Quantitative PCR was analyzed using SYBR Premix Ex
TaqTM II and the ViiA7 real-time PCR system (Applied
Biosystems). U6 small nuclear RNA (snRNA) was used to normalize
let-7g/7i expression levels. Primers for let-7g (cat no.
miRQ0000414-1-1)/7i (cat no. miRQ0000415-1-1) and U6 (cat no.
MQP-0201) snRNA were purchased from RiboBio (Guangzhou, China).
Western blot analysis
Total protein was isolated from cells using cell
lysis buffer (Cell Signaling Technology, Inc.) after transfection
for 48 h. The protein levels were quantified using a DC Protein
Assay (Bio-Rad Laboratories, Hercules, CA, USA). Protein samples
(30 μg) were loaded on a 12% SDS-PAGE gels and electroblotted to
Immun-Blot polyvinylidene fluoride membranes (Millipore, Billerica,
MA, USA). Membranes were blocked and probed with a monoclonal
rabbit anti-human Bcl-xL antibody (1:1,000; Epitomics Inc,
Burlingame, CA, USA), then washed with Tris-buffered saline and
Tween 20 (50 mM Tris, 150 mM NaCl, 0.1% Tween-20; pH 7.6;
Sigma-Aldrich), and incubated with a secondary horseradish
peroxidase-conjugated goat anti-rabbit antibody (1:5,000; Hangzhou
Hua’an Biotechnology Co., Ltd., Hangzhou, China). Protein levels
were normalized to total glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) using a mouse anti-human GAPDH antibody (1:1,000; Abcam,
Cambridge, UK). The intensity of each protein band was quantified
by Quantity One version 4.62 software (Bio-Rad Laboratories).
miRNA target predictions
The algorithms DIANA Lab (http://diana.cslab.ece.ntua.gr/), Pictar (http://pictar.mdc-berlin.de/), and TargetScan
(http://www.targetscan.org/) were used to
predict let-7 family members that could potentially bind to Bcl-xL
mRNA.
Statistical analysis
Data are presented as the mean ± standard deviation
of four independent experiments. Statistical analyses were
performed using Microsoft Excel and SPSS software, version 16.0
(SPSS Inc., Chicago, IL, USA). Quantitative PCR data were analyzed
as follows: U6 snRNA was used to normalize let-7g/i expression
levels. Let-7g/i expression levels were measured using the
threshold cycle (Ct), and the fold-change in expression was
calculated as 2−ΔΔCt. The relative expression of
let-7g/i in hematoma cell lines was calculated using the following
formula: ΔΔCt = (Ctlet-7g/i − CtU6) cancer − (Ctlet-7g/i − CtU6)
L-02. The relative expression of let-7g/i after transfection was
calculated using the equation: ΔΔCt = (Ctlet-7i/g − CtU6)
post-transfection − (Ctlet-7g/i − CtU6) pro-transfection. Factorial
analysis of variance was used to analyze the interaction between
let-7g and let-7i in the EdU retention assay and cell apoptosis
assay. P<0.05 was considered to indicate a statistically
significant difference.
Results
let-7g, let-7i and Bcl-xL expression in
hepatoma cells
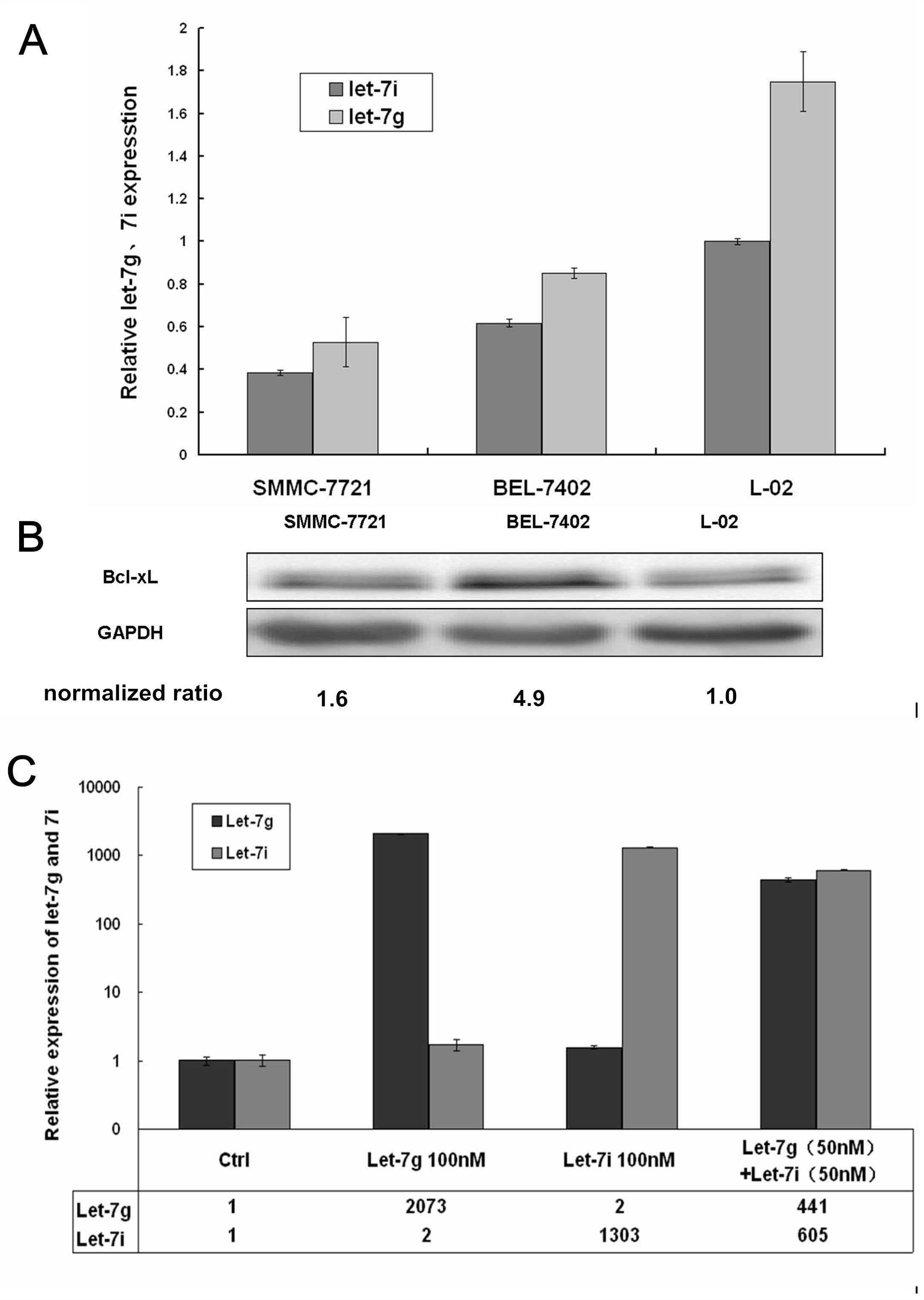
let-7g/i expression was decreased in SMMC-7721 and
BEL-7402 hepatoma cells when compared with the immortalized liver
cell L-02 line (P<0.01) (Fig.
2A).
Bioinformatics analysis (DIANA Lab and PICTAR)
predicted that the anti-apoptotic protein Bcl-xL is a target gene
of let-7g and let-7i, with a potential binding site on the 3′UTR of
Bcl-xL (Fig. 1B). In addition, a
previous study demonstrated that Bcl-xL is the direct target of
let-7c and -7g in Huh7 hepatoma cells (11). In this study, Bcl-xL protein
expression was detected by western blot analysis and it was found
that the protein expression of Bcl-xL was increased in the two
hepatoma cell lines, when compared with the L-02 cell line
(P<0.01) (Fig. 2B).
Expression levels of let-7g and let-7i in
the BEL-7402 cell line following transfection
The fold-changes in let-7g/i levels in the HCC
BEL-7402 cell line were detected following transfection with
agomirs or the negative control. The expression levels of let-7g in
the groups transfected with let-7g or co-transfected with let-7g
and let-7i were 2,073- and 441-fold those of the control group,
respectively. The expression levels of let-7i in the groups
transfected with let-7i and co-transfected with let-7g and let-7i
were 1,303- and 605-fold those of the control group, respectively
(Fig. 2C).
Overexpression of let-7g/i inhibits
hepatoma cell proliferation
The BEL-7402 cells were transfected with let-7g/i
agomir or the negative control to investigate the effects of
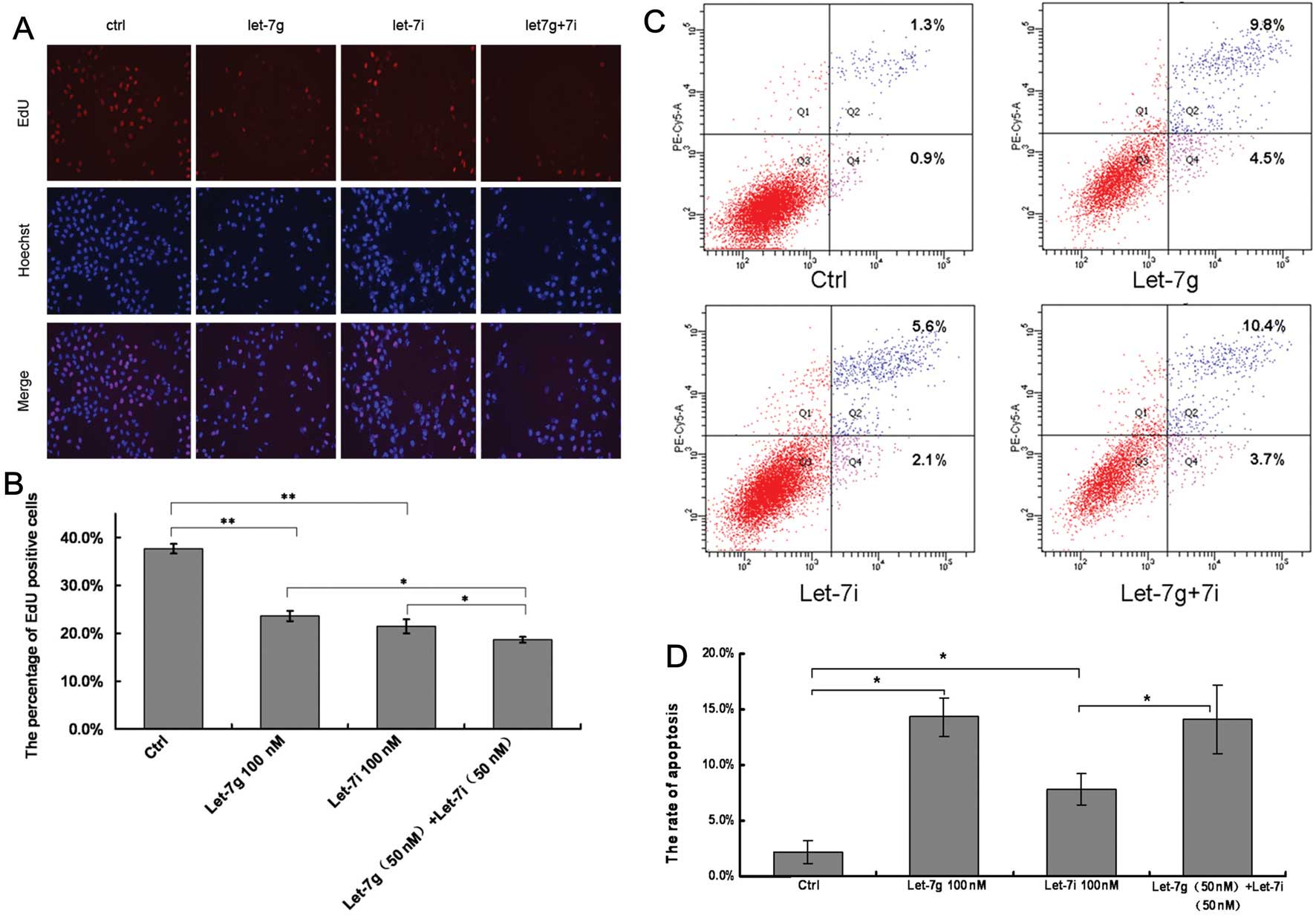
let-7g/i agomir on HCC cell growth. Hoechst staining nuclei in the
experimental groups was dense and contracted (Fig. 3A). DNA replication activity in the
groups of transfected with the negative control, let-7g, let-7i and
let-7g + 7i was 37.7, 23.6, 21.4 and 18.6%, respectively (Fig. 3B). When compared with the control
group, DNA replication activity in the groups transfected with
let-7g or let-7i was significantly decreased (P<0.01). When
compared with the let-7i group or let-7g group, DNA replication
activity of the let-7g + 7i group was inhibited (P<0.05)
(Fig. 3B), indicating that there
may be a combinatorial effect between let-7g and let-7i.
Overexpression of let-7g/i induces
hepatoma cell apoptosis
BEL-7402 cells were transfected with let-7g/i agomir
or the negative control to investigate the effects of let-7g/i on
cell apoptosis. The results revealed that transfection with
let-7g/i agomir increased the percentage of apoptotic cells in the
BEL-7402 cell line (Fig. 3C). The
rates of apoptosis in the groups transfected with let-7g, let-7i
and let-7g + 7i were 14.3, 7.7 and 14.1%, respectively, which were
significantly greater than the rate in the control group (2.2%)
(P<0.05; Fig. 3D).
Co-transfection with Let-7g and let-7i
downregulates the expression level of Bcl-xL
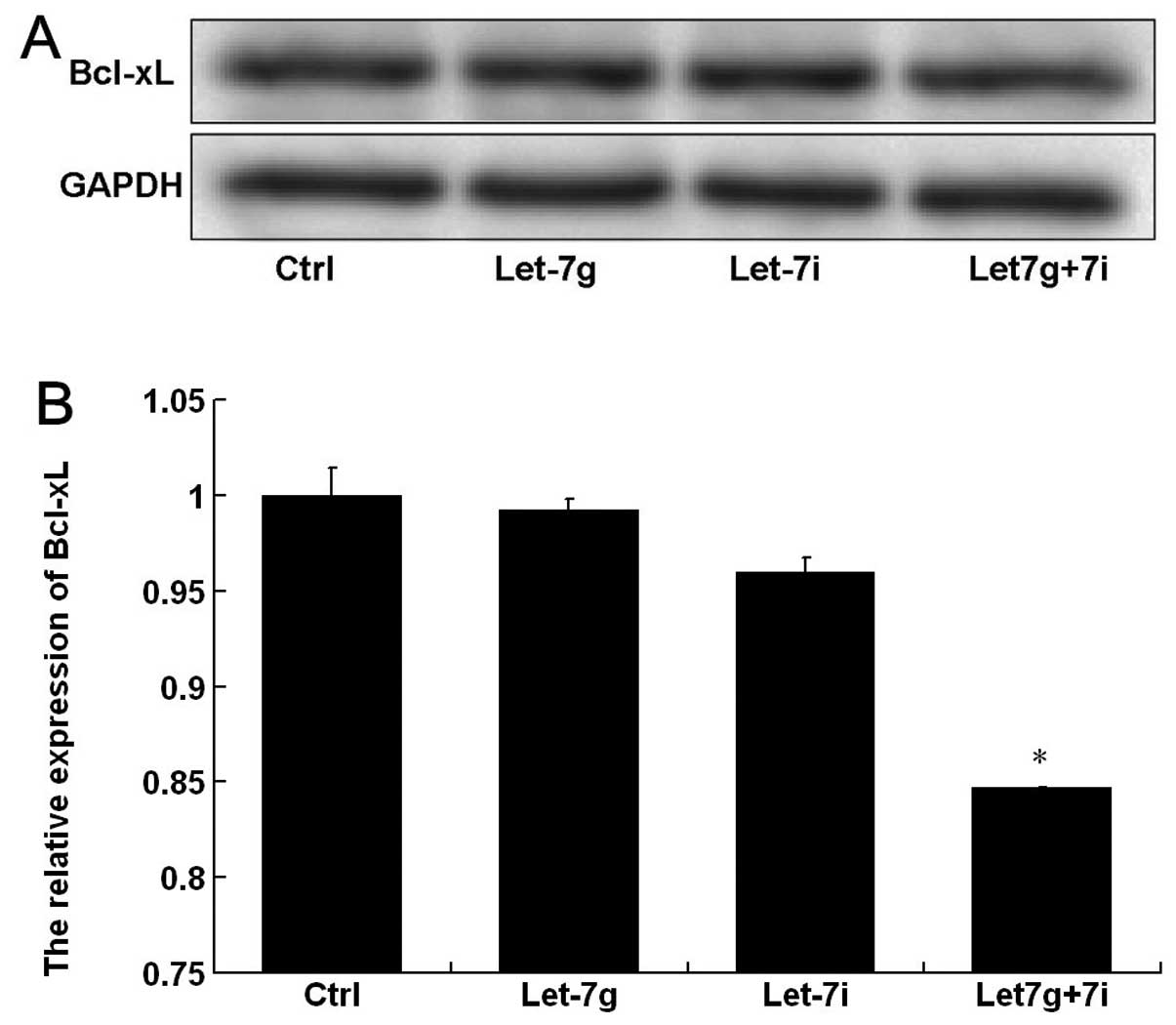
The expression of the anti-apoptotic protein,
Bcl-xL, was analyzed by western blot analysis (Fig. 4A). The gray-value quantitative
analysis showed that Bcl-xL protein expression in BEL-7402 cells
was markedly decreased after co-transfection with let-7g and let-7i
compared with that in the control. However, the protein expression
was not significantly influenced after transfection with let-7g or
let-7i (Fig. 4B), suggesting that
let-7g and let-7i exert a combined effect on Bcl-xL protein in
BEL-7402 cells.
Discussion
The correlation between miRNA mutation or altered
expression and various human cancers indicates that miRNAs may
function as tumor suppressors or oncogenes and, thus, they are
referred to as oncomirs (4). It has
been demonstrated that the let-7 family are important tumor
suppressor genes (17). In this
study, the expression of let-7g/i was found to be significantly
decreased in HCC cells when compared with the L-02 cell line. These
results are consistent with those of Shimizu et al, which
demonstrated that the expression of let-7b, -7g,-7i, -7d, -7a, -7c
and 7e was downregulated in Huh7 cells when compared with normal
hepatocytes (11). A similar result
was found in the present study, however, different methods and cell
lines were used. In addition, the effects of let-7g/i on the
biological behavior of hepatoma cells were investigated, and it was
found that the overexpression of let-7g/i significantly inhibited
the cell proliferation and promoted the cell apoptosis of BEL-7402
cells. miRNAs have been identified as a class of regulatory RNAs in
numerous biological processes (18–20).
The loss of let-7 activity induces abnormal development in
Caenorhabditis elegans (21). In addition, the LIN28/let-7 pathway
exhibits a critical pathobiological role in malignant germ cell
tumors (22). Furthermore, enforced
expression of let-7b inhibited breast cancer cell motility and
affected actin dynamics (13).
In this study, co-transfection with let-7g and
let-7i was found to exhibit an enhanced effect on BEL-7402 cell
proliferation and apoptosis, when compared with the effect of
let-7g or let-7i alone. These results indicate that let-7g and
let-7i exhibit a combinatorial role in suppressing HCC progression.
According to a previous study, the let-7 family members mir-48,
mir-84 and mir-241, function together to control the L2-to-L3
transition, likely by base pairing to complementary sites in the
hbl-1 3′ UTR, indicating that let-7 family miRNAs function in
combination to affect early and late developmental timing decisions
(16). Different target genes may
be regulated by a single miRNA, and multiple miRNAs may also
function together to regulate one or several gene pathways. These
regulatory pathways are hypothesized to affect the development of
cancer (1,23).
Let-7 miRNAs act as tumor suppressors by modulating
major oncogenes, including high mobility group protein (10,24),
ras (25) and caspase-3 (26). Bcl-xL is an important member of the
anti-apoptotic Bcl-2 family, and has been found to be overexpressed
in HCC (27). According to the
bioinformatics (DIANA Lab and PICTAR) prediction, Bcl-xL is a
direct target gene of let-7, which was also confirmed by a previous
molecular study (11). This
previous study demonstrated that the overexpression of let-7c or
let-7g led to a marked decrease in Bcl-xL expression in Huh7 and
HepG2 hepatoma cell lines (11).
The present study also showed that Bcl-xL protein expression in the
BEL-7402 hepatoma cell line was significantly decreased following
combined transfection with let-7g and le-7i. However, the Bcl-xL
protein expression was not significantly influenced by transfection
with let-7g or let-7i alone. These results indicated that let-7g
and let-7i may exhibit a coordinated effect on the Bcl-xL protein.
Two different nucleotides have been identified at the 3′end of
let-7g and let-7i base sequences, resulting in two different
binding sites and modes with Bcl-xL mRNA 3′UTR (Fig. 1B). The two miRNAs augment each other
to regulate the Bcl-xL protein. Therefore, the combinatorial role
of let-7g and let-7i led to the downregulation of Bcl-xL
protein.
In conclusion, let-7g and let-7i exhibit a combined
effect to regulate hepatoma cell proliferation and apoptosis, and
this function is hypothesized to be mediated via the Bcl-xL
protein.
Acknowledgements
This study was supported by the National High-Tech
863 Program (grant no. 2008AA02Z109) and the National Natural
Science Foundation of China (grant no. 81272354).
Abbreviations:
|
miRNA
|
microRNA
|
|
let-7g/i
|
miRNA-let-7g/i
|
|
HCC
|
hepatocellular carcinoma
|
|
UTR
|
untranslated region
|
|
Bcl-xL
|
B-cell lymphoma-extra large
|
References
|
1
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou J, Lin L, Zhou W, et al:
Identification of miRNomes in human liver and hepatocellular
carcinoma reveals miR-199a/b-3p as therapeutic target for
hepatocellular carcinoma. Cancer Cell. 19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akao Y, Nakagawa Y and Naoe T: let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nam EJ, Yoon H, Kim SW, et al: MicroRNA
expression profiles in serous ovarian carcinoma. Clin Cancer Res.
14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng Y, Laser J, Shi G, et al:
Antiproliferative effects by Let-7 repression of high-mobility
group A2 in uterine leiomyoma. Mol Cancer Res. 6:663–673. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimizu S, Takehara T, Hikita H, et al:
The let-7 family of microRNAs inhibits Bcl-xL expression and
potentiates sorafenib-induced apoptosis in human hepatocellular
carcinoma. J Hepatol. 52:698–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu XM, Wu LJ, Xu J, Yang R and Wu FS:
Let-7c microRNA expression and clinical significance in
hepatocellular carcinoma. J Int Med Res. 39:2323–2329. 2011.
View Article : Google Scholar
|
|
13
|
Hu X, Guo J, Zheng L, et al: The
heterochronic microRNA let-7 inhibits cell motility by regulating
the genes in the actin cytoskeleton pathway in breast cancer. Mol
Cancer Res. 11:240–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Doench JG and Sharp PA: Specificity of
microRNA target selection in translational repression. Genes Dev.
18:504–511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abbott AL, Alvarez-Saavedra E, Miska EA,
et al: The let-7 MicroRNA family members mir-48, mir-84, and
mir-241 function together to regulate developmental timing in
Caenorhabditis elegans. Dev Cell. 9:403–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hunter SE, Finnegan EF, Zisoulis DG, et
al: Functional genomic analysis of the let-7 regulatory network in
Caenorhabditis elegans. PLoS Genet. 9:e10033532013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murray MJ, Saini HK, Siegler CA, et al:
LIN28 Expression in malignant germ cell tumors downregulates let-7
and increases oncogene levels. Cancer Res. 73:4872–4884. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bueno MJ, Gomez de Cedron M, Gomez-Lopez
G, et al: Combinatorial effects of microRNAs to suppress the Myc
oncogenic pathway. Blood. 117:6255–6266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsang WP and Kwok TT: Let-7a microRNA
suppresses therapeutics-induced cancer cell death by targeting
caspase-3. Apoptosis. 13:1215–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Watanabe J, Kushihata F, Honda K, et al:
Prognostic significance of Bcl-xL in human hepatocellular
carcinoma. Surgery. 135:604–612. 2004. View Article : Google Scholar : PubMed/NCBI
|