Introduction
FOXL2, a Forkhead box family transcription factor
initially described in Drosophila, is predominantly
expressed in periocular, ovarian and pituitary cells. FOXL2 was
first cloned and localized by Crisponi et al (1) and is mutated in blepharophimosis
ptosis epicanthus inversus syndrome (BPES), a genetic disorder
characterized by eyelid malformations. More than two-thirds of BPES
patients carry intragenic FOXL2 mutations and one-third of
mutations in the FOXL2 coding region are expansions of the
polyalanine tract, from 14 to 24 residues (2,3).
Furthermore, heterozygous mutations in FOXL2 result in premature
ovarian failure and infertility in females (4).
FOXL2 functions as a transcriptional repressor and
requires sumoylation and phosphorylation for its activity (5,6).
Somatic mutations of FOXL2 have been reported to reduce its
activity and may be associated with enhanced cancer cell
proliferation, accelerated cell cycle progression and reduced
sensitivity to apoptosis (7). In
addition, genes that are differentially expressed in ovarian
granulosa cell tumors (GCTs) are significantly enriched for known
FOXL2 target genes, consistent with the prevalence of FOXL2 somatic
mutations in these tumors (8).
MicroRNAs (miRNAs) are short non-coding RNAs which
modulate gene expression by binding to complementary areas in the
3′-untranslated region of protein-coding gene mRNA. miRNAs are
important in maintaining normal physiological conditions in humans,
and abnormal miRNA expression has been associated with numerous
human diseases, including psychiatric disorders and malignant
cancer (9–11). Bioinformatics research has indicated
that all of the miRNAs may target >60% of mammalian
protein-coding genes (12).
The present study used bioinformatic tools to
predict miRNAs that may directly target FOXL2. Among them, miR-30
family members are associated with human ovarian carcinogenesis
(13). Subsequently, dual
luciferase assays and western blotting identified FOXL2 as the
target gene of miR-30a. Furthermore, miR-30a upregulated BCL2A1,
IER3 and cyclin D2 expression by repressing FOXL2 expression.
Materials and methods
Small interfering (si)RNA knockdown
siRNAs against Ago2, miR-30a mimic and miR-30a
inhibitor were purchased from RiboBio Co., Ltd. (Guangzhou, China)
and transfected into COV434 cells at a concentration of 200 nM.
Cell culture
The human granulosa COV43 cells (Shanghai Insitutes
for Biological Sciences, Chinese Academy of Sciences, Shanghai,
China) were cultured in Dulbecco’s modified Eagle medium
supplemented with 10% fetal bovine serum (HyClone Laboratories,
Inc., Logan, UT, USA), 100 IU/ml penicillin and 10 mg/ml
streptomycin. All cells were maintained at 37°C in an atmosphere of
5% CO2.
RNA extraction and detection of miR-30
family member expression
Reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) was performed to determine the relative
expression level of specific miR-30 family members
(miR-30a/b/c/d/e). Total RNA was extracted from COV434 cells using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer’s instructions. The expression levels
of the various miRNAs were detected using TaqMan®
RT-qPCR miRNA assays. Single-stranded complementary DNA was
synthesized using a TaqMan MicroRNA Reverse Transcription kit, and
amplified using TaqMan Universal PCR Master mix and miRNA-specific
TaqMan MGB probes (all TaqMan products were purchased from Applied
Biosystems Life Technologies, Foster City, CA, USA). U6 small
nuclear RNA was used for data normalization. Each sample was
measured in triplicate and the experiment was repeated a minimum of
three times to ensure miRNA detection.
Western blotting
Protein extracts were boiled in
SDS/β-mercaptoethanol (2:1; w/v) sample buffer, and 30 μg samples
were loaded into each lane and separated by electrophoresis on 8%
polyacrylamide gels. The separated proteins were
electrophoretically transferred onto polyvinylidene fluoride
membranes (GE Healthcare Life Sciences, Chalfont, UK), which were
incubated with goat anti-human FOXL2 polyclonal antibody (1:1,000;
cat. no., ab5096; Abcam, Cambridge, MA, USA) and mouse anti-human
AGO2 monoclonal antibody (1:1,000; cat. no., ab57113; Abcam) or
mouse anti-human β-actin monoclonal antibody (1:3,000; cat. no.,
sc-69879; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 1
h at 37°C. The specific protein antibody complex was detected using
horseradish peroxidase-conjugated rabbit anti-goat and rabbit
anti-mouse polyclonal IgG secondary antibody (1:5,000; Santa Cruz
Biotechnology, Inc.) and visualized using an enhanced
chemiluminescence kit (Pierce Manufacturing Inc., Appleton, WI,
USA). The β-actin signal was used as the loading control.
Dual luciferase assay
The full length FOXL2 3′-UTR (1,129 bp) was cloned
into a pGL3 control vector (Promega Corporation, Madison, WI, USA),
downstream of the firefly luciferase coding region, to generate a
luciferase reporter vector. For luciferase reporter assays, COV434
cells were seeded in 48-well plates. An miR-30a mimic or miR-30a
inhibitor were co-transfected with luciferase reporter vectors
(Promega Corporation) using Lipofectamine 2000 (Invitrogen Life
Technologies). Cells were harvested after two days and assayed
using the Dual-Luciferase® Reporter Assay system
(Promega Corporation) to determine the relative luciferase activity
(LUC) of the COV434 cells. pRL-TK containing Renilla luciferase was
co-transfected with the 3′-UTR of FOXL2 for data normalization.
Each treatment was performed in triplicate in three independent
experiments and LUC was expressed as firefly LUC/Renilla LUC.
Statistical analysis
Data were analyzed using SPSS statistical software
(version 16; SPSS, Inc., Chicago, IL, USA). Independent analysis
between the two groups was performed using a t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
FOXL2 expression is regulated by
miRNAs
To explore whether the expression of FOXL2 is
regulated by miRNAs, Ago2, a key component of the RNA-induced
silencing complex, was knocked down in COV434 cells. This knockdown
demonstrated that inactivation of the miRNA system results in
upregulation of FOXL2 expression (Fig.
1A), indicating that miRNAs are involved in the negative
control of FOXL2 expression.
miR-30a represses FOXL2 expression by
binding to 3′-UTR
TargetScan Release 6.2 (http://www.targetscan.org/), an online tool for
predicting the interaction between miRNAs and genes, was used to
probe miRNAs which may suppress FOXL2 expression. Of the candidate
miRNAs, miR-30 family members were reported to be associated with
various types of human cancer, and the expression of miR-30a was
high in COV434 cells, compared with the other miRNAs evaluated
(Fig. 1B). Thus, miR-30a was
selected for further investigation of its role in the repression of
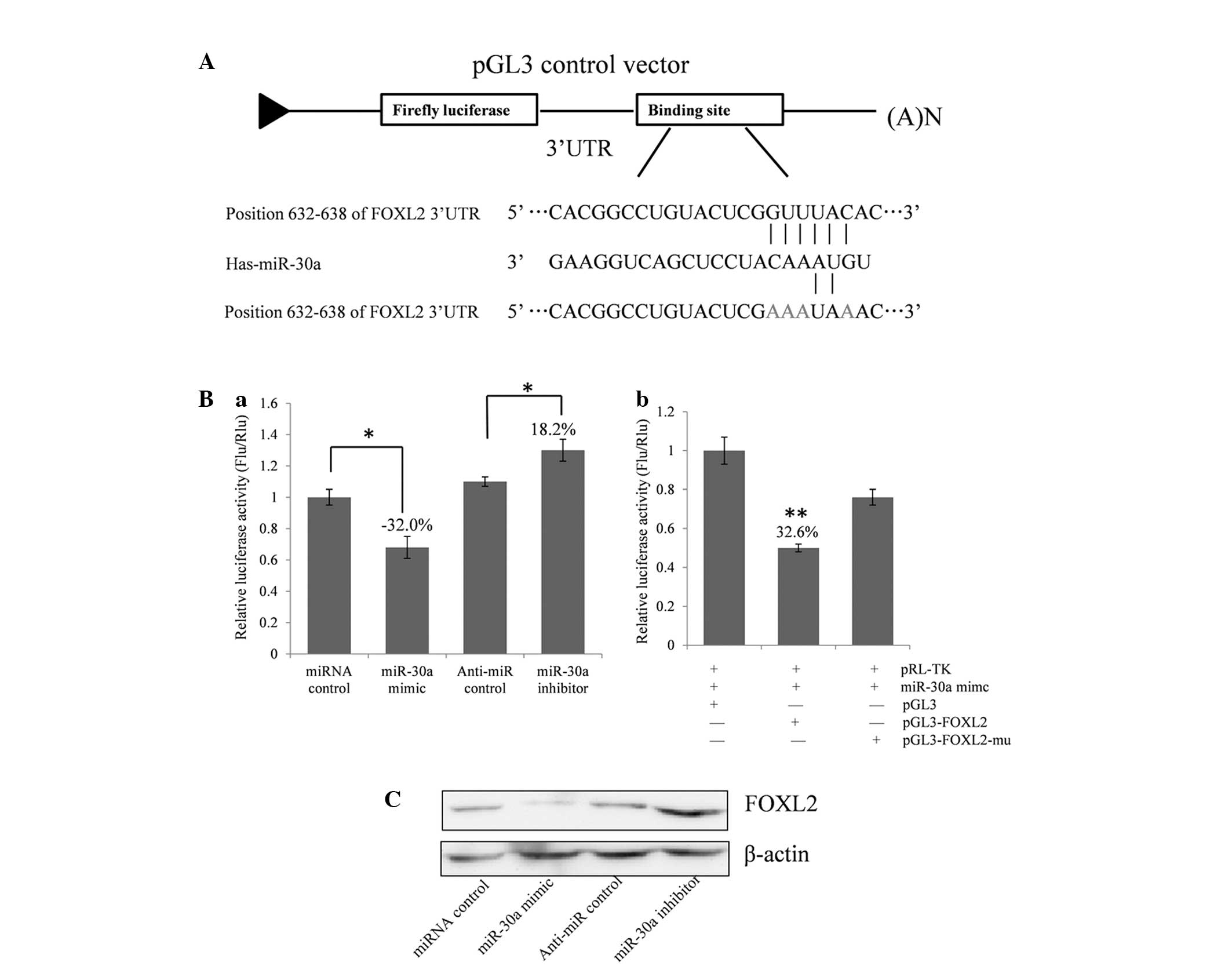
FOXL2 expression. As demonstrated in Fig. 2A, a the full length 3′-UTR of FOXL2
was cloned into the pGL3 control plasmid, downstream of the firefly
luciferase coding region, and a dual luciferase assay was
conducted. COV434 cells were co-transfected with pGL3-FOXL2 and
miR-30a mimics or inhibitors (Fig.
2Ba). The present study identified that luciferase activity was
significantly suppressed by the miRNA control compared with the
miR-30a mimic (~32.0%; P<0.05). Furthermore, luciferase activity
was significantly upregulated by the miR-30a inhibitor compared
with the anti-miR control (~18.2%; P<0.05). These results
indicate that miR-30a targets the 3′-UTR of FOXL2, resulting in
altered translation of the firefly luciferase gene.
A seed sequence mutation clone was used to clarify
the location of the miR-30a binding site (Fig. 2A). A four-nucleotide mutation in the
putative miR-30a binding region of the FOXL2 3′-UTR (termed,
pGL3-FOXL2-Mu) and an empty pGL3 vector were used as the controls.
The histogram in Fig. 2Bb
demonstrates that the relative luciferase activity was reduced by
~32.6% in cells co-transfected with the miR-30a mimic and
pGL3-FOXL2 compared with the miR-30a mimic and pGL3-FOXL2-Mu
(P<0.01). These data indicate that miR-30a may suppress FOXL2
gene expression through binding to the seed sequence at the 3′-UTR
of FOXL2, and that FOXL2 may be a direct target of miR-30a.
miR-30a regulates endogenous FOXL2
expression in COV434 cells
Although FOXL2 was identified as a target gene for
miR-30a, it was unknown whether miR-30a could regulate endogenous
FOXL2 expression. COV434 cells were transfected with miR-30a mimics
or inhibitors to investigate whether the dysregulation of miR-30a
expression affected endogenous FOXL2 expression. Compared with the
corresponding control, the level of FOXL2 protein was significantly
suppressed by miR-30a mimics and upregulated by miR-30a inhibitors
(Fig. 2C).
miR-30a promotes BCL2A1, IER3 and cyclin
D2 gene expression by suppressing FOXL2
The consequences of miR-30a knockdown and
overexpression indicate that miR-30a regulates cell function by
regulating FOXL2. To further investigate this, the effects of
miR-30a treatment on IER3, BCL2A1 and cyclin D2 were investigated.
IER3, BCL2A1 and cyclin D2 mRNA expression were detected using
RT-qPCR, 48 h after transfection with miR-30a. As demonstrated in
Fig. 3, the relative mRNA
expression of IER3, BCL2A1 and cyclin D2 was significantly
increased by 40.2, 75.3 and 43.3%, respectively, following miR-30a
overexpression.
Discussion
FOXL2 is involved in craniofacial and female genital
system development, and FOXL2 mutations can result in the
development of BPES, ovary failure and GCTs (2,14,15).
Studies have indicated that FOXL2 is significantly downregulated in
the COV434 GCT cell line, despite no alterations to the genomic DNA
(16,17). Therefore, the present study aimed to
investigate the negative control system of FOXL2 expression. Since
miRNAs regulate a large proportion of protein-coding genes, Ago2
was initially knocked down to evaluate the effect of miRNAs on
FOXL2 expression. This Ago2 knockdown resulted in a significantly
upregulation of FOXL2 expression. Following prediction using
bioinformatics tools and clarification by dual luciferase assay and
western blotting, miR-30a was identified to repress FOXL2 in COV434
cells. Furthermore, this repression function was reflected in the
upregulation of genes regulated by FOXL2.
As a transcriptional repressor, FOXL2 suppresses
BCL2A1, IER3 and cyclin D2 gene expression in granulosa cells
(18–20). BCL2A1 and IER3 are apoptosis
inhibitors, whereas cyclin D2 is a cell cycle-associated gene which
regulates G1/S transition. High expression levels of these genes
have been observed in ovarian and testicular tumors.
miRNA is a type of post-transcriptional suppressor
and a single miRNA may regulate hundreds of protein-coding genes.
In particular, miR-30a acts as a tumor suppressor in breast cancer,
non-small cell lung cancer and colorectal carcinoma (21–23).
However, the present study identified that miR-30a also acts as an
oncogene by repressing FOXL2 expression in GCTs. In conclusion,
further investigation is required to expand on the present research
and evaluate the function of miR-30a in granulosa cell tumors and
other types of cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81272122) and the
Science and Technology Research Program of Shandong, China (grant
no. jk67).
References
|
1
|
Crisponi L, Uda M, Deiana M, et al: FOXL2
inactivation by a translocation 171 kb away: analysis of 500 kb of
chromosome 3 for candidate long-range regulatory sequences.
Genomics. 83:757–764. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Baere E, Beysen D, Oley C, et al: FOXL2
and BPES: mutational hotspots, phenotypic variability, and revision
of the genotype-phenotype correlation. Am J Hum Genet. 72:478–487.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Baere E, Dixon MJ, Small KW, et al:
Spectrum of FOXL2 gene mutations in
blepharophimosis-ptosis-epicanthus inversus (BPES) families
demonstrates a genotype - phenotype correlation. Hum Mol Genet.
10:1591–1600. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zlotogora J, Sagi M and Cohen T: The
blepharophimosis, ptosis, and epicanthus inversus syndrome:
delineation of two types. Am J Hum Genet. 35:1020–1027.
1983.PubMed/NCBI
|
|
5
|
Benayoun BA, Batista F, Auer J, et al:
Positive and negative feedback regulates the transcription factor
FOXL2 in response to cell stress: evidence for a regulatory
imbalance induced by disease-causing mutations. Hum Mol Genet.
18:632–644. 2009. View Article : Google Scholar
|
|
6
|
Pisarska MD, Kuo FT, Bentsi-Barnes IK,
Khan S and Barlow GM: LATS1 phosphorylates forkhead L2 and
regulates its transcriptional activity. Am J Physiol Endocrinol
Metab. 299:E101–E109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pisarska MD, Barlow G and Kuo FT:
Minireview: roles of the forkhead transcription factor FOXL2 in
granulosa cell biology and pathology. Endocrinology. 152:1199–1208.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Benayoun BA, Anttonen M, L’Hôte D, et al:
Adult ovarian granulosa cell tumor transcriptomics: prevalence of
FOXL2 target genes misregulation gives insights into the pathogenic
mechanism of the p. Cys134Trp somatic mutation. Oncogene.
32:2739–2746. 2013. View Article : Google Scholar
|
|
9
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for Alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu J, Li Y, Wang F, et al: Suppressed
miR-424 expression via upregulation of target gene Chk1 contributes
to the progression of cervical cancer. Oncogene. 32:976–987. 2013.
View Article : Google Scholar
|
|
11
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
13
|
Lee H, Park CS, Deftereos G, et al:
MicroRNA expression in ovarian carcinoma and its correlation with
clinicopathological features. World J Surg Oncol. 10:1742012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corrêa FJ, Tavares AB, Pereira RW and
Abrão MS: A new FOXL2 gene mutation in a woman with premature
ovarian failure and sporadic blepharophimosis-ptosis-epicanthus
inversus syndrome. Fertil Steril. 93:e3–e6. 2010.
|
|
15
|
Kim JH, Yoon S, Park M, et al:
Differential apoptotic activities of wild-type FOXL2 and the
adult-type granulosa cell tumor-associated mutant FOXL2 (C134W).
Oncogene. 30:1653–1663. 2011. View Article : Google Scholar
|
|
16
|
Kalfa N, Fellous M, Boizet-Bonhoure B, et
al: Aberrant expression of ovary determining gene FOXL2 in the
testis and juvenile granulosa cell tumor in children. J Urol.
180:1810–1813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalfa N, Philibert P, Patte C, et al:
Extinction of FOXL2 expression in aggressive ovarian granulosa cell
tumors in children. Fertil Steril. 87:896–901. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
D’Sa-Eipper C and Chinnadurai G:
Functional dissection of Bfl-1, a Bcl-2 homolog: anti-apoptosis,
oncogene-cooperation and cell proliferation activities. Oncogene.
16:3105–3114. 1998. View Article : Google Scholar
|
|
19
|
Wu MX, Ao Z, Prasad KV, Wu R and
Schlossman SF: IEX-1L, an apoptosis inhibitor involved in
NF-kappaB-mediated cell survival. Science. 281:998–1001. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bentsi-Barnes IK, Kuo FT, Barlow GM and
Pisarska MD: Human forkhead L2 represses key genes in granulosa
cell differentiation including aromatase, P450scc, and cyclin D2.
Fertil Steril. 94:353–356. 2010. View Article : Google Scholar :
|
|
21
|
Zhang N, Wang X, Huo Q, et al:
MicroRNA-30a suppresses breast tumor growth and metastasis by
targeting metadherin. Oncogene. 33:3119–3128. 2014. View Article : Google Scholar
|
|
22
|
Jiang BY, Zhang XC, Su J, et al: BCL11A
overexpression predicts survival and relapse in non-small cell lung
cancer and is modulated by microRNA-30a and gene amplification. Mol
Cancer. 12:612013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong M, Bian Z and Wu Z: miR-30a
suppresses cell migration and invasion through downregulation of
PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 31:209–218.
2013. View Article : Google Scholar : PubMed/NCBI
|