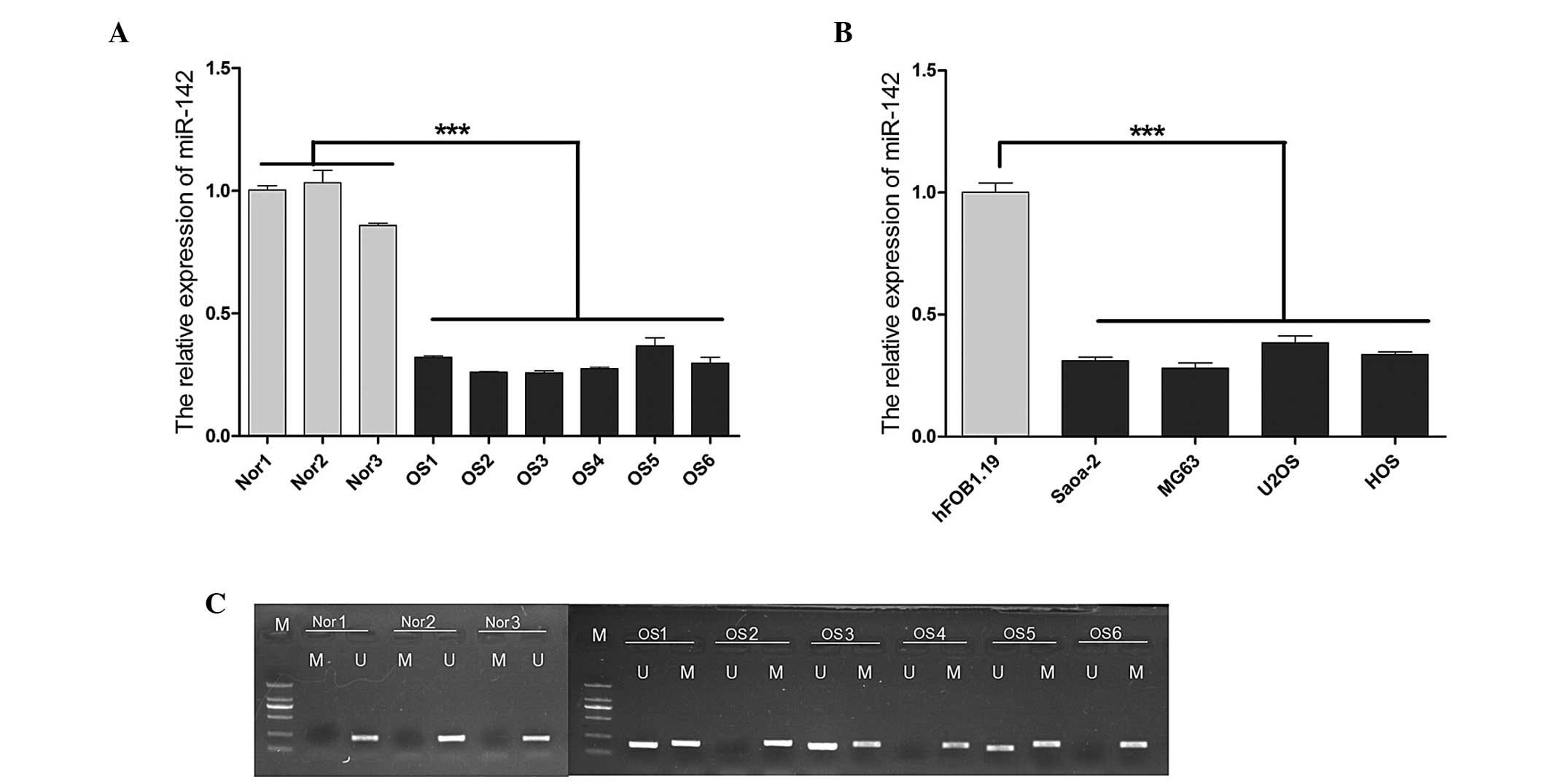
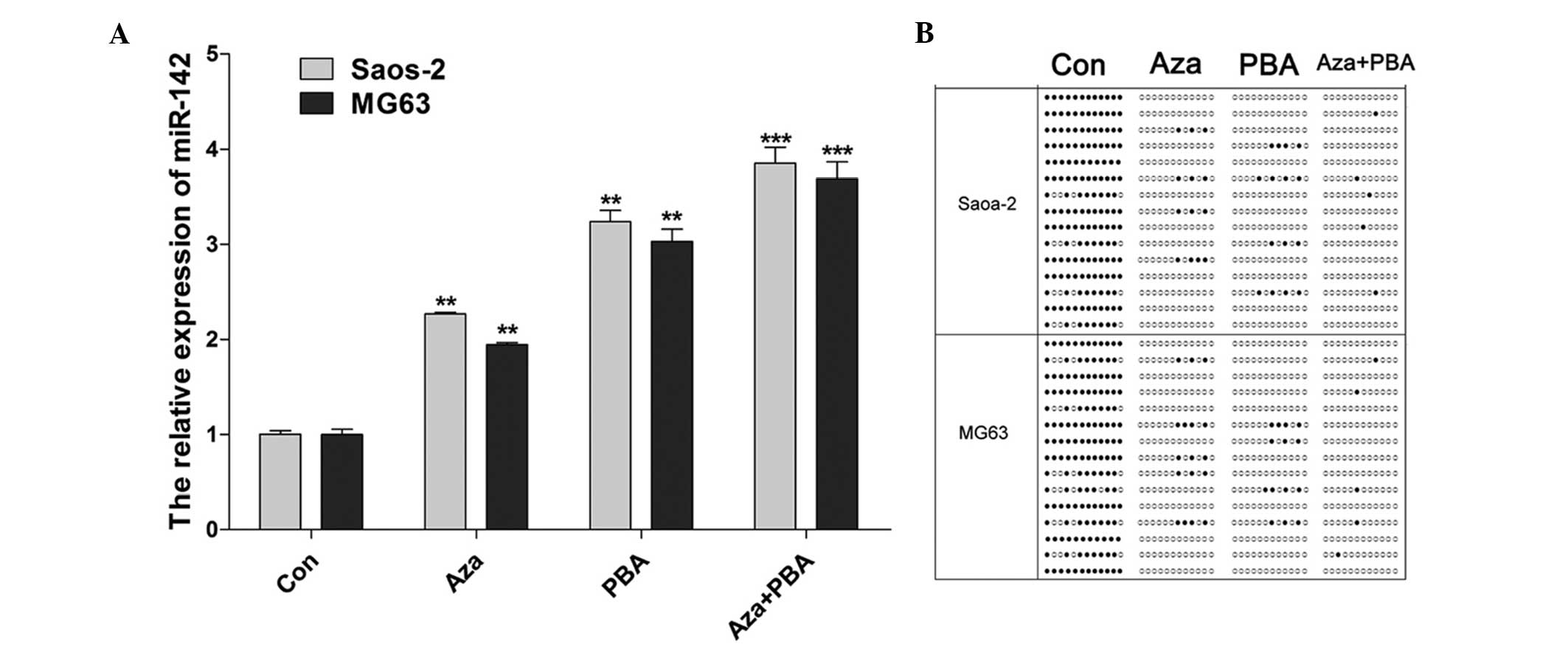
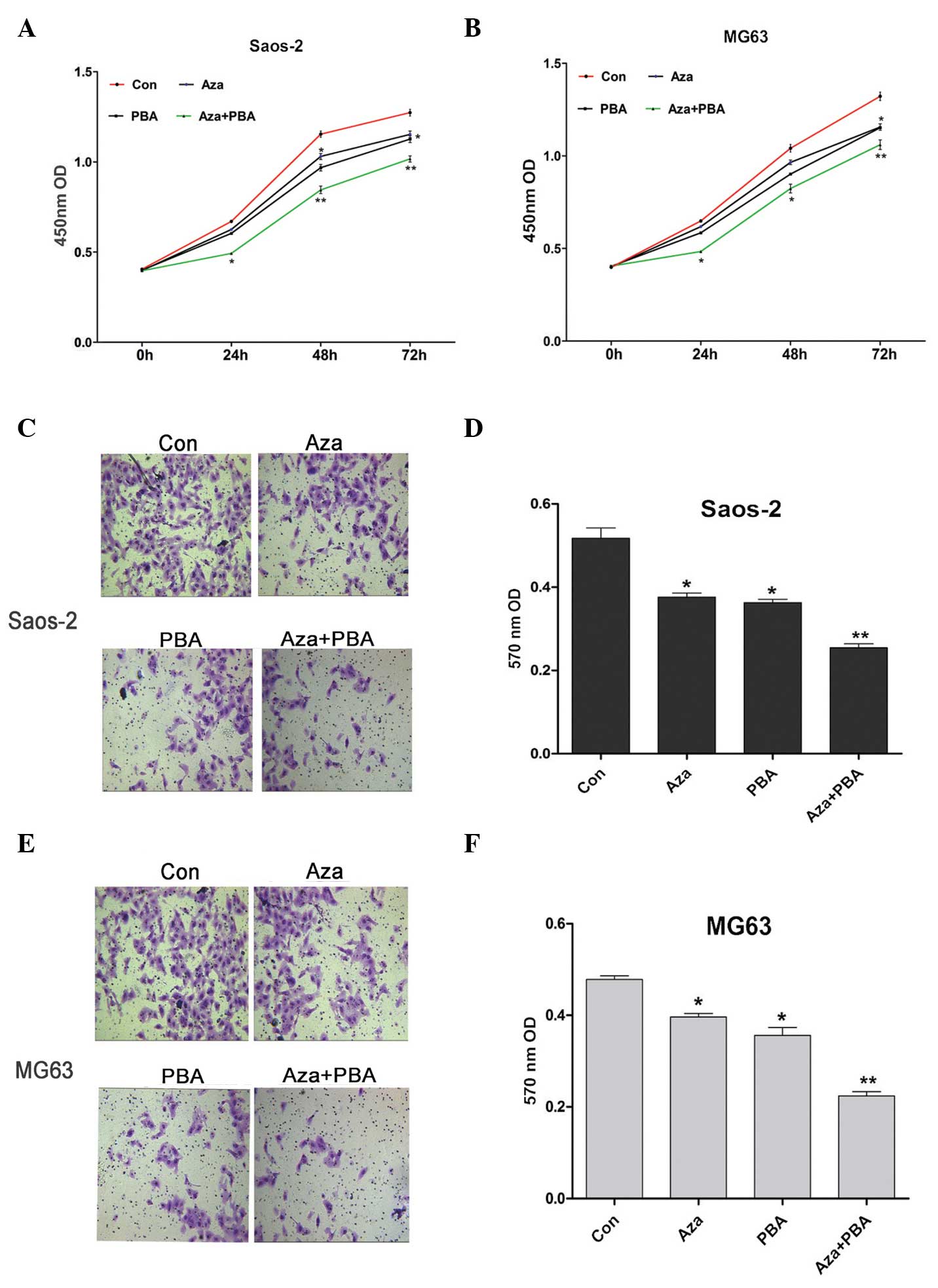
|
1
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma.
2012.3597392012.PubMed/NCBI
|
|
2
|
Cao ZQ, Shen Z and Huang WY: MicroRNA-802
promotes osteosarcoma cell proliferation by targeting p27. Asian
Pac J Cancer Prev. 14:7081–7084. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Diao CY, Guo HB, Ouyang YR, et al:
Screening for metastatic osteosarcoma biomarkers with a DNA
microarray. Asian Pac J Cancer Prev. 15:1817–1822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Benavente CA, McEvoy JD, Finkelstein D, et
al: Cross-speciesgenomic and epigenomic landscape of
retinoblastoma. Oncotarget. 4:844–859. 2013.PubMed/NCBI
|
|
5
|
Jin J, Cai L, Liu ZM and Zhou XS:
miRNA-218 inhibits osteosarcoma cellmigration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewis BP, Burge CB and Bartel DP:
Conserved seedpairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kelly TK, De Carvalho DD and Jones PA:
Epigenetic modifications as therapeutic targets. Nat Biotechnol.
28:1069–1078. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones KB, Salah Z, Del Mare S, et al:
miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Poos K, Smida J, Nathrath M, Maugg D,
Baumhoer D and Korsching E: HowmicroRNA and transcription factor
co-regulatory networks affect osteosarcoma cell proliferation. PLoS
Comput Biol. 9:e10032102013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Png KJ, Yoshida M, Zhang XH, et al:
MicroRNA-335 inhibits tumorreinitiation and is silenced
throughgenetic and epigenetic mechanisms in human breast cancer.
Genes Dev. 25:226–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Das S, Bryan K, Buckley PG, et al:
Modulation of neuroblastoma disease pathogenesis by an extensive
network of epigenetically regulated microRNAs. Oncogene.
32:2927–2936. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He C, Xiong J, Xu X, et al: Functional
elucidation of MiR-34 in osteosarcomacells and primary tumor
samples. Biochem Biophys Res Commun. 388:35–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qian S, Ding JY, Xie R, et al: MicroRNA
expression profile of bronchioalveolar stem cells from mouse lung.
Biochem Biophys Res Commun. 377:668–673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin RJ, Xiao DW, Liao LD, et al:
MiR-142-3p as a potential prognostic biomarker for esophageal
squamous cell carcinoma. J Surg Oncol. 105:175–182. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Namløs HM, Meza-Zepeda LA, Barøy T, et al:
Modulation of the osteosarcoma expression phenotype by microRNAs.
PLoS One. 7:e480862012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Posthuma De Boer J, Witlox MA, Kaspers GJ
and van Royen BJ: Molecular alterations as target for therapy in
metastatic osteosarcoma: a review of literature. Clin Exp
Metastasis. 28:493–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumhoer D, Zillmer S, Unger K, et al:
MicroRNA profiling with correlation to gene expression revealed the
oncogenic miR-17-92 cluster to be up-regulated in osteosarcoma.
Cancer Genet. 205:212–219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lulla RR, Costa FF, Bischof JM, et al:
Identification of differentially expressed microRNAs in
osteosarcoma. Sarcoma. 2011.7326902011.PubMed/NCBI
|
|
21
|
Boeri M, Verri C, Conte D, et al: MicroRNA
signatures intissues and plasma predictdevelopment and prognosis of
computed tomography detected lung cancer. In: Proc Natl Acad Sci
USA. 108. pp. 3713–3718. 2011; View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kaduthanam S, Gade S, Meister M, et al:
Serum miR-142-3p is associated with early relapse in operable lung
adenocarcinoma patients. Lung Cancer. 80:223–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei Z, Xu G, Wang L, et al: MiR-142-3p
represses TGF-β-induced growth inhibition through repression of
TGFβR1 in non-small cell lung cancer. FASEB J. 28:2696–2704. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang XY, Wu MH, Liu F, et al: Differential
miRNAexpression and their target genes between NGX6-positive and
negative colon cancer cells. Mol Cell Biochem. 345:283–290. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
MiR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl).
91:989–1000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee Y, Ahn C, Han J, et al: The nuclear
RNase III Drosha initiates microRNA processing. Nature.
425:415–419. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee Y, Jeon K, Lee JT, Kim S and Kim VN:
MicroRNA maturation: stepwiseprocessing and subcellular
localization. EMBO J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vrba L, Garbe JC, Stampfer MR and Futscher
BW: Epigenetic regulation of normal human mammary cell
type-specific miRNAs. Genome Res. 21:2026–2037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hackett JA and Surani MA: DNA methylation
dynamics during the mammalian life cycle. Philos Trans R Soc Lond B
Biol Sci. 368:201103282013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weber B, Stresemann C, Brueckner B and
Lyko F: Methylation of human microRNA genes innormal and neoplastic
cells. Cell Cycle. 6:1001–1005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bissels U, Wild S, Tomiuk S, et al:
Combined characterization ofmicroRNA and mRNA profiles delineates
early differentiation pathways of CD133+ and
CD34+ hematopoieticstem and progenitor cells. Stem
Cells. 29:847–857. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu L, Cai C, Wang X, Liu M, Li X and Tang
H: MicroRNA-142-3p, a new regulator of RAC1, suppresses
themigration and invasion of hepatocellular carcinoma cells. FEBS
Lett. 585:1322–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang P, Bill K, Liu J, et al: MiR-155 is
a liposarcoma oncogene that targets casein kinase-1α and enhances
β-catenin signaling. Cancer Res. 72:1751–1762. 2012. View Article : Google Scholar : PubMed/NCBI
|