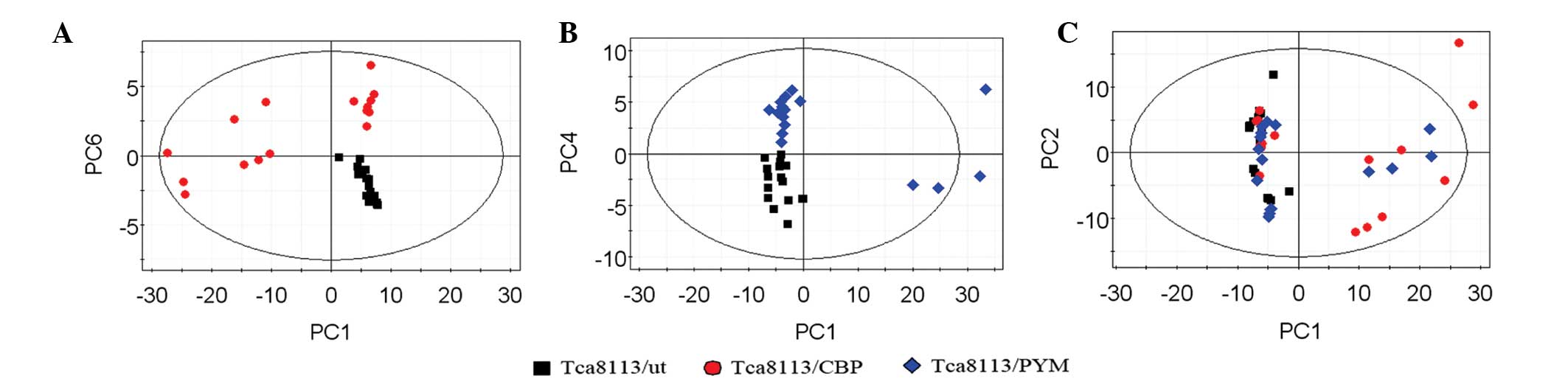
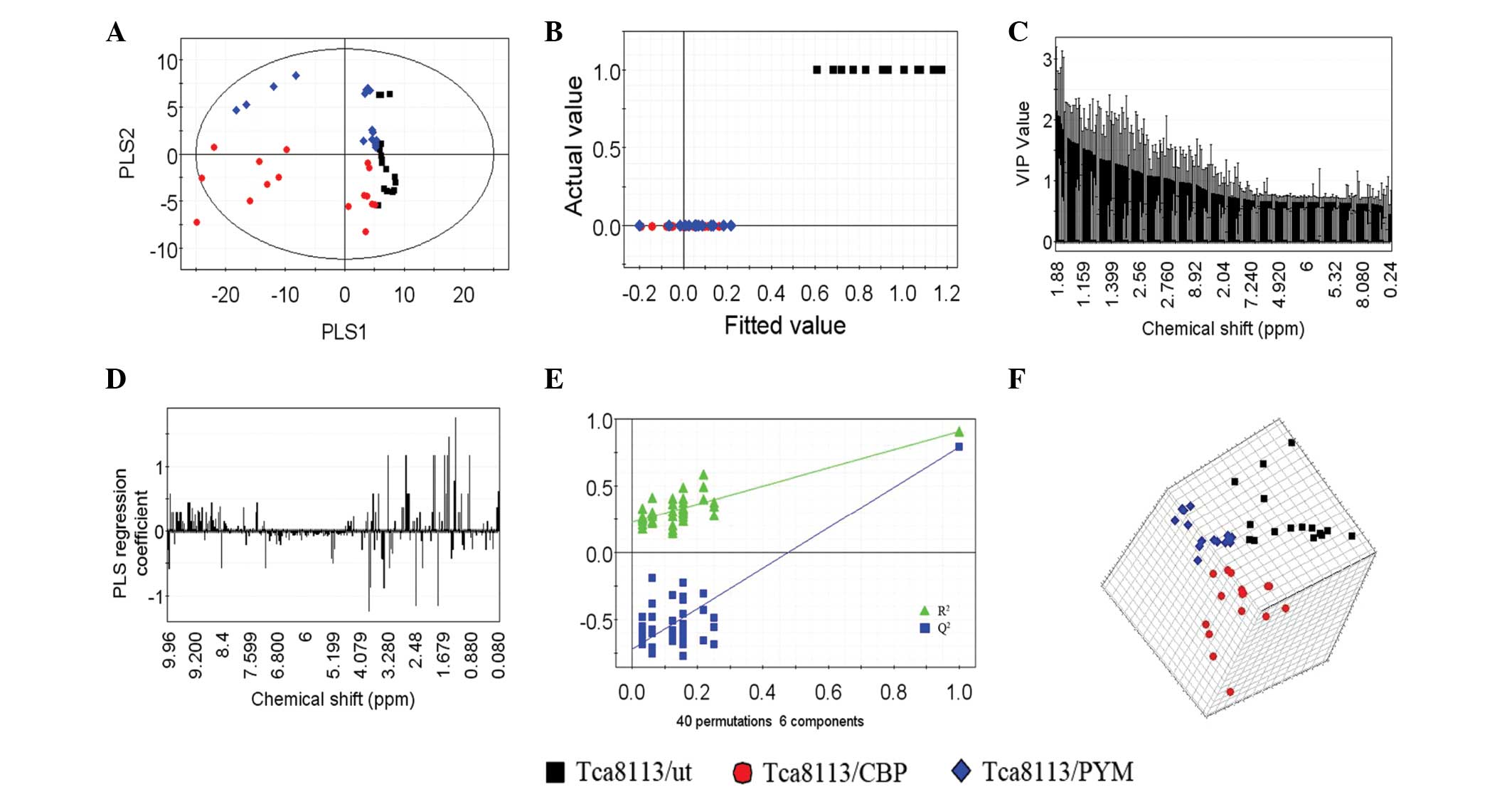
|
1
|
Fardel O, Lecureur V and Guillouzo A: The
P-glycoprotein multidrug transporter. Gen Pharmacol. 27:1283–1291.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstein LJ: MDR1 gene expression in
solid tumours. Eur J Cancer. 32A:1039–1050. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, Lu L, Feng Y, et al: PKD2 mediates
multi-drug resistance in breast cancer cells through modulation of
P-glycoprotein expression. Cancer Lett. 300:48–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teodori E, Dei S, Scapecchi S and
Gualtieri F: The medicinal chemistry of multidrug resistance (MDR)
reversing drugs. Farmaco. 57:385–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan S, Ma D, Ji M, et al: Expression
profile of Notch-related genes in multidrug resistant K562/A02
cells compared with parental K562 cells. Int J Lab Hematol.
32:150–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varma MV, Ashokraj Y, Dey CS and
Panchagnula R: P-glycoprotein inhibitors and their screening: a
perspective from bioavailability enhancement. Pharmacol Res.
48:347–359. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kars MD, Iseri OD, Gündüz U, Ural AU,
Arpaci F and Molnár J: Development of rational in vitro models for
drug resistance in breast cancer and modulation of MDR by selected
compounds. Anticancer Res. 26:4559–4568. 2006.PubMed/NCBI
|
|
8
|
Schmidt C: Metabolomics takes its place as
latest up-and-coming “omic” science. J Natl Cancer Inst.
96:732–734. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ai JY, Smith B and Wong DT: Bioinformatics
advances in saliva diagnostics. Int J Oral Sci. 4:85–87. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
German JB, Bauman DE, Burrin DG, et al:
Metabolomics in the opening decade of the 21st century building the
roads to individualized health. J Nutr. 134:2729–2732.
2004.PubMed/NCBI
|
|
11
|
Lindon JC, Holmes E and Nicholson JK: So
what's the deal with metabonomics? Anal Chem. 75:384A–391A. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ratajczak-Wrona W, Jablonska E, Antonowicz
B, Dziemianczyk D and Grabowska SZ: Levels of biological markers of
nitric oxide in serum of patients with squamous cell carcinoma of
the oral cavity. Int J Oral Sci. 5:141–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goodacre R, Vaidyanathan S, Dunn WB,
Harrigan GG and Kell DB: Metabolomics by numbers: acquiring and
understanding global metabolite data. Trends Biotechnol.
22:245–252. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nicholson JK, Lindon JC and Holmes E:
‘Metabonomics’: understanding the metabolic responses of living
systems to pathophysiological stimuli via multivariate statistical
analysis of biological NMR spectroscopic data. Xenobiotica.
29:1181–1189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bathen TF, Jensen LR, Sitter B, et al:
MR-determined metabolic phenotype of breast cancer in prediction of
lymphatic spread, grade and hormone status. Breast Cancer Res
Treat. 104:181–189. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pec J, Flores-Sanchez IJ, Choi YH and
Verpoorte R: Metabolic analysis of elicited cell suspension
cultures of Cannabis sativa L. by 1H-NMR spectroscopy. Biotechnol
Lett. 32:935–941. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seger C and Sturm S: Analytical aspects of
plant metabolite profiling platforms: current standings and future
aims. J Proteome Res. 6:480–497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim HK, Choi YH and Verpoorte R: NMR-based
metabolomic analysis of plants. Nat Protoc. 5:536–549. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Allwood JW, Clarke A, Goodacre R and Mur
LA: Dual metabolomics: a novel approach to understanding
plant-pathogen interactions. Phytochemistry. 71:590–597. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sheedy JR, Ebeling PR, Gooley PR and
McConville MJ: A sample preparation protocol for 1H nuclear
magnetic resonance studies of water-soluble metabolites in blood
and urine. Anal Biochem. 398:263–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barton RH, Waterman D, Bonner FW, et al:
The influence of EDTA and citrate anticoagulant addition to human
plasma on information recovery from NMR-based metabolic profiling
studies. Mol Biosyst. 6:215–224. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holmes E, Nicholls AW, Lindon JC, et al:
Development of a model for classification of toxin-induced lesions
using 1H NMR spectroscopy of urine combined with pattern
recognition. NMR Biomed. 11:235–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beckonert O, Monnerjahn J, Bonk U and
Leibfritz D: Visualizing metabolic changes in breast-cancer tissue
using 1H-NMR spectroscopy and self-organizing maps. NMR Biomed.
16:1–11. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yokota H, Guo J, Matoba M, Higashi K,
Tonami H and Nagao Y: Lactate, choline and creatine levels measured
by vitro 1H-MRS as prognostic parameters in patients with
non-small-cell lung cancer. J Magn Reson Imaging. 25:992–999. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Griffin JL and Kauppinen RA: Tumour
metabolomics in animal models of human cancer. J Proteome Res.
6:498–505. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tiziani S, Lopes V and Günther UL: Early
stage diagnosis of oral cancer using 1H NMR-based metabolomics.
Neoplasia. 11:269–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lucas LH, Larive CK, Wilkinson PS and Huhn
S: Progress toward automated metabolic profiling of human serum:
comparison of CPMG and gradient-filtered NMR analytical methods. J
Pharm Biomed Anal. 39:156–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Levitt MH: Spin Dynamics. Basics of
Nuclear Magnetic Resonance. 2nd. John Wiley & Sons; New York,
NY: 2008
|
|
29
|
Jackson JE: A User's Guide to Principal
Components. John Wiley & Sons, Inc.; New York, NY: pp. 20–150.
1991
|
|
30
|
Zhou J, Xu B, Huang J, et al: 1H NMR-based
metabonomic and pattern recognition analysis for detection of oral
squamous cell carcinoma. Clin Chim Acta. 401:8–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gavaghan CL, Wilson ID and Nicholson JK:
Physiological variation in metabolic phenotyping and functional
genomic studies: use of orthogonal signal correction and PLS-DA.
FEBS Lett. 530:191–196. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rocha CM, Barros AS, Gil AM, et al:
Metabolic profiling of human lung cancer tissue by 1H high
resolution magic angle spinning (HRMAS) NMR spectroscopy. J
Proteome Res. 9:319–332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang H, Wang Y, Nicholson JK and Lindon
JC: Use of relaxation-edited one-dimensional and two-dimensional
nuclear magnetic resonance spectroscopy to improve detection of
small metabolites in blood plasma. Anal Biochem. 325:260–272. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ala-Korpela M: 1H NMR spectroscopy of
human blood plasma. Prog Nucl Magn Reson Spectrosc. 27:475–554.
1995. View Article : Google Scholar
|
|
35
|
Wang L, Chen J, Chen L, et al: 1H-NMR
based metabonomic profiling of human esophageal cancer tissue. Mol
Cancer. 12:252013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lenz EM, Bright J, Wilson ID, Morgan SR
and Nash AF: A 1H NMR-based metabonomic study of urine and plasma
samples obtained from healthy human subjects. J Pharm Biomed Anal.
33:1103–1115. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nicholson JK, Foxall PJ, Spraul M, Farrant
RD and Lindon JC: 750 MHz 1H and 1H-13C NMR spectroscopy of human
blood plasma. Anal Chem. 67:793–811. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ulrich EL, Akutsu H, Doreleijers JF, et
al: BioMagResBank. Nucleic Acids Res. 36:(Database). D402–D408.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wishart DS, Tzur D, Knox C, et al: HMDB:
The human metabolome database. Nucleic Acids Res. 35:(Database).
D521–D526. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Briske-Anderson MJ, Finley JW and Newman
SM: The influence of culture time and passage number on the
morphological and physiological development of Caco-2 cells. Proc
Soc Exp Biol Med. 214:248–257. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Siissalo S, Laitinen L, Koljonen M, et al:
Effect of cell differentiation and passage number on the expression
of efflux proteins in wild type and vinblastine-induced Caco-2 cell
lines. Eur J Pharm Biopharm. 67:548–554. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goldsmith P, Fenton H, Morris-Stiff G,
Ahmad N, Fisher J and Prasad KR: Metabonomics: A useful tool for
the future surgeon. J Surg Res. 160:122–132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Holmes E, Foxall PJ, Nicholson JK, et al:
Automatic data reduction and pattern recognition methods for
analysis of 1H nuclear magnetic resonance spectra of human urine
from normal and pathological states. Anal Biochem. 220:284–296.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fauvelle F, Dorandeu F, Carpentier P, et
al: Changes in mouse brain metabolism following a convulsive dose
of soman: a proton HRMAS NMR study. Toxicology. 267:99–111. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schroeder FC: Small molecule signaling in
Caenorhabditis elegans. ACS Chem Biol. 1:198–200. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schroeder FC, Gibson DM, Churchill AC, et
al: Differential analysis of 2D NMR spectra: New natural products
from a pilot-scale fungal extract library. Angew Chem Int Ed Engl.
46:901–904. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bédouet L, Rusconi F, Rousseau M, et al:
Identification of low molecular weight molecules as new components
of the nacre organic matrix. Comp Biochem Physiol B Biochem Mol
Biol. 144:532–543. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vinay P, Prud'Homme M, Vinet B, et al:
Acetate metabolism and bicarbonate generation during hemodialysis:
10 years of observation. Kidney Int. 31:1194–1204. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhu Y, Eiteman MA, Lee SA and Altman E:
Conversion of glycerol to pyruvate by Escherichia coli using
acetate- and acetate/glucose-limited fed-batch processes. J Ind
Microbiol Biotechnol. 37:307–312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Meng PH, Raynaud C, Tcherkez G, et al:
Crosstalks between myo-inositol metabolism, programmed cell death
and basal immunity in Arabidopsis. PLoS One. 4:e73642009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Downes CP, Gray A and Lucocq JM: Probing
phosphoinositide functions in signaling and membrane trafficking.
Trends Cell Biol. 15:259–268. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Malan TP Jr and Porreca F: Lipid mediators
regulating pain sensitivity. Prostaglandins Other Lipid Mediat.
77:123–130. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Loewus FA and Murthy PP: Myo-Inositol
metabolism in plants. Plant Sci. 150:1–19. 2000. View Article : Google Scholar
|