Introduction
Osteosarcoma (OS) is the most common primary
malignant bone tumor amongst children and adolescents, and is
localized to the metaphysis of the long bones (1). Through a combination of chemotherapy and
advanced surgery, the 5-year survival rate for patients with OS has
significantly improved, to 70% (2).
However, a considerable proportion of OS patients possess a
significant risk of local relapse or distant metastasis, even
subsequent to neoadjuvant chemotherapy and beneficial surgery
(3). Local and metastatic relapses
have been consistently demonstrated to markedly reduce survival
(4,5).
Although numerous studies have focused on improving the treatment
of OS in order to enhance patient prognosis, early detection of
cancer remains highly recommended for the improvement of survival
rates (6).
MicroRNAs (miRNAs), initially discovered in
Caenorhabditis elegans as products of the Lin-4 gene
(7), are a family of small
non-protein coding RNAs that regulate a wide variety of genes at
the post-transcriptional level. miRNAs have a critical role in
numerous biological and pathological processes, including cell
proliferation, differentiation and apoptosis (8,9). It was
reported that miRNAs modulate almost 60% of protein-coding genes in
humans by base-pairing with the 3′-untranslated regions of their
target miRNAs (10), resulting in
messenger RNA (mRNA) degradation or translation inhibition. In
addition to this silencing effect, certain miRNAs are able to
activate gene expression (11).
Attention has turned to the examination of the role of miRNAs in
carcinogenesis. In cancer, miRNAs may function as oncogenes and/or
tumor suppressors through various mechanisms, including deletions,
amplifications or mutations in miRNA loci, as well as epigenetic
changes, dysregulation of transcription factors targeting specific
miRNAs or inhibition of miRNA processing (12).
Following the initial demonstration that elevated
serum levels of miRNA-21 were associated with relapse-free survival
of diffuse large B-cell lymphoma patients (13), several circulating miRNAs were
identified as potential diagnostic and prognostic biomarkers for
various types of cancer, including nasopharyngeal carcinoma
(14) and glioma (15), as well as breast (6,16,17), gastric (18,19),
prostate (20,21), pancreatic (22) and colorectal cancer (23). As a tumor screening marker,
circulating miRNAs have several advantages, including the
availability of serum samples and their stability during storage
relative to that of tissue samples. Serum miRNAs are particularly
stable, even following exposure to harsh conditions, for example
high/low pH, boiling and multiple freeze-thaw cycles (11,20).
The aim of the present study was to identify
potential circulating miRNA biomarkers for the early diagnosis and
relapse prediction of osteosarcoma.
Materials and methods
Patients
Serum samples were collected from 20 consecutive OS
patients at the Second Xiangya Hospital of Central South University
(Changsha, China) between 2012 and 2013. All patients exhibited a
positive diagnosis for OS based on pathological analysis of
biopsies or surgical samples. All OS samples were collected prior
to any therapeutic interventions. The 20 gender and age-matched
controls were also recruited from the Second Xiangya Hospital, and
were confirmed to be free of any other types of cancer or chronic
disease. The demographics and clinicopathological characteristics
of the patients are indicated in Table
I. The present study was approved by the Ethics Committee of
the Second Xiangya Hospital. Furthermore, informed consent was
obtained from all participants included in the project prior to
blood sampling.
 | Table I.Demographic and clinical features of
OS patients and healthy individuals. |
Table I.
Demographic and clinical features of
OS patients and healthy individuals.
| Variable | OS patients | Controls | P-value |
|---|
| Age, years | 13±4.38 | 14.3±4.5 | 0.360 |
| Gender, n |
|
|
|
|
Male | 13 | 12 | 0.744 |
|
Female | 7 | 8 |
|
| Tumor location,
n |
|
|
|
|
Femur | 13 |
|
|
|
Tibia | 5 |
|
|
|
Others | 2 |
|
|
| Enneking stage,
n |
|
|
|
|
IIA | 7 |
|
|
|
IIB | 11 |
|
|
|
III | 2 |
|
|
RNA isolation
Peripheral blood samples (5 ml) were collected from
all participants according to protocols approved by the Second
Xiangya Hospital. Whole blood samples were stored at room
temperature for 60 min then centrifuged at 1000 × g for 10 min at
4°C. The serum samples were stored in phased liquid nitrogen or
used for RNA isolation directly following collection. Total RNA was
isolated from 250 µl human serum using TRIzol-LS Reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. A total of 5 µl artificial microRNA
(cel-miR-39; RiboBio, Guangzhou, China) was added to each sample
prior to the isolation procedure to serve as an internal
control.
miRNA profiling
The expression levels of 168 unique human miRNAs,
previously selected from an earlier study (3) and the Exiqon company database
(http://www.exiqon.com/plate-layout-files), were
detected using the miRCURY LNA Universal reverse transcription (RT)
microRNA polymerase chain reaction (PCR) system and ready to use
Serum/Plasma Focus microRNA PCR Panel (V3.M; Exiqon, Kangchen,
China). Each Exiqon miRNA quantitative PCR panel included 168
target miRNA probes, 5 internal control primer sets and 2 reference
miRNA probes (miR-103-3p and miR-191-5p). The PCR amplification was
performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10
sec and 60°C for 60 sec and subsequent melting curve analysis. The
expression levels of miRNAs were normalized to the internal
controls and evaluated using the ΔΔCt method, where ΔCt was
calculated by subtracting the average Ct values of reference miRNA
from the average Ct value of the miRNA of interest.
RT-qPCR
RT and qPCR kits produced specifically for accurate
miRNA analysis (GeneCopoeia, Inc., Rockville, MD, USA) were used to
evaluate the expression of miRNAs in serum samples. Equal
quantities of RNA (1000 ng/reaction) were reverse-transcribed. The
25 µl RT reactions were performed using an All-in-One™ miRNA
RT-qPCR Detection kit (catalog no. AOMD-Q050; GeneCopoeia, Inc.)
and incubated for 60 min at 37°C, 5 min at 85°C and then maintained
at 4°C. RT products were subsequently diluted 3-fold to a total of
75 µl. For PCR analysis, 2 µl diluted RT products were used as
templates in 20 µl reaction mixtures containing primers for each
miRNA, according to the manufacturer's instructions. All reactions
were run on the 7900HT Fast Real-Time PCR System (Life
Technologies, Grand Island, NY, USA) using the following
conditions: 95°C for 10 min, followed by 50 cycles at 95°C for 15
sec, 60°C for 20 sec and 72°C for 15 sec. Each sample was run in
duplicate for analysis. The relative miRNA quantities in serum from
participants were determined using the comparative Ct method, and
the miRNA levels were normalized to cel-miR-39. Fold-change of
miRNA expression between groups was determined using the ΔΔCt
equation, where ΔCt was calculated by subtracting the Ct values of
cel-miR-39 from the average Ct value of the miRNA of interest.
Statistical analysis
Data is presented as the mean ± standard deviation.
Differences between groups were analyzed using a two-tailed
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference. Receiver operating
characteristic (ROC) and the area under ROC curve (AUC) were used
to estimate the diagnostic value of candidate miRNAs in OS. Cut-off
values for the expression of each miRNA were defined according to
the Youden index from the ROC curve. All statistical analyses were
performed by using GraphPad PRISM software, version 5 (GraphPad
Software, La Jolla, CA, USA).
Results
miRNA profiling of pre-therapeutic OS
patients
Three samples from patients with pre-therapeutic OS
and three healthy individual samples were selected for the
identification of differentially expressed miRNAs. A volcano plot
was constructed to present the results of miRNAs profiling, which
are presented in Fig. 1. In the plot,
the black line indicates a fold-change of 1 and the pink lines
indicate a 1.8 fold-change in gene expression, while the blue line
indicates the threshold P-value of 0.05 for the t-test. Fourteen
candidate miRNAs were selected using the following inclusion
criteria: i) The Ct value of each miRNA was <35; ii) the
fold-difference between the two groups was >1.8; and iii) the
value of P was <0.05. Due to the small size of the sample for
screening, a 1.8-fold change was used as the cut-off value to avoid
eliminating potentially significant biomarkers (3). The identified miRNAs have been reported
in previous studies as being differentially expressed in OS or
other tumors. Information regarding these 14 miRNAs is summarized
in Table II. The initial selection
of 14 miRNAs was further validated in the larger 20 patient and 20
control cohorts. The following 14 miRNAs were shown to be
differentially expressed: miR-451a, miR-551b-3p, miR-20a-5p,
miR-34a-5p, miR-2110, miR-95-3p, miR-16-5p, miR-186-5p, miR-320a,
miR-106a-5p, miR-25-3p, miR-223-5p, miR-139-5p and miR-425-5p.
 | Table II.Differentially expressed serum
microRNAs in osteosarcoma, identified by reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Differentially expressed serum
microRNAs in osteosarcoma, identified by reverse
transcription-quantitative polymerase chain reaction.
| MicroRNA |
Fold-changea | Ct
valueb | P-value |
|---|
| miR-451a | −2.40 | 23.31±0.25 | 0.0191 |
| miR-551b-3p | −4.94 | 34.79±0.49 | 0.0070 |
| miR-20a-5p | −2.00 | 25.53±0.16 | 0.0055 |
| miR-34a-5p |
2.48 | 31.63±0.42 | 0.0057 |
| miR-2110 | −2.70 | 32.45±0.37 | 0.0096 |
| miR-95-3p |
2.48 | 33.63+0.03 | 0.0011 |
| miR-16-5p | −1.96 | 21.61±0.05 | 0.0364 |
| miR-186-5p | −1.97 | 30.52±0.26 | 0.0044 |
| miR-320a |
1.97 | 27.15±0.13 | 0.0029 |
| miR-106a-5p | −1.91 | 25.36±0.26 | 0.0122 |
| miR-25-3p | −1.89 | 25.36±0.14 | 0.0023 |
| miR-223-5p | −1.91 | 31.18±0.14 | 0.0228 |
| miR-139-5p | −1.84 | 29.05±0.13 | 0.0305 |
| miR-425-5p | −1.90 | 26.38±0.37 | 0.0368 |
Validation of candidate miRNAs
To confirm the results of the miRNA profiling, miRNA
levels in serum were examined by RT-qPCR in the 20 pre-therapeutic
OS patients and 20 healthy controls. The 14 differentially
expressed miRNAs were validated in the larger cohort by RT-qPCR;
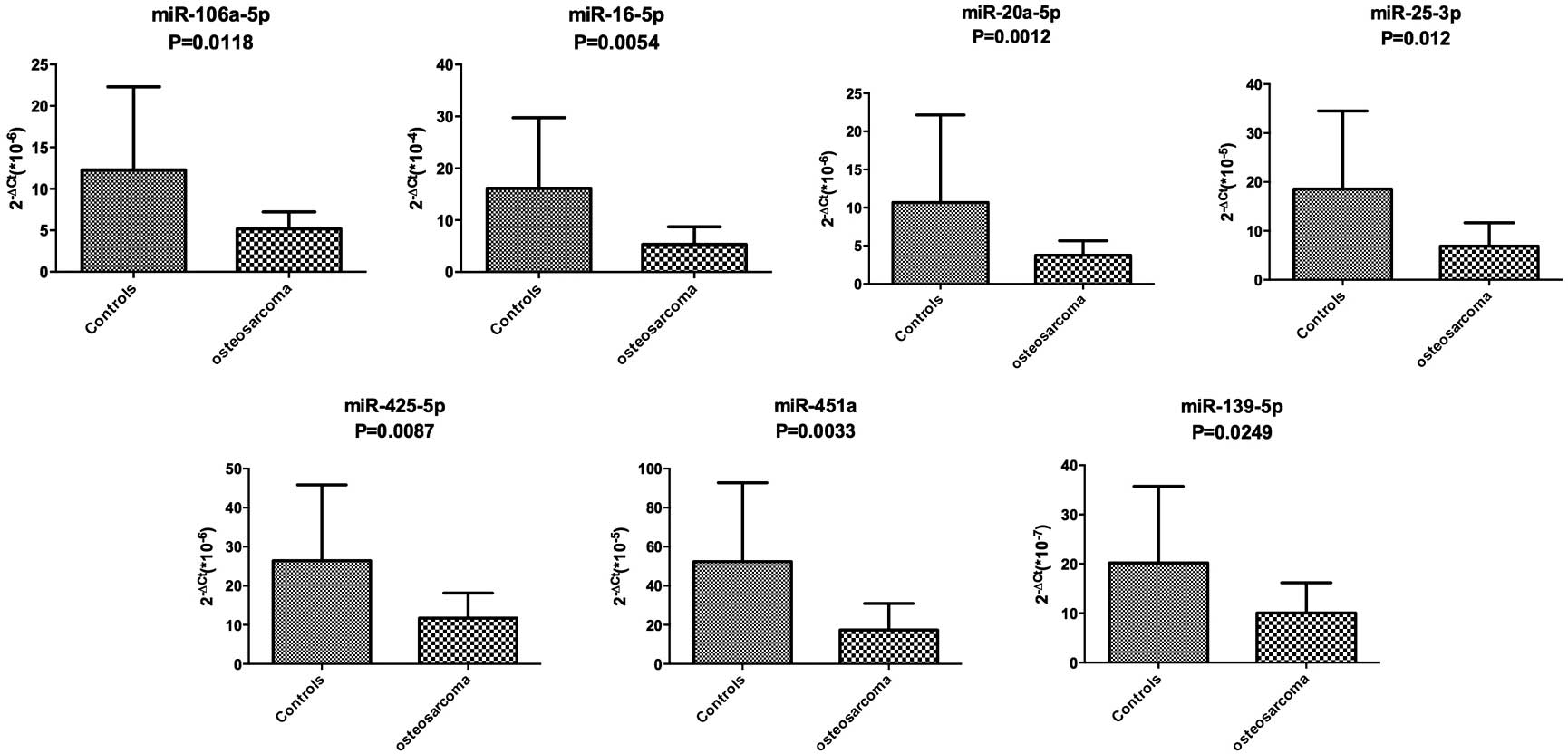
however, only 7 miRNAs demonstrated a statistically significant
difference. The data indicating that the expression of 7 miRNAs was
downregulated in the pre-therapeutic OS group were validated by
RT-qPCR analysis when compared with that of the control group
(Fig. 2). The data was analyzed using
the ΔΔCt equation as previously described.
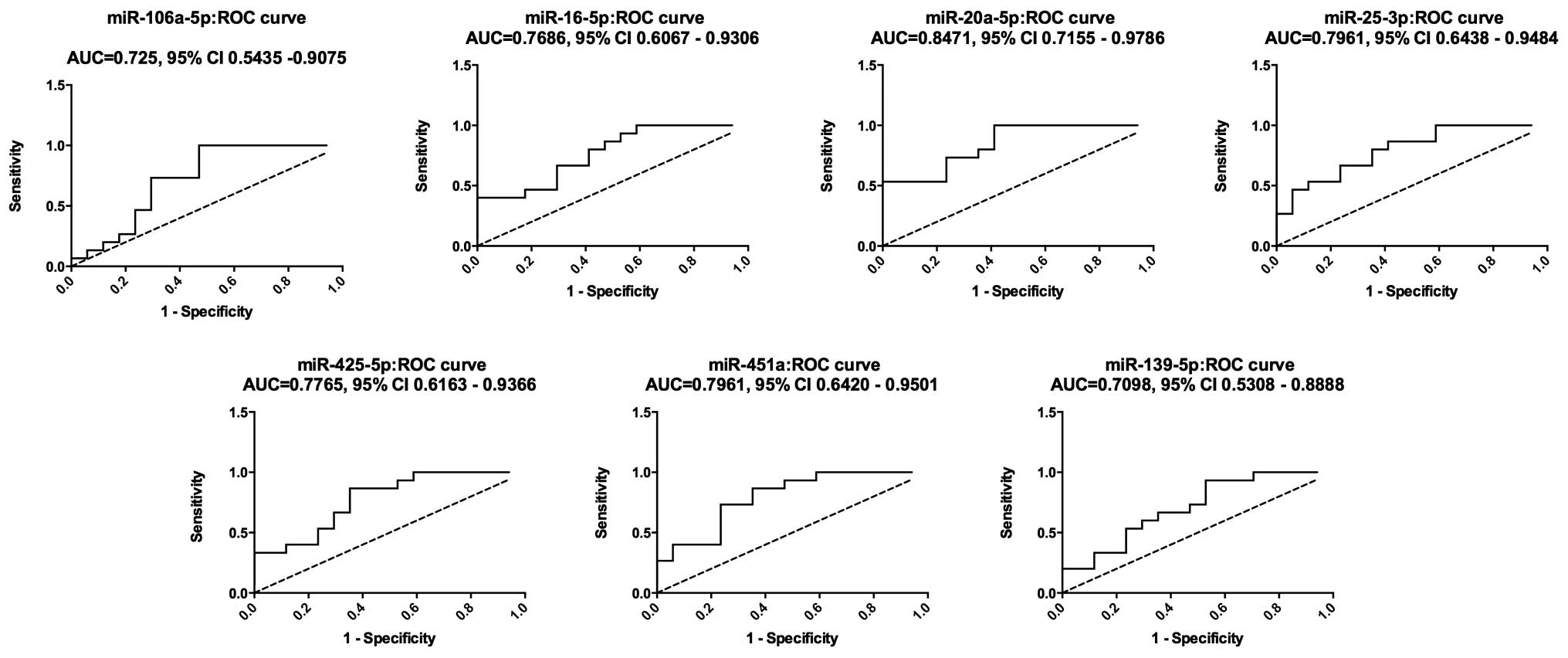
An ROC curve was constructed to estimate the
sensitivity and specificity of these 7 miRNAs as a means of
discriminating OS patients from healthy controls. The AUC for the
expression of these 7 miRNAs in serum for OS diagnosis were 0.7255
[95% confidence interval (CI), 0.5435–0.9075], 0.7686 (95% CI,
0.6067–0.9306), 0.8471 (95% CI, 0.7155–0.9786) and 0.7961 (95% CI,
0.6438–0.9484) for miR-106a-5p, miR-16-5p, miR-20a-5p and
miR-25-3p, respectively; while the AUC for miR-425-5p, miR-451a and
miR-139-5p were 0.7765 (95% CI, 0.6163–0.9366), 0.7961 (95% CI,
0.6420–0.9501) and 0.7098 (95% CI, 0.5308–0.8888), respectively
(Fig. 3). These results indicated
that these 7 miRNAs may be used as diagnostic biomarkers, with the
ability to resolve OS patients from the healthy cohort.
In addition, in order to confirm the prognostic
value of the 7 miRNAs at early stages of OS, the serum levels of
these 7 miRNAs were analyzed in patients with OS at different
Enneking stages. No significant differences were noted between
miRNA expression in OS patients at Enneking IIA and Enneking IIB.
Due to the small sample size of Enneking III (2 samples),
statistical analysis between Enneking IIA/B and Enneking III was
not performed.
Discussion
OS is the most common primary malignant bone tumor
amongst adolescents and young adults (2,24).
However, the molecular mechanisms underlying disease development
have remained elusive. Typically, diagnosis of OS is initiated by
patient complaints, followed by imaging tests, including bone
X-ray, magnetic resonance imaging (MRI) and computed tomography,
with a final diagnosis often requiring confirmation by biopsy
(2,24). This series of examinations/tests are
time-consuming and expensive, particularly in developing countries.
Furthermore, the majority of patients with OS are diagnosed at an
advanced stage. The patients included in the present study all
exhibited OS beyond Enneking stage IIA. The identification of novel
screening biomarkers represents a promising technique for
facilitation of the early detection of OS, which may improve
treatment outcomes.
The present study demonstrated the utility of miRNA
profiling as a means of diagnosing OS. Previous studies reported
the expression of miR-9, miR-99, miR-195, miR-148a and miR-181a to
be elevated in OS cell lines, while miR-143, miR-145, miR-335 and
miR-539 were decreased (25). Another
group reported elevated levels of miR-181a, miR-181b, and miR-181c
in human osteosarcoma tissues, while miR-16, miR-29b and miR-142-5p
were downregulated (26). Multiple
target genes of miRNAs and associated downstream signaling pathways
have been identified, which are correlated with the pathogenesis
and progression of OS. For example, miR-221 stimulates cell
survival and cisplatin resistance by targeting the phosphatase and
tensin homolog (PTEN) gene in the phosphoinositide 3-kinase/Akt
pathway (27). Furthermore, increased
levels of miR-128 are negatively correlated with PTEN levels, and
increased proliferation of MG63 and U2OS OS cells (28). In contrast to these oncogene-like
miRNA functions, miR-335 and miR-340 function as tumor suppressors
by targeting the Rho-associated, coiled-coil containing protein
kinase 1 gene and, thereby, inhibit OS cell migration and invasion
(29,30). miR-126 inhibits the proliferation of
OS cells by targeting sirt1, a histone deacetylase (31). A previous study by our group confirmed
that miR-125, which is typically downregulated in OS samples,
negatively regulates signal transducer and activator of
transcription 3, which suppresses proliferation and migration of OS
cells (32).
Based on the differential expression patterns of
miRNAs in OS, numerous miRNAs have been identified as potentially
valuable diagnostic biomarkers. Compared with their tissue
counterparts, the potential significance of circulating miRNAs in
OS diagnosis is poorly understood. In the present study,
miR-106a-5p, miR16-5p, miR-20a-5p, miR-425-5p, miR451a, miR-25-3P
and miR139-5p were demonstrated to be downregulated in the serum of
patients with OS when compared with that of healthy controls.
These 7 miRNAs may also be involved in the
pathogenesis of other cancers. miR-106a was previously shown to be
downregulated in glioma, colon cancer, squamous cell carcinoma and
astrocytoma, and functions as a tumor suppressor by targeting
solute carrier family 2 (facilitated glucose transporter), member 3
(33), E2F transcription factor 1
(34) and Fas-activated
serine/threonine kinase (35).
Similarly, miR-20a is downregulated in hepatocellular carcinoma
(HCC) and correlated with HCC recurrence and poor prognosis
(36). In addition, miR-425 is able
to reduce proliferation, impair tumorigenesis and metastasis, and
increase expression of epithelial markers in aggressive breast
cancer cells by targeting SATB homeobox 1, CCND2 and Fascin
actin-bundling protein 1 (37). The
tumor suppressive role of miR-16 was also confirmed by in
vitro and in vivo functional experiments associated with
OS (26). In these studies, miR-16,
which is reduced in OS, was shown to inhibit cell proliferation by
targeting the insulin-like growth factor 1 receptor and
Raf1-mitogen activated protein kinase kinase 1/2-extracellular
signal-regulated kinase 1/2 pathways (38). Furthermore, miR-451 has been shown to
act as a tumor suppressor in various human malignancies, including
nasopharyngeal carcinoma (39),
colorectal carcinoma (40), lung
cancer (41), renal cell carcinoma
(42) and gastric cancer (43). Cisplatin sensitivity was increased by
overexpression of miR-451 in lung cancer cells (41). Another downregulated miRNA, miR-25,
may function as a tumor suppressor that inhibits cell proliferation
and migration by targeting Smad 7 in colon cancer (44) and EZH2 in anaplastic thyroid carcinoma
(45). Finally, studies have
suggested a tumor suppressor role for miR-139-5p, which is
downregulated in various types of cancer, including endometrial
serous adenocarcinoma (46), breast
carcinoma (47), glioblastoma
(48), bladder cancer (49), basal cell carcinoma (50) and esophageal squamous cell carcinoma
(51).
Consistent with the results of prior studies, the
present findings confirmed that the expression of these 7 miRNAs
were reduced in the serum of OS patients, which is concurrent with
their function as tumor suppressor genes. However, an oncogenetic
function was proposed for a number of these miRNAs, based on their
ability to target tumor suppressor genes. The high expression of
miR-106a was previously demonstrated to indicate a high risk of
tumor penetration in advanced gastric carcinoma (52) and mediate proliferation and tumor
differentiation in ovarian cancer through direct targeting of
retinoblastoma-like protein 2, a member of the retinoblastoma tumor
suppressor family (53). High
expression levels of miR-20a are associated with lymph node
metastasis in advanced gastric carcinoma (52). Recently, a report indicated that
miR-20 was able to downregulate Fas expression, thus contributing
to the metastatic potential of OS cells (54). Furthermore, miR-425 was detected at
high levels in gastric cancer (55).
Hummel et al (56) revealed
that miR-425 was also upregulated in esophageal carcinoma cells
following short- or long-term treatment with cisplatin. miR-25,
which is overexpressed in ovarian cancer, gastric cancer and
esophageal squamous cell carcinoma, promotes tumorigenesis and
metastasis by targeting Bim, reversion-inducing cysteine-rich
protein with Kazal motifs and cadherin 1, respectively (57–59).
In view of the controversial roles of miRNAs in
various tumors, it is important for discrepancies between studies
to be clarified. Heterogeneity of primary tumors may be an
explanation for the differential expression profiles of identical
miRNAs in various tumors. It was also reported that the levels of
circulating miRNAs may differ from those in tissues (60,61), which
may be explained by the differential secretory mechanisms and/or
stability of miRNAs in blood (12,62). Thus,
circulating miRNAs may not necessarily reflect changes in
expression within the tumor tissue. In addition, the extraction
quantification methods used may contribute to the conflicting
results. In addition, to the best of our knowledge, there is no
commonly accepted internal control for this type of analysis,
therefore the various artificial miRNAs or endogenous housekeeping
genes used for normalization may affect the final results.
Although the results presented in the present study
are promising, several limitations should be acknowledged: i) The
OS patient sample size was relatively small and therefore
validation in larger patient cohorts is required; ii) research on
the association between serum miRNA levels and patient survival
remains a critical future goal; and iii) the association between
tissue and serum miRNA levels requires further investigation.
In conclusion, the present study indicated that
changes in the serum profile of 7 miRNAs are associated with OS.
The results indicated that these 7 miRNAs may be used as potential
circulating biomarkers in the future and provide novel
opportunities for early blood screening for OS.
Acknowledgements
The present study was supported by the Scientific
Innovation Research Program for College Graduates in Hunan province
(grant no. CX2012B096) and grants from the National Natural Science
Foundation of China (grant nos. 81372871 and 81302339).
References
|
1
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hameed M and Dorfman H: Primary malignant
bone tumors - recent developments. Semin Diagn Pathol. 28:86–101.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma. 2012:3597392012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weeden S, Grimer RJ, Cannon SR, Taminiau
AH and Uscinska BM: The effect of local recurrence on survival in
resected osteosarcoma. Eur J Cancer. 37:39–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harting MT, Blakely ML, Jaffe N, Cox CS
Jr, Hayes-Jordan A, Benjamin RS, Raymond AK, Andrassy RJ and Lally
KP: Long-term survival after aggressive resection of pulmonary
metastases among children and adolescents with osteosarcoma. J
Pediatr Surg. 41:194–199. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soifer HS, Rossi JJ and Saetrom P:
MicroRNAs in disease and potential therapeutic applications. Mol
Ther. 15:2070–2079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma R, Jiang T and Kang X: Circulating
microRNAs in cancer: Origin, function and application. J Exp Clin
Cancer Res. 31:382012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumour-associated microRNAs in
serum of patients with diffuse large B-cell lymphoma. Br J
Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng X, Xiang J, Wu M, Xiong W, Tang H,
Deng M, Li X, Liao Q, Su B, Luo Z, et al: Circulating miR-17,
miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers
in nasopharyngeal carcinoma. PLoS One. 7:e463672012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Q, Li P, Li A, Jiang W, Wang H, Wang
J and Xie K: Plasma specific miRNAs as predictive biomarkers for
diagnosis and prognosis of glioma. J Exp Clin Cancer Res.
31:972012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma
ES, Pang R, Chua D, Chu KM, Law WL, et al: Circulating microRNAs as
specific biomarkers for breast cancer detection. PLoS One.
8:e531412013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H
and Hu C: Circulating microRNA-92a and microRNA-21 as novel
minimally invasive biomarkers for primary breast cancer. J Cancer
Res Clin Oncol. 139:223–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY
and Zhu ZG: MiRNA-199a-3p: A potential circulating diagnostic
biomarker for early gastric cancer. J Surg Oncol. 108:89–92. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Valladares-Ayerbes M, Reboredo M,
Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M,
Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, et al:
Circulating miR-200c as a diagnostic and prognostic biomarker for
gastric cancer. J Transl Med. 10:1862012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sita-Lumsden A, Dart DA, Waxman J and
Bevan CL: Circulating microRNAs as potential new biomarkers for
prostate cancer. Br J Cancer. 108:1925–1930. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morimura R, Komatsu S, Ichikawa D,
Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H,
Okamoto K, et al: Novel diagnostic value of circulating miR-18a in
plasma of patients with pancreatic cancer. Br J Cancer.
105:1733–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Menéndez P, Padilla D, Villarejo P,
Palomino T, Nieto P, Menéndez JM and Rodríguez-Montes JA:
Prognostic implications of serum microRNA-21 in colorectal cancer.
J Surg Oncol. 108:369–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fox MG and Trotta BM: Osteosarcoma: Review
of the various types with emphasis on recent advancements in
imaging. Semin Musculoskelet Radiol. 17:123–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu H, Zhang Y, Cai XH, Huang JF and Cai L:
Changes in microRNA expression in the MG-63 osteosarcoma cell line
compared with osteoblasts. Oncol Lett. 4:1037–1042. 2012.PubMed/NCBI
|
|
26
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen L, Chen XD and Zhang YH: MicroRNA-128
promotes proliferation in osteosarcoma cells by downregulating
PTEN. Tumour Biol. 35:2069–2074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Zhao W and Fu Q: miR-335
suppresses migration and invasion by targeting ROCK1 in
osteosarcoma cells. Mol Cell Biochem. 384:105–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou X, Wei M and Wang W: MicroRNA-340
suppresses osteosarcoma tumor growth and metastasis by directly
targeting ROCK1. Biochem Biophys Res Commun. 437:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu JQ, Liu P, Si MJ and Ding XY:
MicroRNA-126 inhibits osteosarcoma cells proliferation by targeting
Sirt1. Tumour Biol. 34:3871–3877. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu LH, Li H, Li JP, Zhong H, Zhang HC,
Chen J and Xiao T: miR-125b suppresses the proliferation and
migration of osteosarcoma cells through down-regulation of STAT3.
Biochem Biophys Res Commun. 416:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dai DW, Lu Q, Wang LX, Zhao WY, Cao YQ, Li
YN, Han GS, Liu JM and Yue ZJ: Decreased miR-106a inhibits glioma
cell glucose uptake and proliferation by targeting SLC2A3 in GBM.
BMC Cancer. 13:4782013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Díaz R, Silva J, García JM, Lorenzo Y,
García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F and
Domínguez G: Deregulated expression of miR-106a predicts survival
in human colon cancer patients. Genes Chromosomes Cancer.
47:794–802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang
Q, Zhang Y, Wang R, Xue L, Wang S, et al: miR-106a-5p inhibits the
proliferation and migration of astrocytoma cells and promotes
apoptosis by targeting FASTK. PLoS One. 8:e723902013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Cancer Res. 32:212013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Di Leva G, Piovan C, Gasparini P, Ngankeu
A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder
H, et al: Estrogen mediated-activation of miR-191/425 cluster
modulates tumorigenicity of breast cancer cells depending on
estrogen receptor status. PLoS Genet. 9:e10033112013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen L, Wang Q, Wang GD, Wang HS, Huang Y,
Liu XM and Cai XH: miR-16 inhibits cell proliferation by targeting
IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS
Lett. 587:1366–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu N, Jiang N, Guo R, Jiang W, He QM, Xu
YF, Li YQ, Tang LL, Mao YP, Sun Y and Ma J: MiR-451 inhibits cell
growth and invasion by targeting MIF and is associated with
survival in nasopharyngeal carcinoma. Mol Cancer. 12:1232013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li HY, Zhang Y, Cai JH and Bian HL:
MicroRNA-451 inhibits growth of human colorectal carcinoma cells
via downregulation of Pi3k/Akt pathway. Asian Pac J Cancer Prev.
14:3631–3634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bian HB, Pan X, Yang JS, Wang ZX and De W:
Upregulation of microRNA-451 increases cisplatin sensitivity of
non-small cell lung cancer cell line (A549). J Exp Clin Cancer Res.
30:202011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Redova M, Poprach A, Nekvindova J, Iliev
R, Radova L, Lakomy R, Svoboda M, Vyzula R and Slaby O: Circulating
miR-378 and miR-451 in serum are potential biomarkers for renal
cell carcinoma. J Transl Med. 10:552012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bandres E, Bitarte N, Arias F, Agorreta J,
Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ,
et al: MicroRNA-451 regulates macrophage migration inhibitory
factor production and proliferation of gastrointestinal cancer
cells. Clin Cancer Res. 15:2281–2290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H,
Wang W, Zhang L, Zhang X, Tang Q, et al: MicroRNA-25 functions as a
potential tumor suppressor in colon cancer by targeting Smad7.
Cancer Lett. 335:168–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Esposito F, Tornincasa M, Pallante P,
Federico A, Borbone E, Pierantoni GM and Fusco A: Down-regulation
of the miR-25 and miR-30d contributes to the development of
anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J
Clin Endocrinol Metab. 97:E710–E718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li RY, Chen LC, Zhang HY, Du WZ, Feng Y,
Wang HB, Wen JQ, Liu X, Li XF, Sun Y, et al: MiR-139 inhibits Mcl-1
expression and potentiates TMZ-induced apoptosis in glioma. CNS
Neurosci Ther. 19:477–483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S and Jung K: miRNA profiling identifies candidate miRNAs for
bladder cancer diagnosis and clinical outcome. J Mol Diagn.
15:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang M, Liu R, Sheng J, Liao J, Wang Y,
Pan E, Guo W, Pu Y and Yin L: Differential expression profiles of
microRNAs as potential biomarkers for the early diagnosis of
esophageal squamous cell carcinoma. Oncol Rep. 29:169–176.
2013.PubMed/NCBI
|
|
52
|
Kim BH, Hong SW, Kim A, Choi SH and Yoon
SO: Prognostic implications for high expression of oncogenic
microRNAs in advanced gastric carcinoma. J Surg Oncol. 107:505–510.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Z, Gersbach E, Zhang X, Xu X, Dong R,
Lee P, Liu J, Kong B, Shao C and Wei JJ: miR-106a represses the Rb
tumor suppressor p130 to regulate cellular proliferation and
differentiation in high-grade serous ovarian carcinoma. Mol Cancer
Res. 11:1314–1325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang G, Nishimoto K, Zhou Z, Hughes D and
Kleinerman ES: miR-20a encoded by the miR-17-92 cluster increases
the metastatic potential of osteosarcoma cells by regulating Fas
expression. Cancer Res. 72:908–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M, Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
57
|
Zhang H, Zuo Z, Lu X, Wang L, Wang H and
Zhu Z: MiR-25 regulates apoptosis by targeting Bim in human ovarian
cancer. Oncol Rep. 27:594–598. 2012.PubMed/NCBI
|
|
58
|
Zhao H, Wang Y, Yang L, Jiang R and Li W:
MiR-25 promotes gastric cancer cells growth and motility by
targeting RECK. Mol Cell Biochem. 385:207–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang
Z, Tan F, Tan X, Zhou F, Sun J, et al: MicroRNA-25 promotes cell
migration and invasion in esophageal squamous cell carcinoma.
Biochem Biophys Res Commun. 421:640–645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cookson VJ, Bentley MA, Hogan BV, Horgan
K, Hayward BE, Hazelwood LD and Hughes TA: Circulating microRNA
profiles reflect the presence of breast tumours but not the
profiles of microRNAs within the tumours. Cell Oncol (Dordr).
35:301–308. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Boeri M, Verri C, Conte D, Roz L, Modena
P, Facchinetti F, Calabrò E, Croce CM, Pastorino U and Sozzi G:
MicroRNA signatures in tissues and plasma predict development and
prognosis of computed tomography detected lung cancer. Proc Natl
Acad Sci USA. 108:3713–3718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Brase JC, Wuttig D, Kuner R and Sültmann
H: Serum microRNAs as non-invasive biomarkers for cancer. Mol
Cancer. 9:3062010. View Article : Google Scholar : PubMed/NCBI
|