Introduction
Osteosarcoma (OS) is the most common primary bone
malignancy, which mainly occurs in children and young adolescents
(1–3).
OS is a complex tumor, characterized by numerous chromosomal
alterations and extensive gene mutations (4,5). Numerous
molecular studies of OS have been undertaken in recent years, the
results of which have provided insight into the molecular
pathogenesis of OS (4–9). However, the fundamental molecular
mechanisms underlying the occurrence and development of this
sarcoma remain to be fully elucidated. Despite significant advances
in the development of multimodality treatments comprising wide
tumor excision with aggressive adjuvant chemotherapy, the prognosis
of patients with recurrent or metastatic OS remains poor (10,11).
Therefore, the identification of novel molecular biomarkers to
facilitate the early diagnosis and therapy of OS is required, in
order to improve the clinical outcome of patients with OS (12).
MicroRNAs (miRNAs) are small non-coding RNAs of ~22
nucleotides, which have emerged as a major class of regulatory
genes in animals and plants (13,14).
miRNAs are estimated to regulate >30% of the human genome, and
are therefore involved in diverse functions, including development,
cell differentiation, regulation of the cell cycle and apoptosis
(15). A number of studies have
demonstrated that miRNA alterations are involved in the development
of human cancer (16,17). miRNA genes were frequently
demonstrated to be located in cancer-associated genomic regions or
fragile sites, suggesting that miRNAs in the genome may be
extensively involved in cancer (18).
In addition, miRNAs may function as oncogenes or tumor suppressors
depending on the nature of their targets (19–21).
Overexpressed miRNAs in various types of cancer,
including the miR-17-92 cluster, which comprises 7 miRNAs and is
located in intron 3 of the C13orf25 gene at 13q31.3, have been
reported to function as oncogenes and accelerate tumor development
(22,23). Tumor-suppressive miRNAs, for example
the let-7 family, are located in fragile regions of the human
genome, and their loss is indicative of poor prognosis in various
types of human cancer (24).
Accumulating evidence suggests that miRNA expression profiling may
be used in the classification of human cancers, indicating the
potential of miRNAs as a novel diagnostic and prognostic tools for
various types of cancer (25).
Several recent studies have identified a number of
dysregulated miRNAs in OS (26–29). Maire
et al (30) performed miRNA
expression profiling of seven OS samples and identified several
aberrantly expressed miRNAs. Lulla et al (29) identified 22 differentially expressed
miRNAs in OS tumor samples, compared with normal osteoblasts.
However, given the number of aberrant miRNAs identified thus far,
it was suggested that there may be additional miRNAs involved in
OS, which remain to be identified. Recent advances in
high-throughput deep sequencing have markedly increased the speed
of the search for cancer-associated miRNAs (31,32).
High-throughput deep sequencing indicates the expression levels of
each miRNA in the miRNome, and is thus one of the most effective
and accurate approaches for evaluation of global miRNA expression
levels (33,34). To the best of our knowledge, deep
sequencing of the miRNome associated with OS has not previously
been performed, and such comprehensive analysis may provide insight
into the molecular mechanisms of OS.
The present study aimed to comprehend current
literature by complete profiling of OS miRNA expression patterns,
and further evaluating significantly differentially expressed
miRNAs and their effects on human OS cells in vitro.
Materials and methods
Patients and samples
The primary OS samples and matched noncancerous bone
tissue samples used for the high-throughput deep sequencing
experiments were obtained from two patients with OS undergoing
surgical resection at Shenzhen Second People's Hospital (Shenzhen,
China). Collected samples were flash frozen in liquid nitrogen
following surgery. All patients were informed about the aims of the
specimen collection and provided written informed consent. For the
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) verification, 32 OS samples were obtained from an archive
of formalin-fixed, paraffin-embedded (FFPE) diagnostic tissues in
the Pathology Department at Shenzhen Second People's Hospital,
collected between 1990 and 2013. All tumor samples were high-grade
OS of stage IIA or IIB, according to the Enneking system (35). The mean age of the patients was 21
years (range, 18–36 years), and 59% were male. All diagnoses were
confirmed by an experienced pathologist. Specimens from 36 normal
muscles of patients who had undergone orthopedic surgery were
collected and immediately stored in liquid nitrogen prior to use as
the negative controls. The study was approved by the ethics
committee of Shenzhen Second People's Hospital.
Identification of differentially
expressed miRNAs
High-throughput deep sequencing was performed using
the Illumina Cluster Station and Genome Analyzer II (Illumina Inc.,
San Diego, CA, USA). Small RNA library construction, sequencing and
bioinformatics analysis was conducted as previously described
(36,37). The miRNA expression levels were
compared between OS samples and paired normal bone tissues to
detect the differentially expressed miRNAs. The expression levels
of miRNAs in two samples were first normalized to obtain the
expression of transcripts per million, and the fold-change and
P-values were then calculated from the normalized expression level.
In general, if the adjusted P-values were <0.01 based on the
Benjamini and Hochberg multiple testing correction (38) and there was a ≥2-fold change (OS
samples/paired normal bone tissues) in the normalized expression,
the miRNA was considered to be significantly differentially
expressed.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) of
miRNAs
FFPE samples were cut into 10-µm sections. Total RNA
was isolated from FFPE samples using the Qiagen RNeasy FFPE
protocol (Qiagen, Inc., Valencia, CA, USA). For surgical resection
specimens, total RNA was extracted using the mirVana miRNA
isolation kit (Ambion, Austin, TX, USA) according to the
manufacturer's instructions.
RT-qPCR analysis of mature miR-33a-5p was performed
in triplicate using the TaqMan MicroRNA assay kit (Ambion)
according to the manufacturer's instructions. The RT reaction
mixture was comprised of 10 ng total RNA, 1 mM deoxynucleotide
triphosphates, 50 U Multiscribe Reverse Transcriptase, 1.5 µl 10X
RT buffer, 0.188 µl RNase inhibitor and 3 µl 5X TaqMan MicroRNA RT
primer for each reaction (15 µl). The RT reaction was performed
under the following conditions: 16°C for 30 min; 42°C for 30 min
and 85°C for 5 min, prior to holding at 4°C. Following RT, the
complementary DNA products of the RT reaction were diluted 15
times. PCR was conducted using 1.33 µl of the diluted product in 20
µl PCR reaction mixture, comprising 1 µl TaqMan MicroRNA Assay and
10 µl TaqMan Universal PCR Master mix. Subsequently, amplification
was performed under the following conditions: 95°C for 10 min,
followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Relative
expression was calculated using the comparative C(T) method
(39) and normalized to the
expression of RNU6B (Ambion).
miRNAs identified by high-throughput deep sequencing
were validated using the miScript PCR System (Qiagen, Inc.,
Gaithersburg, MD, USA) according to the manufacturer's
instructions. The RT reaction mixtures with the miScript II RT kit
(Qiagen) contained 1 µg total RNA, 4 µl 5X miScript HiSpec buffer,
2 µl 10X miScript nucleics mix and 2 µl miScript reverse
transcriptase mix for each reaction (20 µl). RT was performed under
the following conditions: 37°C for 60 min, followed by 95°C for 5
min. Subsequently, the cDNA products of the RT reaction were
diluted 15 times. PCR was performed with 1.5 µl of the diluted
products in 20 µl PCR reaction mixture containing 10 µl 2X
QuantiTect SYBR Green PCR master mix, 2 µl 10X miScript universal
primer, 2 µl 10X miScript primer assay. Amplification was performed
under the following conditions: 95°C for 15 min, followed by 40
cycles at 94°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec. All
reactions were performed in triplicate. Relative expression was
calculated using the comparative C(T) method and normalized to the
expression of RNU6B.
Cell culture
The human U2-OS and MG-63 OS cell lines were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). U2-OS and MG-63 cells were cultured
in McCoy's 5A media (modified with tricine; (Gibco Life
Technologies, Grand Island, NY, USA) and minimum essential medium
supplemented with 10% fetal bovine serum (Gibco), respectively.
Cells were incubated at 37°C in a 5% CO2 atmosphere.
miR-33a-5p precursor transfection
The miR-33a-5p precursor and random sequence
CY3-labeled miR-Scramble were synthesized by Ambion. U2-OS and
MG-63 cells were counted and plated at a density of
4×105 cells/well in 6-well plates for overnight
incubation prior to transfection with 100 nM miR-33a-5p precursor
or miR-Scramble using Lipofectamine® 2000 (Invitrogen Life
Technologies, CA, USA) according to the manufacturer's
instructions. Transfection efficiency was estimated by CY3-labeled
miR-Scramble using a fluorescence microscope (Axio Observer A1;
Zeiss, Jena, Germany).
Cell proliferation assays
The effect of miR-33a-5p on cell proliferation was
measured by WST-1 assay. Cells were counted and plated at a density
of 3×103 cells/well in 96-well plates in triplicates.
Cell viability was determined at 24, 48 and 72 h post-transfection.
Spectrophotometry (Beckman DU spectrophotometer, Beckman-Coulter,
Brea, CA, USA) was performed at λ=450 nm and λ ref=630 nm following
incubation with 10 µl WST-1 (Roche Diagnostics, New York, NY, USA)
for 2 h. Cell proliferation was evaluated using a colony formation
assay. Briefly, cells were seeded in six-well plates
(0.5×103 cells/well) and cultured for two weeks.
Colonies were fixed with methanol for 10 min and stained with 1%
crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 1 min.
Visible colonies (defined as containing >50 cells) in 10 random
fields were manually counted. Each cell group was measured in
triplicate.
Statistical analysis
miR-33a-5p expression in OS samples and normal bone
or muscle tissues were compared using the Mann-Whitney U test.
Correlation was evaluated using Pearson's Correlation Coefficient.
A comparison of means among two or more groups was performed using
one-way analysis of variance or Student's t-test. All numerical
data are expressed as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were performed using GraphPad Prism 5.0
(GraphPad Software, Inc., La Jolla, CA, USA) and SPSS software
version 11 (SPSS, Inc., Chicago, IL, USA).
Results
miRNAs differentially expressed in OS
and normal bone tissues
To investigate the expression profiles of miRNAs in
OS and adjacent normal bone tissues, high-throughput deep
sequencing was used to compare their expression levels.
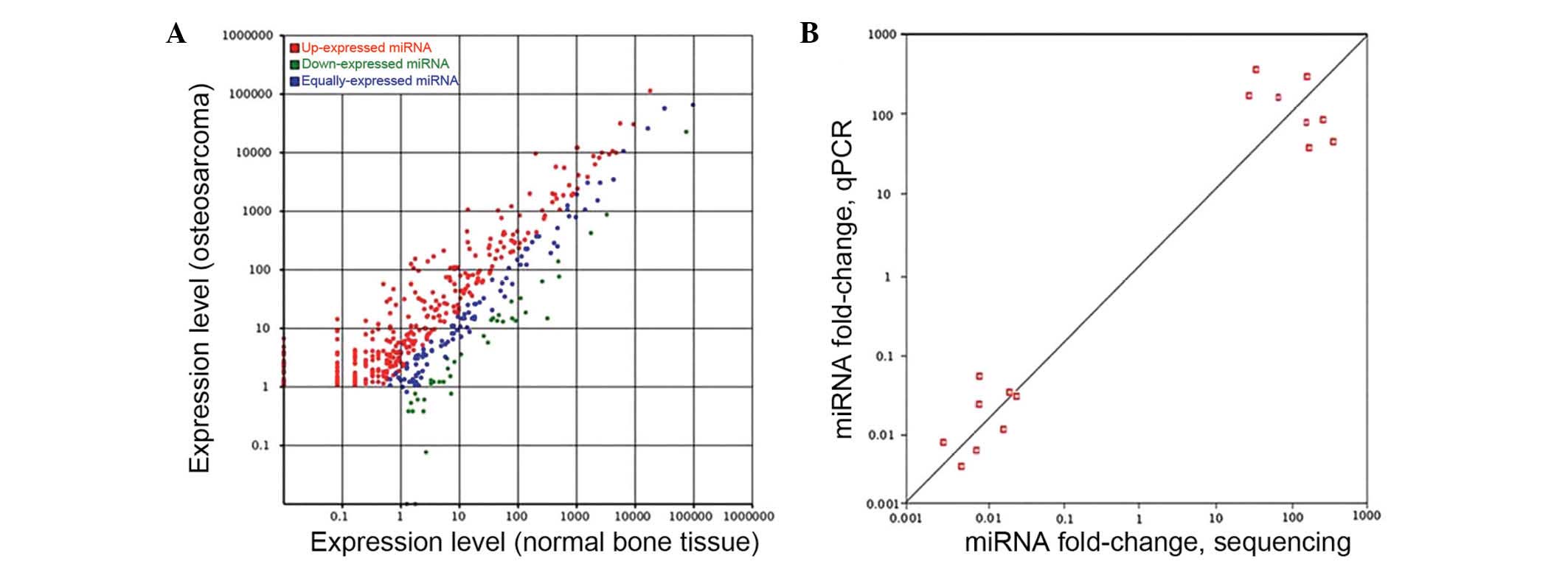
High-throughput deep sequencing revealed a series of miRNAs with
altered expression in OS tissues: 310 miRNAs were significantly
overexpressed and 41 miRNAs were significantly downregulated
(>2-fold; adjusted P<0.05), compared with those of normal
tissues (Fig. 1A). Among these
differentially expressed miRNAs, a total of 47 miRNAs with
>32-fold elevated expression levels and 17 miRNAs with
expression levels reduced >4-fold were identified (Table I).
 | Table I.Significantly differentially
expressed known miRNAs in osteosarcoma. |
Table I.
Significantly differentially
expressed known miRNAs in osteosarcoma.
| miRNA ID | Log2,
fold-changea | Adjusted
P-value |
|---|
| Upregulated |
|
|
|
hsa-miR-512-3p | 9.39 |
2.80×10−62 |
|
hsa-miR-377-5p | 8.90 |
1.95×10−9 |
|
hsa-miR-433-3p | 8.60 |
1.54×10−5 |
|
hsa-miR-1323 | 8.45 |
4.72×10−5 |
|
hsa-miR-337-3p | 8.11 |
5.30×10−22 |
|
hsa-miR-485-3p | 8.07 |
8.00×10−56 |
|
hsa-miR-6503-5p | 8.07 |
8.08×10−7 |
|
hsa-miR-656-3p | 7.99 | <0 |
|
hsa-miR-411-3p | 7.94 |
6.82×10−8 |
|
hsa-miR-494-3p | 7.85 |
1.98×10−4 |
|
hsa-miR-4709-5p | 7.81 |
7.79×10−4 |
|
hsa-miR-508-3p | 7.76 |
1.23×10−6 |
|
hsa-miR-187-3p | 7.54 |
6.42×10−12 |
|
hsa-miR-370-5p | 7.54 |
4.12×10−9 |
|
hsa-miR-105-3p | 7.42 |
1.08×10−105 |
|
hsa-miR-873-3p | 7.37 |
3.22×10−4 |
|
hsa-miR-1185-5p | 7.07 |
1.36×10−51 |
|
hsa-miR-541-3p | 7.07 |
7.67×10−6 |
|
hsa-miR-885-5p | 7.07 |
3.50×10−16 |
|
hsa-miR-329-5p | 6.99 |
7.26×10−4 |
|
hsa-miR-337-5p | 6.99 |
1.84×10−3 |
|
hsa-miR-219a-1-3p | 6.90 |
2.34×10−13 |
|
hsa-miR-329-3p | 6.90 | <0 |
|
hsa-miR-134-3p | 6.81 | <0 |
|
hsa-miR-134-5p | 6.80 |
1.83×10−22 |
|
hsa-miR-654-5p | 6.77 | <0 |
|
hsa-miR-758-3p | 6.77 |
1.44×10−5 |
|
hsa-miR-487b-3p | 6.72 |
2.48×10−6 |
|
hsa-miR-20b-3p | 6.71 |
3.18×10−292 |
|
hsa-miR-380-3p | 6.71 |
3.27×10−5 |
|
hsa-miR-654-3p | 6.42 |
4.30×10−31 |
|
hsa-miR-432-5p | 6.35 |
1.67×10−16 |
|
hsa-miR-105-5p | 6.25 |
3.64×10−22 |
|
hsa-miR-409-3p | 6.21 |
2.26×10−3 |
|
hsa-miR-1269b | 5.97 |
9.01×10−4 |
|
hsa-miR-493-3p | 5.93 |
5.29×10−97 |
|
hsa-miR-431-3p | 5.73 | <0 |
|
hsa-miR-127-3p | 5.57 |
3.27×10−11 |
|
hsa-miR-409-5p | 5.56 |
5.01×10−3 |
|
hsa-miR-370-3p | 5.54 |
5.75×10−7 |
|
hsa-miR-767-5p | 5.54 |
2.20×10−7 |
|
hsa-miR-410-3p | 5.54 |
9.27×10−3 |
|
hsa-miR-493-5p | 5.51 | <0 |
|
hsa-miR-487a-3p | 5.50 |
1.24×10−8 |
|
hsa-miR-520a-3p | 5.41 |
4.47×10−10 |
|
hsa-miR-381-3p | 5.24 |
4.87×10−14 |
|
hsa-miR-149-5p | 5.22 |
1.42×10−3 |
| Downregulated |
|
|
|
hsa-miR-33a-5p | −7.46 | <0 |
|
hsa-miR-551b-3p | −6.98 |
6.63×10−5 |
|
hsa-miR-3613-5p | −5.17 |
1.53×10−3 |
|
hsa-miR-144-3p | −4.47 | <0 |
|
hsa-miR-190a-5p | −3.28 |
6.73×10−83 |
|
hsa-miR-335-5p | −2.90 |
1.59×10−9 |
|
hsa-miR-144-5p | −2.75 |
9.27×10−13 |
|
hsa-miR-224-3p | −2.71 |
1.72×10−140 |
|
hsa-miR-193a-3p | −2.46 |
1.63×10−55 |
|
hsa-miR-19a-3p | −2.46 |
4.56×10−6 |
|
hsa-miR-33b-5p | −2.26 | <0 |
|
hsa-miR-452-3p | −2.17 |
9.84×10−289 |
|
hsa-miR-29c-3p | −2.12 |
1.43×10−18 |
|
hsa-miR-101-3p | −2.11 | <0 |
|
hsa-miR-2467-5p | −2.10 |
5.55×10−10 |
|
hsa-miR-378a-5p | −2.08 |
1.13×10−5 |
|
hsa-miR-145-5p | −2.04 |
1.22×10−7 |
To further validate these differentially expressed
miRNAs, eight miRNAs identified by high-throughput deep sequencing
were re-examined by RT-qPCR. The eight miRNAs selected included the
most upregulated miRNAs (miR-512-3p, miR-377-5p, miR-433-3p and
miR-1323) and most downregulated miRNAs (miR-33a-5p, miR-551b-3p,
miR-3613-5p and miR-144-3p) in OS. As illustrated in Fig. 1B, the Illumina deep sequencing data
correlated with the RT-qPCR results (r=0.805; P<0.001),
indicating the reliability of sequencing-based expression
analysis.
miR-33a-5p expression is decreased in
paraffin-embedded OS samples
Subsequently, miR-33a-5p was further analyzed, as
this was the most downregulated miRNA identified in the OS samples.
To verify the expression levels of miR-33a in OS, miR-33a-5p
expression levels were determined in 32 paraffin-embedded human OS
samples and 36 normal muscle tissues by TaqMan RT-qPCR. As shown in
Fig. 2A, miR-33a-5p expression was
significantly downregulated in paraffin-embedded OS tissues,
compared with that of normal muscle tissue (P=0.0238). These
results suggested that miR-33a-5p may have a role in the
pathogenesis of OS.
miR-33a-5p inhibits OS cell
proliferation
To investigate the functional role of miR-33a-5p in
OS, human U2-OS and MG-63 OS cell lines were transfected with 100
nM chemically synthesized miR-33a-5p precursor, which mimics
endogenous mature miR-33a-5p function. Cells transfected with 100
nM miR-Scramble (scrambled oligonucleotides) were used as the
control. Transfection efficiency of miRNA in these two cell lines
was estimated by CY3-labeled miR-Scramble (>80%; data not show).
Additionally, 24 h post transfection, miR-33a-5p expression levels
were evaluated by RT-qPCR. The results demonstrated that miR-33a-5p
mimic enhanced miR-33a-5p expression by ~152-fold (P<0.001) in
U2-OS cells and ~341-fold in MG-63 cells (P<0.001), compared
with the scramble-transfected group (Fig.
2B). These results indicated that the miR-33a-5p precursor was
able to effectively increase miR-33a-5p expression in U2-OS and
MG-63 cells.
Following 48, 72 and 96 h of incubation, U2-OS and
MG-63 cells overexpressing miR-33a-5p exhibited decreased cell
proliferation, as compared with miR-Scramble-transfected cells,
respectively (P<0.05; Fig. 3A).
Consistent with these results, in the WST-1 assay, cells
transfected with miR-33a-5p demonstrated formation of significantly
fewer colonies than those of cells transfected with the
miR-Scramble (P<0.001; Fig.
3B).
Discussion
In the present study, miRNAs that were up- or
downregulated in OS, as compared with matched noncancerous bone
tissues, were detected through high-throughput deep sequencing. The
results demonstrated that the expression levels of 310 miRNAs were
increased and 41 miRNAs were decreased in the OS tissues. Among
these, miR-512-3p, miR-377-5p, miR-433-3p and miR-1323 were the
greatest upregulated miRNAs, whereas miR-33a-5p, miR-551b-3p,
miR-3613-5p and miR-144-3p were the most decreased miRNAs in OS.
These miRNAs were re-examined by RT-qPCR analysis and the results
correlated with those of the sequencing analysis. Specifically,
miR-33a-5p was decreased most in OS, which suggested that
miR-33a-5p may have a role in the pathogenesis of OS.
To the best of our knowledge, the role of miR-33a-5p
in OS has not previously been reported. miR-33a-5p has recently
emerged as a key regulator of metabolism, and was shown to regulate
cholesterol and lipid metabolism (40,41).
miR-33a is downregulated in lung cancer cells and functions as a
potent tumor suppressor, which decreases osteolytic bone metastasis
via suppression of parathyroid hormone-related protein (42). miR-33a also functions as a tumor
suppressor miRNA through its capacity to downregulate the
expression of oncogenic kinase Pim-1 in K562 lymphoma and colon
carcinoma (43,44). miR-33 family members have been
associated with modulation of the expression of various genes
involved in cell cycle regulation and proliferation (45,46).
miR-33 decreases cellular proliferation and cell cycle progression
via inhibition of cyclin-dependent kinase 6 and cyclin D1 (45,46). In
the present study, miR-33a-5p was demonstrated to be downregulated
in OS, while overexpression of miR-33a-5p by transfection,
significantly attenuated OS cell growth in vitro.
The aberrant expression of miRNAs may occur via a
number of mechanisms, for example by deletion in fragile regions of
the genome containing cancer-suppressing miRNAs, as a result of
inherent or spontaneous mutations in miRNA genes or following
methylation of miRNA promoters (47–50).
However, the mechanism by which miR-33a-5p is downregulated in OS
remains to be elucidated.
In conclusion, the results of the present study
demonstrated that multiple miRNAs are aberrantly expressed in human
OS. Among these miRNAs, miR-33a-5p is significantly downregulated
in the majority of OS tissues. miR-33a-5p demonstrated tumor
suppressive abilities in vitro by inhibiting OS cell
proliferation, which suggested that miR-33a-5p may have a tumor
suppressor function in human OS. These results provide support for
the rescue of miR-33a-5p expression via gene therapy, and
demonstrated the potential use of miR-33a-5p as diagnostic marker
or therapeutic tool for the treatment of human OS.
Acknowledgements
The authors would like to thank their colleagues for
their insight and technical support. The present study was
supported by the National Natural Scientific Foundation of China
(no. 81171447), the China Postdoctoral Science Foundation (no.
2013M531896) and the Shenzhen Science and Technology Foundation
(nos. GJHZ20130412153906739 and ZDSY20120614154551201).
References
|
1
|
Sweetnam R: Osteosarcoma. Br J Hosp Med.
28:116–121. 1982.
|
|
2
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma and Ewing's sarcoma: National Cancer
Data Base Report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dorfman HD and Czerniak B: Bone cancers.
Cancer. 75 (1 Suppl):S203–S210. 1995. View Article : Google Scholar
|
|
4
|
Tarkkanen M, Karhu R, Kallioniemi A,
Elomaa I, Kivioja AH, Nevalainen J, Böhling T, Karaharju E,
Hyytinen E, Knuutila S, et al: Gains and losses of DNA sequences in
osteosarcomas by comparative genomic hybridization. Cancer Res.
55:1334–1338. 1995.PubMed/NCBI
|
|
5
|
AlRomaih K, Bayani J, Vorobyova J,
Karaskova J, Park PC, Zielenska M and Squire JA: Chromosomal
instability in osteosarcoma and its association with centrosome
abnormalities. Cancer Genet Cytogenet. 144:91–99. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ragland BD, Bell WC, Lopez RR and Siegal
GP: Cytogenetics and molecular biology of osteosarcoma. Lab Invest.
82:365–373. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kansara M and Thomas DM: Molecular
pathogenesis of osteosarcoma. DNA Cell Biol. 26:1–18. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tabone MD, Kalifa C, Rodary C, Raquin M,
ValteauCouanet D and Lemerle J: Osteosarcoma recurrences in
pediatric patients previously treated with intensive chemotherapy.
J Clin Oncol. 12:2614–2620. 1994.PubMed/NCBI
|
|
11
|
KempfBielack B, Bielack SS, Jurgens H,
Branscheid D, Berdel WE, Exner GU, Göbel U, Helmke K, Jundt G,
Kabisch SF, et al: Osteosarcoma relapse after combined modality
therapy: An analysis of unselected patients in the Cooperative
Osteosarcoma Study Group (COSS). J Clin Oncol. 23:559–568. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davis AM, Bell RS and Goodwin PJ:
Prognostic factors in osteosarcoma: A critical review. J Clin
Oncol. 12:423–431. 1994.PubMed/NCBI
|
|
13
|
ArteagaVazquez M, CaballeroPerez J and
Vielle-Calzada JP: A family of microRNAs present in plants and
animals. Plant Cell. 18:3355–3369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B, Wang Q and Pan X: MicroRNAs and
their regulatory roles in animals and plants. J Cell Physiol.
210:279–289. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death and tumorigenesis. Br J Cancer. 96
(Suppl):R40–R44. 2007.PubMed/NCBI
|
|
16
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Voorhoeve PM: MicroRNAs: Oncogenes, tumor
suppressors or master regulators of cancer heterogeneity? Biochim
Biophys Acta. 1805:72–86. 2010.PubMed/NCBI
|
|
20
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Diosdado B, van de Wiel MA, Terhaar Sive
Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B and Meijer
GA: MiR-17-92 cluster is associated with 13q gain and c-myc
expression during colorectal adenoma to adenocarcinoma progression.
Br J Cancer. 101:707–714. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu J, Getz G, Miska EA, AlvarezSaavedra E,
Lamb J, Peck D, SweetCordero A, Ebert BL, Mak RH, Ferrando AA, et
al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang W, Gao B, Fu P, Xu S, Qian Y and Fu
Q: The miRNAs in the pathgenesis of osteosarcoma. Front Biosci
(Landmark Ed). 18:788–794. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma. 2012:3597392012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lulla RR, Costa FF, Bischof JM, Chou PM,
de F Bonaldo M, Vanin EF and Soares MB: Identification of
differentially expressed microRNAs in osteosarcoma. Sarcoma.
2011:7326902011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maire G, Martin JW, Yoshimoto M,
ChiltonMacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leidinger P, Keller A, Borries A,
Reichrath J, Rass K, Jager SU, Lenhof HP and Meese E:
High-throughput miRNA profiling of human melanoma blood samples.
BMC Cancer. 10:2622010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gillis AJ, Stoop HJ, Hersmus R, Oosterhuis
JW, Sun Y, Chen C, Guenther S, Sherlock J, Veltman I, Baeten J, et
al: High-throughput microRNAome analysis in human germ cell
tumours. J Pathol. 213:319–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schotte D, Akbari Moqadam F,
Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R and den Boer
ML: Discovery of new microRNAs by small RNAome deep sequencing in
childhood acute lymphoblastic leukemia. Leukemia. 25:1389–1399.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leptidis S, El Azzouzi H, Lok SI, de Weger
R, Olieslagers S, Kisters N, Silva GJ, Heymans S, Cuppen E,
Berezikov E, et al: A deep sequencing approach to uncover the
miRNOME in the human heart. PLoS One. 8:e578002013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Enneking WF, Spanier SS and Goodman MA: A
system for the surgical staging of musculoskeletal sarcoma. Clin
Orthop Relat Res. 153:106–120. 1980.PubMed/NCBI
|
|
36
|
Zhang J, Luo X, Li H, Deng L and Wang Y:
Genome-wide uncovering of STAT3-mediated miRNA expression profiles
in colorectal cancer cell lines. Biomed Res Int.
2014:1871052014.PubMed/NCBI
|
|
37
|
Zhang J, Wang Y, Zhen P, Luo X, Zhang C,
Zhou L, Lu Y, Yang Y, Zhang W and Wan J: Genome-wide analysis of
miRNA signature differentially expressed in doxorubicin-resistant
and parental human hepatocellular carcinoma cell lines. PLoS One.
8:e541112013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yekutieli D and Benjamini Y:
Resampling-based false discovery rate controlling multiple test
procedures for correlated test statistics. J Stat Plan Inference.
82:171–196. 1999. View Article : Google Scholar
|
|
39
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goedeke L and Fernández-Hernando C:
microRNAs: A connection between cholesterol metabolism and
neurodegeneration. Neurobiol Dis. 72:48–53. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Goedeke L, ValesLara FM, Fenstermaker M,
CireraSalinas D, ChamorroJorganes A, Ramírez CM, Mattison JA, de
Cabo R, Suárez Y and Fernández-Hernando C: A regulatory role for
microRNA 33* in controlling lipid metabolism gene expression. Mol
Cell Biol. 33:2339–2352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuo PL, Liao SH, Hung JY, Huang MS and Hsu
YL: MicroRNA-33a functions as a bone metastasis suppressor in lung
cancer by targeting parathyroid hormone related protein. Biochim
Biophys Acta. 1830:3756–3766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thomas M, LangeGrunweller K, Weirauch U,
Weirauch U, Gutsch D, Aigner A, Grünweller A and Hartmann RK: The
proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 31:918–928.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ibrahim AF, Weirauch U, Thomas M,
Grünweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Iwakiri Y: A role of miR-33 for cell cycle
progression and cell proliferation. Cell Cycle. 11:1057–1058. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
CireraSalinas D, Pauta M, Allen RM,
Salerno AG, Ramírez CM, ChamorroJorganes A, Wanschel AC, Lasuncion
MA, MoralesRuiz M, Suarez Y, et al: Mir-33 regulates cell
proliferation and cell cycle progression. Cell Cycle. 11:922–933.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wong KY, Yu L and Chim CS: DNA methylation
of tumor suppressor miRNA genes: A lesson from the miR-34 family.
Epigenomics. 3:83–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dela Cruz F and Matushansky I: MicroRNAs
in chromosomal translocation-associated solid tumors: Learning from
sarcomas. Discov Med. 12:307–317. 2011.PubMed/NCBI
|