Introduction
Cervical cancer is the second most common cause of
cancer-associated mortality in women worldwide (1), and its global incidence increased at an
annual rate of 0.6% between 1980 and 2010 (2). Invasion and metastasis are the primary
causes of treatment failure and subsequent mortality in patients
with cervical cancer (3). Recurrence
and metastasis of cervical carcinoma to other sites, including the
lymph nodes (4), bones (5), lungs (6)
and liver (7) may also occur.
Therefore, the inhibition of metastasis is an auxiliary strategy
for curing patients of cancer. However, the molecular alterations
that drive invasion and metastasis in cervical cancer are not well
established. Identifying these molecular mechanisms may provide
insights into potential targets for diagnosis and therapy.
Twist-related protein 1 (Twist1) is a class II
member of the highly conserved family of basic helix-loop-helix
transcription factors. Twist1 is overexpressed in a number of types
of cancer, and correlates with low E-cadherin expression, high
cancer aggressiveness and poor patient survival rates (8,9). The role
of Twist1 in tumor invasion and metastasis has been attracting
increasing interest. Previous studies have demonstrated that
carcinoma invasion and metastasis are driven by a process termed
epithelial-mesenchymal transition (EMT) (10), which is a process whereby epithelial
cells acquire mesenchymal properties and override senescence
(11). Twist1 overexpression promotes
metastasis in vivo by inducing EMT (8). Suppression of Twist1 by small
interfering RNA in prostate cancer cells induced the expression of
epithelial components and specifically inhibited their capacity for
invasion and metastasis (12,13). Furthermore, Twist1 expression also
enhances cell migration and invasion in gastric cancer in
vitro and in vivo (14).
Therefore, targeting Twist1 may be a novel therapeutic approach for
the treatment of cervical cancer.
The present study targeted Twist1 in human cervical
cancer HeLa cells and reduced it's expression levels using a short
hairpin (sh)RNA lentivirus (LV-sh-Twist1) and assessed the
resulting effects on migration and invasion of the cells.
Materials and methods
Cell culture
The human cervical cancer cell line HeLa was
maintained in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA),
supplemented with 10% fetal bovine serum (FBS) (MinHai
Bio-Engineering, Lanzhou, China), 1% penicillin-streptomycin
(Invitrogen Life Technologies, Carlsbad, CA, USA) in a humidified
incubator at 37°C, 5% CO2 atmosphere and 95% air.
Lentiviral shRNA production and stable
knockdown of Twist1
The GIPZ lentiviral shRNAmir-GFP system (Open
Biosystems, Lafayette, CO, USA) was used to knockdown human Twist1
in HeLa cells. Lentiviruses (LVs) that co-express green fluorescent
protein (GFP) and shRNAs targeting Twist1, and with expression of
GFP and empty vector control were used to infect HeLa cells. The
detailed method for making an shRNA-based stable knockdown of
Twist1 cell lines is described in a previous study (15). The LVs produced were titered and
stored according to the manufacturer's instructions.
MTT assay
HeLa cells were cultured in 96-well plates at a
density of 1×105 cells per well overnight and infected
with LV-sh-Twist1. After 0, 24, 48 and 72 h, 50 ml MTT (1 mg/ml)
from Sigma-Aldrich (St Louis, MO, USA) was added to the cell media.
After 4 h, the MTT was discarded and 150 ml dimethyl sulfoxide was
loaded into each well. The spectrophotometric absorbance of the
samples was measured using a microplate reader (Model 680; Bio-Rad
Laboratories, Inc., Richmond, CA, USA) at 570 nm with a reference
wavelength of 655 nm. The percentage of cell survival was
calculated using the following formula: Cell viability =
(absorbance value of infected cells / absorbance value of
uninfected control cells) × 100. Six duplicate wells were measured
at each concentration, and each experiment was performed at least
three times.
Apoptosis assay
At 48 h after transfection, the HeLa cells were
collected and washed twice with cold phosphate-buffered saline,
resuspended in 400 µl Annexin V-fluorescein isothiocyanate (FITC)
binding buffer at a density of 1×106 cells/ml. The cells
were stained with 5 µl Annexin V-FITC and 10 µl propidium iodide
from an Apoptosis Detection kit (Jingmei Biotech, Shanghai, China)
following the manufacturer's instructions. The cells were then
subjected to flow cytometry (BD Biosciences, San Jose, CA, USA) to
detect cell apoptosis. This experiment was conducted 3 times.
Cell migration and invasion Transwell
assays
In vitro cell invasion assays were carried
out in Matrigel-based Transwell plates with slight modifications.
HeLa cells (500 µl of 1×105 cells/ml in 0.1% FBS +
RPMI-1640) were seeded into the Matrigel™-coated upper chambers of
an 8 µm pore-sized polycarbonate membrane (Corning Costar,
Cambridge, MA, USA). The lower compartments were filled with medium
supplemented with 20% FBS. After 24 h, the cells that were present
on the other side were stained with crystal violet dye and counted
under a light microscope (DP70; Olympus, Melville, NY, USA). The
number of cells were determined in eight random fields. The
experiment was repeated 3 times for each group. Cell migration
assays were carried out in a similar way but without Matrigel™.
Migration or invasion of the cells through the chamber to the
underside of the filter was assessed as described previously
(16).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNAs were prepared from the treated cells
using TRIzol® reagent (Invitrogen Life Technologies). qPCR was
carried out using the ABI 7300 real-time PCR system (Applied
Biosystems, Foster City, CA, USA) and performed by SYBR® Green dye
according to the manufacturer's instructions. Oligonucleotide
primers were synthesized by Sangon Biotech Co., Ltd (Shanghai,
China). Sequences of primers used in the qPCR analysis are
presented in Table I. The qPCR
results were calculated using the 2−ΔΔCT method as
described previously (17).
 | Table I.List of reverse
transcription-polymerase chain reaction primers used in the present
study. |
Table I.
List of reverse
transcription-polymerase chain reaction primers used in the present
study.
| Gene name | Sequence | Product length
(bp) |
|---|
| β-actin | F:
5′TGACGTGGACATCCGCAAAG3′ | 205 |
|
| R:
5′CTGGAAGGTGGACAGCGAGG3′ |
|
| E-cadherin | F:
5′TGCCGCCATCGCTTACAC3′ | 179 |
|
| R:
5′TGCTTAACCCCTCACCTTGA3′ |
|
| Vimentin | F:
5′AAATGGCTCGTCACCTTCG3′ | 186 |
|
| R:
5′GGGTATCAACCAGAGGGAGTG3′ |
|
| Fibronectin | F:
5′TGCCAACCTTTACAGACCTATCC3′ | 122 |
|
| R:
5′GAAATGTGAGATGGCTGTGGTG3′ |
|
Western blot analysis
Whole cell proteins were separated on 10% sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) at
6–8 V/cm and transferred to polyvinylidene difluoride membranes
using an electrotransfer system (Bio-RadLaboratories, Inc.). The
membranes were then blocked in 5% non-fat milk diluted in
Tris-buffered saline. The filters were hybridized with polyclonal
rabbit anti-Twist1 (cat no. sc-15393; Santa Cruz Biotechnology,
Santa Cruz, CA, USA), mouse anti-E-cadherin (cat no. 14472s),
rabbit anti-matrix metalloproteinase-9 (MMP-9; cat no. 3852s),
anti-MMP-2 (cat no. 4022s), rabbit anti-vimentin (cat no. 12826;
Cell Signaling Technology, Beverly, MA, USA) and rabbit
anti-fibronectin (cat no. ab2413; Abcam, Cambridge, UK) antibodies
diluted to 1:1,000 at 4°C overnight, followed by incubation with
horseradish peroxidase-conjugated goat anti-mouse IgG or goat
anti-rabbit IgG secondary antibodies (Santa Cruz Biotechnology;
1:4,000 dilution) for 1 h at room temperature. Mouse anti-GAPDH
(cat no. sc-365062; 1:1,000 dilution; Santa Cruz Biotechnology) was
used as a loading control. Antibody-antigen complexes were detected
with electrochemiluminescence reagents (GE Healthcare, Freiburg,
Germany) and the protein bands were quantified by densitometry for
subsequent analysis (ImageJ software; National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was conducted using SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA). Data from at
least three separate experiments are presented as the mean ±
standard error of the mean. Paired Student's t-tests were
used to determine any significant differences. P<0.05 was
considered to indicate a statistically significant difference.
Results
LV-sh-Twist1 inhibits cervical cancer
cell viability and induces cell apoptosis
To determine the function of Twist1 in cervical
cancer cells, Twist1 was knocked down in HeLa cells using
LV-sh-Twist1, which was selected from three candidates by qPCR
(data not shown). The effects of LV-sh-Twist1 on Twist1 expression
levels were examined by western blotting in cervical cancer HeLa
cells. The results demonstrated that transfection of LV-sh-Twist1
resulted in the downregulation of Twist1 (Fig. 1A and B). The expression of Twist1 was
reduced in HeLa cells at 24, 48 and 72 h after LV-sh-Twist1
transfection and that the level of inhibition increased in a
time-dependent manner (Fig. 1A). The
expression level of Twist1 was decreased at 48 h after LV-sh-Twist1
transfection compared with LV-GFP (Fig.
1B).
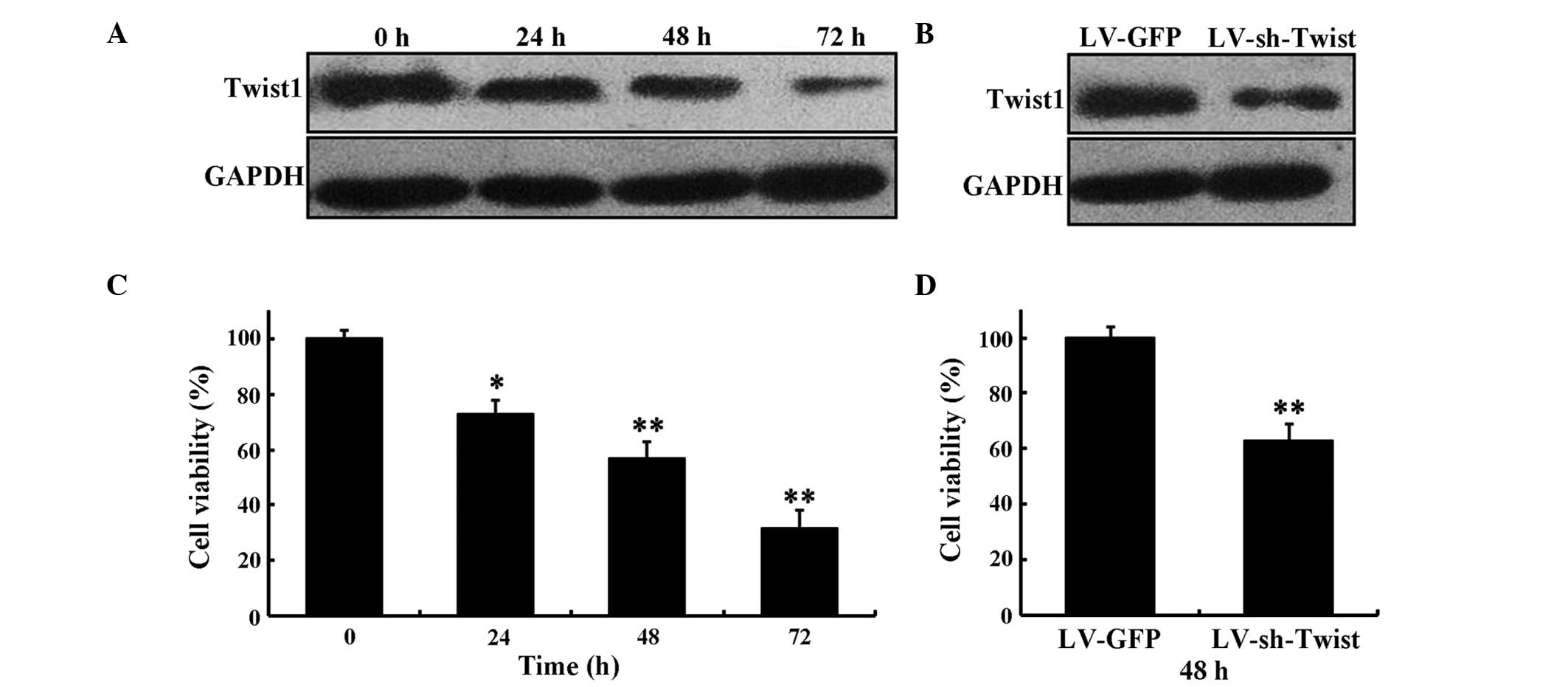 | Figure 1.Inhibitory effect of LV-sh-Twist1 on
the survival rate of HeLa cells. (A) The expression levels of
Twist1 by LV-sh-Twist1 in HeLa cells at 0, 24, 48 and 72 h,
respectively. (B) The expression levels of Twist1 at 48 h after
transfection with LV-sh-Twist1 or LV-GFP. (C) HeLa cells were
treated with LV-sh-Twist1. MTT assay was performed to determine
cell viability rates at 0, 24, 48 and 72 h, respectively. (D) At 48
h after transfection with LV-sh-Twist1 or LV-GFP, the viability of
HeLa cells was assessed using MTT assay. Data are presented as the
mean ± standard deviation from three independent experiments.
*P<0.05 and **P<0.01 compared with control. Twist1,
Twist-related protein 1; sh, short hairpin; LV, lentivirus; GFP,
green fluorescent protein. |
MTT assay was used to determine the effect of
LV-sh-Twist1 on HeLa cell proliferation. The present results
revealed that the knockdown of Twist1 evoked a marked inhibition
effect on cell proliferation after 24, 48 and 72 h of LV-sh-Twist1
transfection in HeLa cells (73.2±5.3%, P<0.05; 57.4±6.1%,
P<0.01; and 32±6.7%, P<0.01, respectively; Fig. 1C), whereas the cell viability in the
LV-sh-Twist1 transfection group was reduced significantly compared
with the LV-GFP group at 48 h (63.5±6.6%, P<0.01; Fig. 1D).
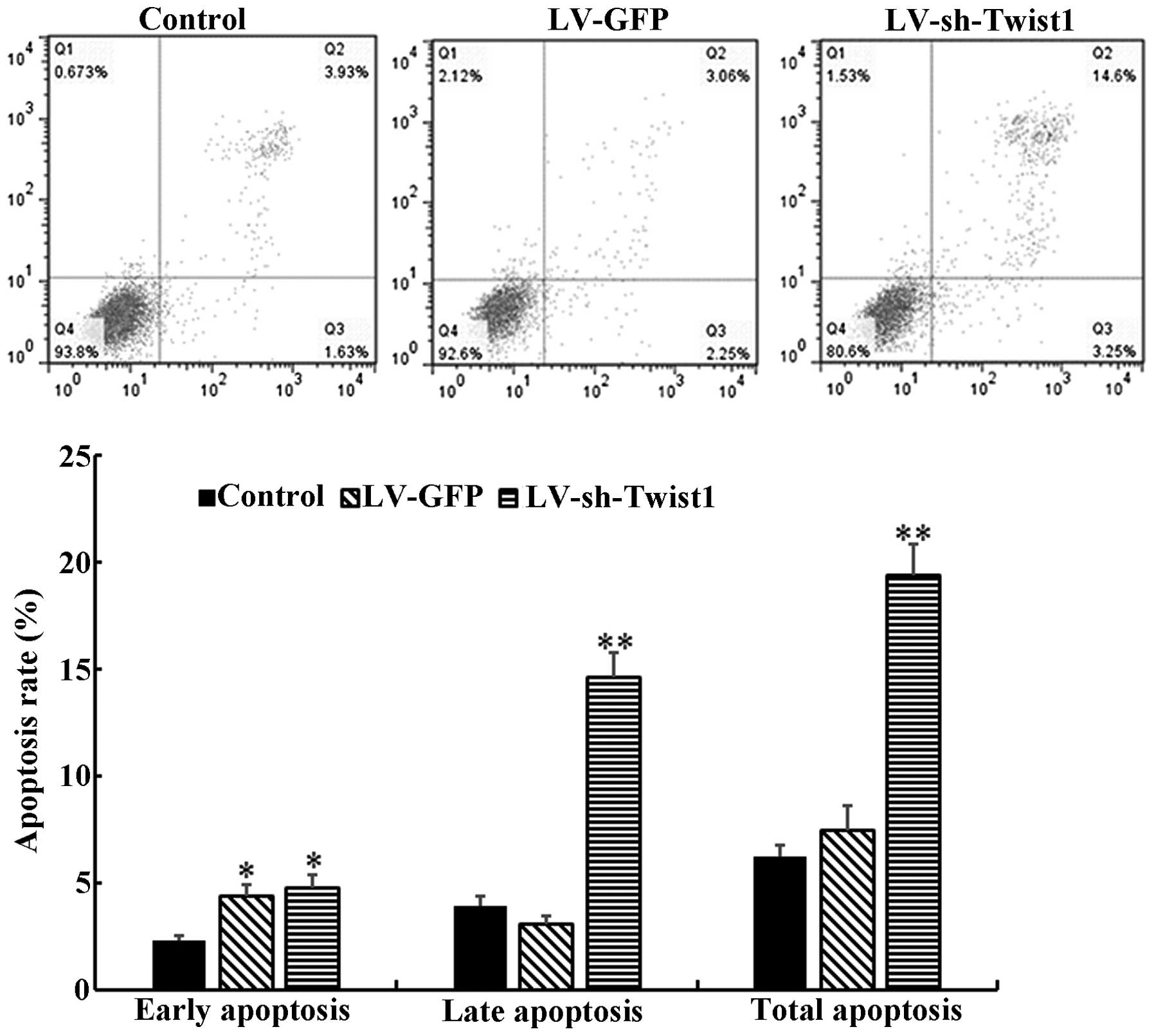
As it has been demonstrated that cell apoptosis
serves a considerable role in the progression and development of
tumors (18), the present study
further explored whether the cell proliferation inhibition observed
was due to the induction of apoptosis. Flow cytometry was used to
determine the level of cell apoptosis at 48 h after control, LV-GFP
or LV-sh-Twist1 transfection. The results demonstrated that the
total cell apoptosis rate of the LV-sh-Twist1 transfection group
was significantly increased compared with the control group
(19.4±1.5 vs. 6.2±0.6%, P<0.01; Fig.
2). The proportions of early and late cell apoptosis rates were
consistent with the total cell apoptosis rates.
Altered expression of Twist1
influences migration and invasion in cervical cancer cells
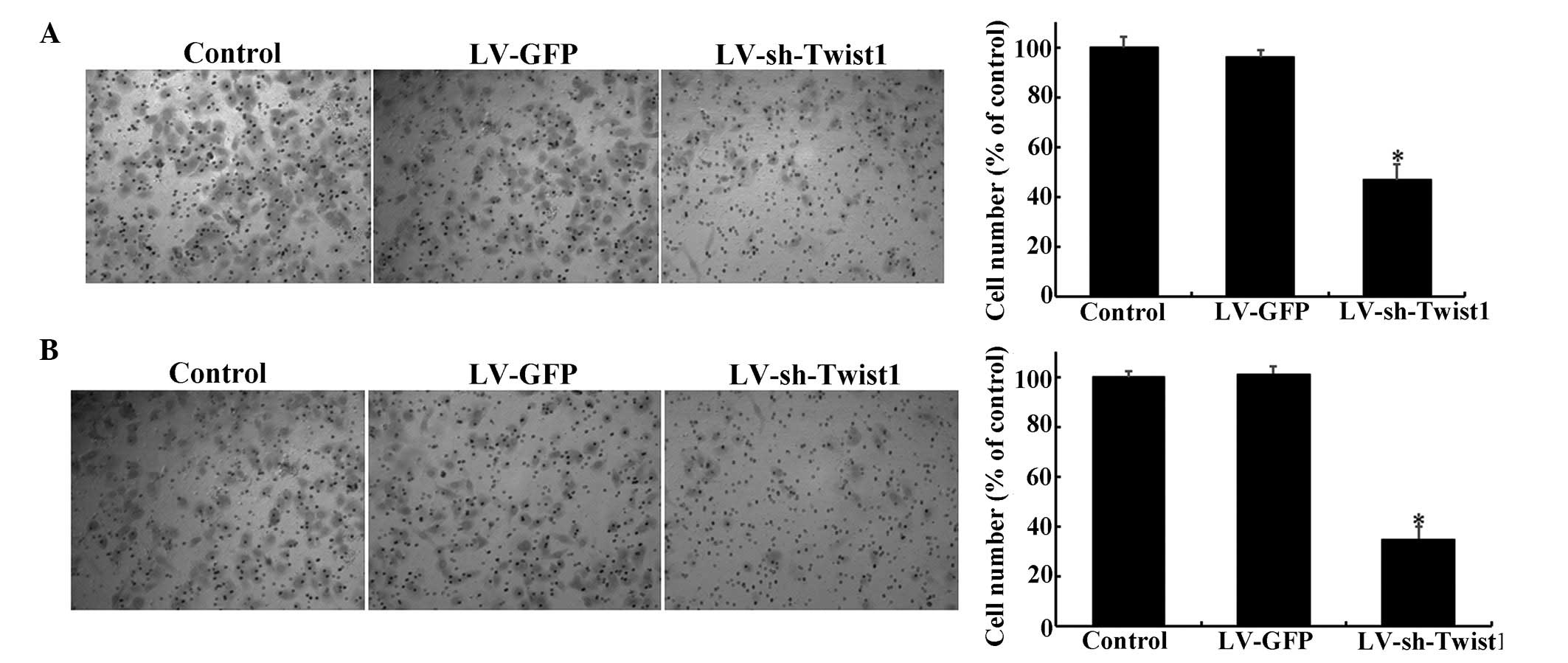
The present study further examined whether the
migration and invasion of cervical cancer cells is attenuated by
the reduction of Twist1 expression via LV-mediated shRNA.
Transwell® assays were employed to investigate whether the
migration and invasion of cervical cells were affected by
LV-sh-Twist1. Notably, when Twist1 was downregulated by
LV-sh-Twist1 transfection, the migration of HeLa cells was reduced
by ~53% compared with the control group (Fig. 3A; P<0.05). Similar results were
obtained from invasion assays, which showed a reduction of ~5%
compared with the control group (Fig.
3B; P<0.05).
LV-sh-Twist1 inhibits EMT of cervical
cells
As numerous studies have revealed that EMT is
closely associated with the migration and invasion of cancer cells
(19,20), the present study investigated the
effect of LV-sh-Twist1 transfection on EMT processes in cervical
cells by examining the changes in E-cadherin, fibronectin, MMP-9,
MMP-2 and vimentin expression. RT-qPCR results demonstrated that
the expression of the epithelial cell marker, E-cadherin, was
significantly elevated in HeLa cells 48 h after LV-sh-Twist1
transfection (P<0.05). As hypothesized, the expression levels of
mesenchymal cell markers (fibronectin and vimentin) were
significantly reduced in the LV-sh-Twist1 transfection group
(P<0.05; Fig. 4). In addition,
similar results were observed from western blot analysis; the
expression levels of fibronectin, vimentin, MMP-9 and MMP-2 were
downregulated, whereas E-cadherin was upregulated in the
LV-sh-Twist1 transfection group (Fig.
4). These results provide evidence for a potential role of EMT
during the effects of Twist1 downregulation, which reduces the
migration and invasion of cells in cervical cancer.
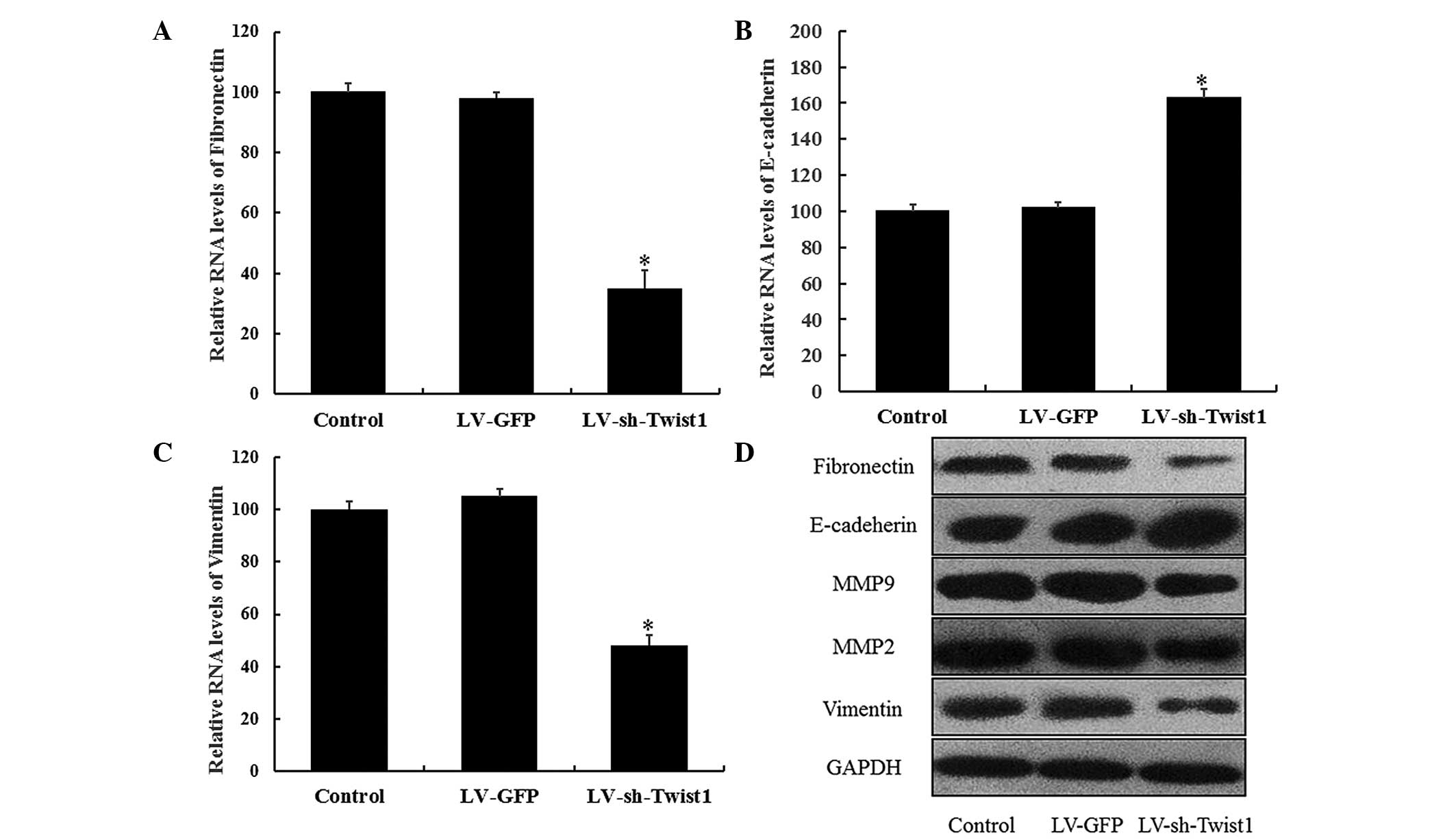 | Figure 4.Inhibition of Twist1 expression
regulates the epithelial and mesenchymal markers in HeLa cells.
HeLa cells were transfected with control, LV-GFP or LV-sh-Twist1
and incubated for 48 h. mRNA expression of (A) E-cadherin, (B)
vimentin and (C) fibronectin were quantified by reverse
transcription quantitative polymerase chain reaction. Each
expression was normalized to that of β-actin, and the relative
expression levels compared with the sample of each control solvent
are shown. Data are presented as the mean ± standard error of the
mean of three different experiments. *P<0.05 compared with
control. (D) Whole-cell extracts were analyzed by SDS-PAGE and
western blot analysis with specified antibodies. Fibronectin,
E-cadherin, MMP-9, MMP-2 and vimentin proteins were determined and
GAPDH was selected as the endogenous control. Densitometry was used
to quantify and analyze the data. Twist1, Twist-related protein 1;
MMP, matrix metalloproteinase; sh, short hairpin; LV, lentivirus;
GFP, green fluorescent protein. |
Discussion
Cervical cancer remains the only major gynecological
malignancy that is clinically staged. At present, no accurate,
efficient techniques for indicating prognosis and diagnosing
parametrium invasion and lymph node metastasis are available for
the selection of the most suitable treatment (21). The identification and functional
characterization of molecules critically involved in prognosis,
parametrium invasion and lymph node metastasis may reveal targets
for diagnostic and therapeutic applications.
The majority of cancer-related mortality events
occur as a result of metastasis rather than the original tumor;
therefore, inhibiting cancer cell metastasis is a crucial aspect of
cancer prevention. Previous studies observed that Twist1 expression
promoted the migration and invasion capabilities of human breast
cancer and hepatocellular carcinoma cells (22,23), which
are essential for tumor metastasis (8). Furthermore, Twist1 expression in
cervical cancer is associated with poor disease outcome (24). These results indicate that Twist1 is
closely correlated with the invasion of cervical cancer cells. The
objective of the present study was to investigate whether the
migration and invasion of cervical cancer cells was regulated by
Twist1, and if this was the case, which molecular mechanisms and
signaling pathways were involved. To investigate the therapeutic
relevance of inhibiting Twist1 in cervical cancer, Twist1
expression was knocked down using shRNA and the effects on cell
invasion and migration were assessed. The results of the present
study indicated that specific inhibition of Twist1 expression
resulted in marked reductions in cervical cancer cell invasion
in vitro. These findings are consistent with the
pro-invasive functions of Twist1 in cervical cancer and support the
therapeutic potential of inhibiting Twist1 or Twist1-mediated EMT
to inhibit cervical cancer cell invasion and migration. The present
data demonstrated that the inhibition of Twist1 expression resulted
in a notable reduction in cervical cancer cell growth and an
increased cell apoptosis rate. These results indicate that the
inhibition of Twist1 may have therapeutic potential, resulting in
the targeting of cervical cancer cell invasiveness that contributes
to tumor growth, progression and treatment resistance. To further
address this potential, ongoing and future studies should address
the effects of Twist1 inhibition in cervical cancer cells on tumor
growth, invasion and response to therapy in vivo. The
present study revealed that the downregulation of Twist1 by
LV-sh-Twist1 transfection attenuates the cell migration and
invasion abilities of cervical cancer cells through the reversal of
EMT or suppression of MMP expression.
EMT is characterized by increased migratory
features, reduced epithelial cell adhesion, loss of cytoskeleton
components and acquisition of mesenchymal components (25), which is also critical for cancer
metastasis. Induction of EMT may result in cancer cells invading
the surrounding stroma, and to intravasation, dissemination and
colonization of distant sites. Thus, reversal of EMT is considered
to be an effective strategy against cancer metastasis (26). In the present study, downregulation of
Twist1 by LV-sh-Twist1 was demonstrated to be capable of reversing
the process of EMT by reducing the expression of mesenchymal
markers vimentin and fibronectin, and increasing the expression of
the epithelial marker E-cadherin. These findings indicate that the
inhibition of invasion by LV-sh-Twist1 in HeLa cells may be partly
attributed to the reversal of EMT.
MMPs serve a critical role in cancer invasion,
migration, metastasis and tumorigenesis. Blocking tumor cell
expression of MMPs significantly reduces tumor invasion and
metastasis (27). MMP-2 and −9 are
major components of the extracellular matrix and basement membrane.
A number of human tumors have been reported to be associated with
increased expression of MMP-2 and −9 (28), and tumor aggressiveness has been found
to significantly correlate with increased levels of MMP-2 and −9 in
prostate (29) and breast (30) cancer. Cytokines and inhibitors
regulate MMP-2 and −9 expression in cervical and ovarian cancer
cells (31). In the present study,
knockdown of Twist1 reduced expression of MMP-2 and −9 in the
cervical cancer cell line.
In conclusion, the present study provides
experimental evidence that knockdown of Twist1 by LV-sh-Twist1
suppresses cell migration and invasion in cervical cancer, which in
turn drives EMT. Therefore, the results demonstrate an effect of
Twist1 on cervical cancer cell invasion and metastasis, which may
lead to the identification of novel diagnostic markers and
therapeutic targets, and thus aid the understanding of the
mechanisms behind cervical cancer metastasis.
Acknowledgements
The present study was supported by grants from the
Project of the Science and Technology Bureau Foundation of Chengdu
City (no. 0GGYB452SF-023) and the Health and Family Planning
Commission Foundation of Sichuan Province (no. 100368).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Forman D, Mathers CD and Bray F:
Breast and cervical cancer in 187 countries between 1980 and 2010.
Lancet. 379:1390–1391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waggoner SE: Cervical cancer. Lancet.
361:2217–2225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takeuchi H, Kitajima M and Kitagawa Y:
Sentinel lymph node as a target of molecular diagnosis of lymphatic
micrometastasis and local immunoresponse to malignant cells. Cancer
Sci. 99:441–450. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thanapprapasr D, Nartthanarung A,
Likittanasombut P, Ayudhya NI Na, Charakorn C, Udomsubpayakul U,
Subhadarbandhu T and Wilailak S: Bone metastasis in cervical cancer
patients over a 10-year period. Int J Gynecol Cancer. 20:373–378.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamamoto K, Yoshikawa H, Shiromizu K,
Saito T, Kuzuya K, Tsunematsu R and Kamura T: Pulmonary
metastasectomy for uterine cervical cancer: A multivariate
analysis. Ann Thorac Surg. 77:1179–1182. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park JY, Lim MC, Lim SY, Bae JM, Yoo CW,
Seo SS, Kang S and Park SY: Port-site and liver metastases after
laparoscopic pelvic and para-aortic lymph node dissection for
surgical staging of locally advanced cervical cancer. Int J Gynecol
Cancer. 18:176–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin Q, Xu Y, He T, Qin C and Xu J: Normal
and disease-related biological functions of Twist1 and underlying
molecular mechanisms. Cell Res. 22:90–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Campisi J: Senescent cells, tumor
suppression and organismal aging: Good citizens, bad neighbors.
Cell. 120:513–522. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alexander NR, Tran NL, Rekapally H,
Summers CE, Glackin C and Heimark RL: N-cadherin gene expression in
prostate carcinoma is modulated by integrin-dependent nuclear
translocation of Twist1. Cancer Res. 66:3365–3369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C,
Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, et al: Up-regulation
of TWIST in prostate cancer and its implication as a therapeutic
target. Cancer Res. 65:5153–5162. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo GQ, Li JH, Wen JF, Zhou YH, Hu YB and
Zhou JH: Effect and mechanism of the Twist gene on invasion and
metastasis of gastric carcinoma cells. World J Gastroenterol.
14:2487–2493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu J, Qin L, He T, Qin J, Hong J, Wong J,
Liao L and Xu J: The twist/mi2/nurd protein complex and its
essential role in cancer metastasis. Cell Res. 21:275–289. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Komiya E, Ohnuma K, Yamazaki H, Hatano R,
Iwata S, Okamoto T, Dang NH, Yamada T and Morimoto C: CD26-mediated
regulation of periostin expression contributes to migration and
invasion of malignant pleural mesothelioma cells. Biochem Biophys
Res Commun. 447:609–615. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bu Q, Hu Z, Chen F, Zhu R, Deng Y, Shao X,
Li Y, Zhao J, Li H, Zhang B, et al: Transcriptome analysis of long
non-coding RNAs of the nucleus accumbens in cocaine-conditioned
mice. J Neurochem. 123:790–799. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Condeelis J and Segall JE: Intravital
imaging of cell movement in tumours. Nat Rev Cancer. 3:921–930.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rogers L, Siu SS, Luesley D, Bryant A and
Dickinson HO: Radiotherapy and chemoradiation after surgery for
early cervical cancer. Cochrane Database Syst Rev.
5:CD0075832012.PubMed/NCBI
|
|
22
|
Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD
and Wang LH: Twist transcriptionally up-regulates AKT2 in breast
cancer cells leading to increased migration, invasion and
resistance to paclitaxel. Cancer Res. 67:1979–1987. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsuo N, Shiraha H, Fujikawa T, Takaoka
N, Ueda N, Tanaka S, Nishina S, Nakanishi Y, Uemura M, Takaki A, et
al: Twist expression promotes migration and invasion in
hepatocellular carcinoma. BMC Cancer. 9:2402009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shibata K, Kajiyama H, Ino K, Terauchi M,
Yamamoto E, Nawa A, Nomura S and Kikkawa F: Twist expression in
patients with cervical cancer is associated with poor disease
outcome. Ann Oncol. 19:81–85. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liotta LA, Tryggvason K, Garbisa S, Hart
I, Foltz CM and Shafie S: Metastatic potential correlates with
enzymatic degradation of basement membrane collagen. Nature.
284:67–68. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bérubé M, Deschambeault A, Boucher M,
Germain L, Petitclerc E and Guérin SL: MMP-2 expression in uveal
melanoma: Differential activation status dictated by the cellular
environment. Mol Vis. 11:1101–1111. 2005.PubMed/NCBI
|
|
29
|
Nemeth JA, Yousif R, Herzog M, Che M,
Upadhyay J, Shekarriz B, Bhagat S, Mullins C, Fridman R and Cher
ML: Matrix metalloproteinase activity, bone matrix turnover and
tumor cell proliferation in prostate cancer bone metastasis. J Natl
Cancer Inst. 94:17–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jones JL, Glynn P and Walker RA:
Expression of MMP-2 and MMP-9, their inhibitors and the activator
MT1-MMP in primary breast carcinomas. J Pathol. 189:161–168. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Roomi MW, Monterrey JC, Kalinovsky T, Rath
M and Niedzwiecki A: In vitro modulation of MMP-2 and MMP-9 in
human cervical and ovarian cancer cell lines by cytokines, inducers
and inhibitors. Oncol Rep. 23:605–614. 2010.PubMed/NCBI
|