Introduction
Adenocarcinoma is a malignant tumor of the glands of
epithelial tissues that grows invasively and is not easy to resect
(1). The rate of lymph node
metastasis is relatively high in adenocarcinoma and may reach 36%.
Relapse occurs easily, which leads to a poor prognosis (2). Previous studies have demonstrated that
the differentially-expressed genes and miRNAs of adenocarcinoma may
affect the development, transfer and treatment of cancer (3–7). In
addition, there is a complex regulatory network between the TFs,
miRNAs, and target and host genes in adenocarcinoma (3). Through these studies, a novel method of
studying cancer may be produced. The present study aimed to
investigate the elements and regulatory associations between the
elements in adenocarcinoma to increase knowledge with regards to
the pathogenesis, development and gene therapy of
adenocarcinoma.
TFs are a type of protein that promotes or
suppresses the transcription of genes by binding to the upstream
regions of genes (8). TFs may
regulate the transcription of genes individually or in combination
with other proteins (8).
MicroRNAs (miRNAs) are short non-coding RNA
sequences that demonstrate regulatory functions (8). At present, the biological function of
certain miRNAs has been confirmed, which has revealed the notable
regulatory role played by miRNA in the growth, differentiation and
apoptosis of cells and the developmental process of disease
(4). A decrease in miRNA levels is
often observed in human cancers, indicating that miRNA may possess
an intrinsic function of tumor suppression (9). In addition, miRNAs participate in the
adjustment of cancer development by adjusting the target genes of
the miRNA, which has been assessed in previous studies (4,10–12). O'Donnell et al revealed that
the mir-17-92 gene may be associated with tumors, and also
identified that mir-17-92 regulates MYC by regulating E2F1
(4).
Host genes are the genes that code for miRNAs. A
considerable number of miRNAs have been identified in the introns
of host genes, and these miRNAs were termed intronic miRNAs
(5). The intronic miRNAs are
transcribed in parallel with the host genes (5,6). Intronic
miRNAs and the host genes usually act as potential partners to
achieve biological function and affect the alteration of pathways
(7). These previous studies have
indicated that miRNA and the miRNA host genes may affect the
development of cancer.
The presence of regulatory associations between TFs,
miRNAs, and the target and host genes of miRNA in adenocarcinoma
has been determined from the aforementioned understanding. These
elements may affect the development of cancer, and a considerable
number of studies have investigated these regulatory associations
(3–7).
From these previous studies, it has been revealed that miRNAs
regulate TFs and TFs regulate miRNAs (13). It has also been demonstrated that TFs
and miRNA may regulate genes (8) and
miRNA may regulate target genes (10). In addition, the data of numerous
studies have been combined to form various databases, including
TransmiR (13), computational
predicted methods (14),
experimentally validated databases (15,16),
miRBase (17), the KEGG pathway
database (18), the miR2Disease
database (19) and the GeneCards
database (20). In these databases, a
large amount of data may be found, which was the basis for the
present study.
In the present study, TFs, miRNAs, and the host and
target genes of miRNAs in adenocarcinoma were collected and
analyzed. The aim of the present study was to identify the networks
surrounding various elements in adenocarcinoma and to analyze these
networks. Data were manually collected for adenocarcinoma,
consisting of the differentially-expressed genes and miRNA,
associated genes and miRNA, and the host and target genes in
adenocarcinoma. The regulatory associations between the elements in
adenocarcinoma were also recorded. This data formed the basis of
follow-up assessments. Subsequent to the collection of various
data, this data was used to construct three networks, which
consisted of the differentially-expressed, associated and global
networks. However, the global network was so complex that no useful
data was obtained. The differentially-expressed and associated
networks were considered to be more notable compared with the
global network in the present study. In these networks, the key
elements and pathways in adenocarcinoma were identified. Finally,
the similarities and differences of the three level networks were
compared and analyzed. Key elements and pathways in adenocarcinoma
were then identified.
Materials and methods
Material collection and data
processing
Collection of data
Initially, 3 tables, which consisted of the target
genes, TFs and host genes of human miRNAs, were identified and
summarized. The experimentally validated dataset obtained from
Tarbase 5.0 and miRTarBase, TransmiR (13), miRBase (17) and National Center for Biotechnology
Information (NCBI) was found. In order to increase the convenience
of the use of the collected data, the official marks to symbolize
miRNAs and genes were used. These official marks were obtained from
the NCBI database. Following the collection of these data, three
tables were developed, which were important components of the
present study. The genes and miRNAs were then identified, as they
were required to construct the three networks in the present
study.
Differentially-expressed genes
The differentially-expressed genes in adenocarcinoma
were obtained from the NCBI snp database (21), KEGG pathway database (18) and relevant literature.
Differentially-expressed miRNAs
The Harbin Institute of Technology miR2Disease
database (19) is a manually created
database of differentially-expressed miRNAs in various human
diseases. This database was used in the present study to identify
differentially-expressed miRNAs in adenocarcinoma. In addition,
differentially-expressed miRNAs and associated miRNAs in
adenocarcinoma were identified using the relevant literature
(20,22–29).
Following the collection of these data, differentially-expressed
miRNAs from miR2Disease and the relevant literature were summarized
as differentially-expressed miRNAs in adenocarcinoma. The
differentially-expressed and associated miRNAs in adenocarcinoma
were summarized from the relevant literature, and were classified
as the associated miRNAs in adenocarcinoma.
Associated genes
The associated gene network in adenocarcinoma
consisted of 4 components. Firstly, certain
differentially-expressed genes in adenocarcinoma were identified
using the NCBI snp database, KEGG pathway database and relevant
literature (30–34). These genes formed a component of the
associated gene network in adenocarcinoma. Secondly, genes were
identified in adenocarcinoma using the GeneCards database (35). The genes with a relevance score
>0.8, according to the GeneCards database, were extracted as a
component of the associated gene network for adenocarcinoma.
Thirdly, 1,000-nt promoter region sequences of the target genes of
differentially-expressed genes were obtained from the University of
California, Santa Cruz database (36). The P-match method, which combines
pattern matching and weight matrix approaches, was then used to
identify transcription factor binding sites (TFBSs) in 1,000-nt
promoter region sequences and to map TFBSs onto the promoter region
of target genes. This method identified certain associated genes
that corresponded with miRNAs through target genes. These
associated genes were a component of the associated gene network in
adenocarcinoma. Finally, the last component of the associated gene
network in adenocarcinoma was identified using the relevant
literature (37–47). The four aforementioned components were
then summarized as the associated gene network in
adenocarcinoma.
Construction of the three
networks
Following the collection of various data regarding
adenocarcinoma, the data were used to construct three level
networks, which consisted of the differentially-expressed,
associated and global networks. Firstly, the global network was
constructed according to the TFs, miRNAs, target genes and host
genes in adenocarcinoma and the regulatory associations between
these elements. Secondly, the differentially-expressed miRNAs in
adenocarcinoma and the global network were used to identify the
regulatory associations between the differentially-expressed miRNAs
and the host genes. Differentially-expressed genes and miRNAs and
the global network were used to find the regulatory associations
between the differentially-expressed genes and
differentially-expressed miRNAs. The differentially-expressed
network consisted of these regulatory associations. Finally, in the
same way, the associated genes and miRNAs were used to construct
the associated network of adenocarcinoma.
Results
Differentially-expressed network of
adenocarcinoma
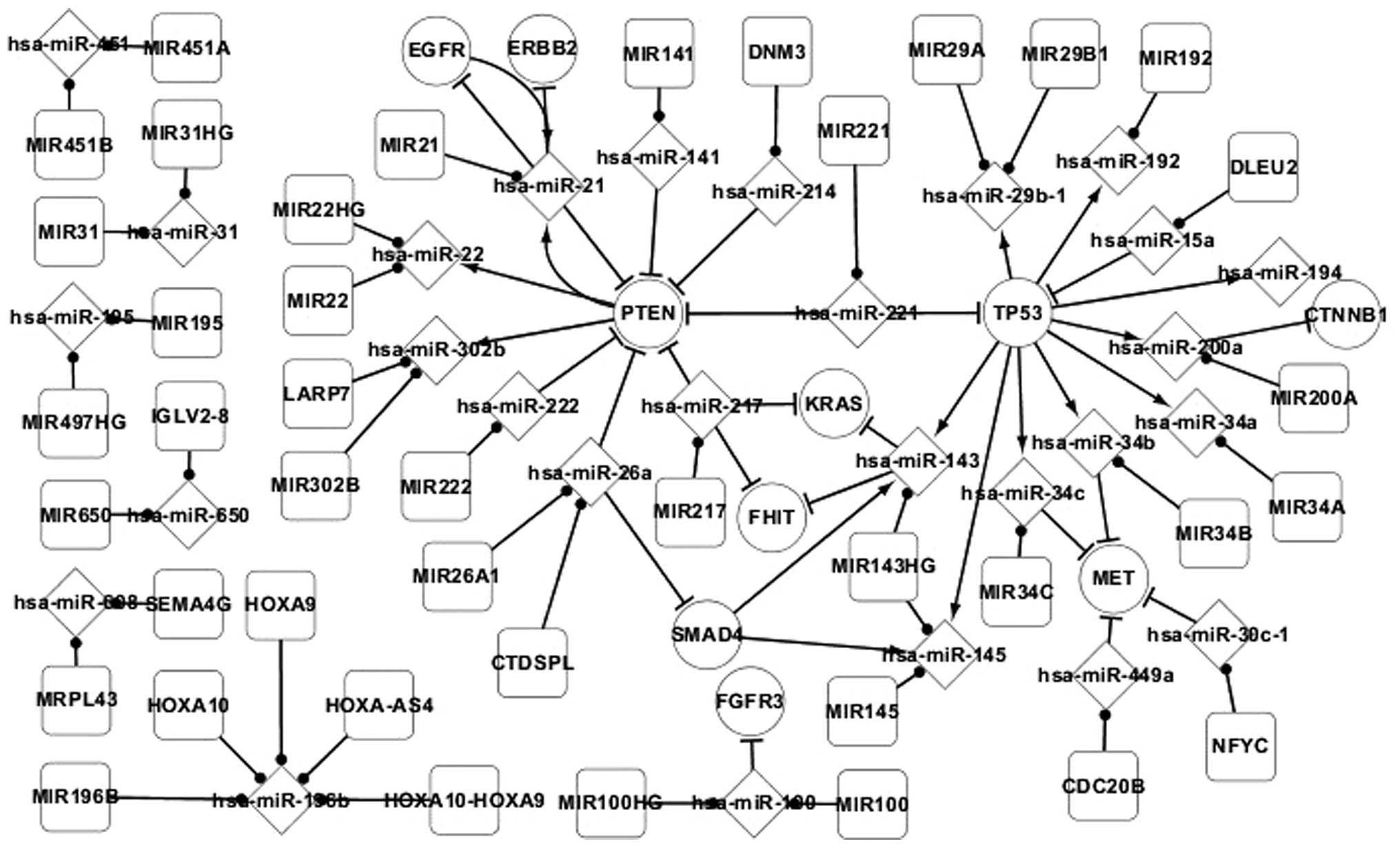
Fig. 1 presents the
differentially-expressed network of adenocarcinoma and reports the
regulatory associations between the differentially-expressed genes
and miRNAs, and target and host genes in adenocarcinoma. In this
network, all the elements were differentially-expressed, with the
exception of the host genes of miRNAs, and all the pathways between
these elements have been experimentally validated. In total, 4 TFs,
consisting of EGFR, phosphatase and tensin homolog (PTEN), SMAD4
and TP53, were identified in the differentially-expressed network
of adenocarcinoma. The regulatory associations between these TFs
and miRNAs are important. Therefore, these TFs and the
corresponding regulatory associations with miRNAs were focused
on.
In the differentially-expressed network, TP53 was
found to directly regulate 9 miRNAs, consisting of hsa-miR-29b-1,
hsa-miR-192, hsa-miR-194, hsa-miR-200a, hsa-miR-34a, has-miR-34b,
hsa-miR-34c, hsa-miR-145 and hsa-miR-143. TP53 may also regulate an
additional 4 genes, consisting of CTNNB1, MET, FHIT and KRAS,
indirectly by regulating these 9 miRNAs. This reveals that genes
may regulate other genes by regulating miRNA. TP53 regulates
hsa-miR-143 and hsa-miR-145, which are regulated by SMAD4. SMAD4 is
regulated by hsa-miR-26a. hsa-miR-26a and other miRNAs regulate the
expression of PTEN. PTEN regulates 3 miRNAs, consisting of
hsa-miR-21, hsa-miR-22 and hsa-miR-302b. These phenomena reveal
that miRNAs may regulate genes individually or in combination with
other miRNAs, and miRNAs may regulate other miRNAs by regulating
genes. In addition, self-regulating associations exist between the
EGFR and PTEN genes and hsa-miR-21. Therefore, PTEN and EGFR may
indirectly regulate each other by regulating hsa-miR-21.
Regulatory associations between
differentially-expressed genes and differentially-expressed miRNAs
in adenocarcinoma are reported clearly in this network. Out of the
three networks, the differentially-expressed network is the most
notable network, as understanding the pathogenesis of
adenocarcinoma is of considerable use. In addition, the
differentially-expressed network possesses a more noteworthy
significance, as it is known that the most important elements in
adenocarcinoma are those in the differentially-expressed network.
When the number of genes and miRNAs in the differentially-expressed
network are at a normal level, individuals do not develop
adenocarcinoma. If the number of genes and miRNAs in the
differentially-expressed network are at an abnormal level, it is
possible that adenocarcinoma may develop. Once the number of key
elements in the differentially-expressed network of an
adenocarcinoma patient is controlled properly, in order to return
the number to a normal level, the differentially-expressed network
may return to the normal state through the pathways in the network.
Therefore, adenocarcinoma may be successfully treated. This forms
the principle of gene therapy.
The differentially-expressed network in the present
study revealed that TP53, PTEN and SMAD4 are extremely important
TFs. These TFs regulate differentially-expressed genes and the
majority of the differentially-expressed miRNAs in adenocarcinoma
directly or indirectly. This provides a theoretical basis for
studies investigating gene therapy as a treatment method for
adenocarcinoma. Careful investigation of the
differentially-expressed network and an understanding of the
regulatory associations between the elements in adenocarcinoma may
result in the successful treatment of adenocarcinoma by
appropriately controlling the levels of key elements in the
differentially-expressed network.
Associated network of
adenocarcinoma
The method used to construct the
differentially-expressed network was also used to construct the
associated network of adenocarcinoma. The associated network
revealed differentially-expressed genes and miRNAs, associated
genes and miRNAs, and target and host genes in adenocarcinoma, in
addition to the regulatory associations between these elements. The
elements and pathways in the differentially-expressed network are
included in the associated network.
In the associated network, there were 23 associated
TFs in addition to the 4 differentially-expressed TFs, 18
associated miRNAs in addition to the 71 differentially-expressed
miRNAs, and numerous additional pathways to those included in the
differentially-expressed network. One example is the
self-regulating associations between the MYC and E2F1 genes and
hsa-miR-17 in the associated network. Therefore, MYC and E2F1 may
regulate each other. Differentially-expressed genes and miRNAs play
an important role in the development of adenocarcinoma, but these
associated genes and associated miRNAs may also affect the
pathogenesis and development of adenocarcinoma. Thus, the
construction and investigation of the associated network may aid
the understanding of the pathogenesis of adenocarcinoma.
Global network of adenocarcinoma
Genes, miRNAs, target genes and host genes and the
regulatory associations between these elements were used to
construct the global network. The global network contains all the
elements and pathways that were included in the
differentially-expressed or associated networks. The global network
is the most complex network out of the three networks.
In order to describe the networks of adenocarcinoma
more clearly, the upstream and downstream information of the
important elements was extracted from this network.
Regulatory associations between
differentially-expressed genes
The predecessor and successor nodes of the
differentially-expressed genes in adenocarcinoma were extracted
from the three networks. These extracted genes revealed that the
predecessor and successor nodes surrounding
differentially-expressed genes demonstrate evident ladder
characteristics in the three networks. This clearly demonstrated
the regulatory associations between the differentially-expressed
genes in adenocarcinoma.
In total, 30 differentially-expressed genes were
identified in adenocarcinoma using the aforementioned method (data
not shown). Overall, 12 genes did not possess adjacent nodes. Those
genes that possessed adjacent nodes were analyzed in the present
study. In total, 4 differentially-expressed TFs, which are
extremely important elements in adenocarcinoma, were identified.
Each demonstrated 6 types of adjacent node, with 3 types of
predecessor nodes and 3 types of successor nodes. Only PTEN was
focused on as an example, however.
PTEN demonstrated significant features in the three
networks, as reported in Table I. In
the differentially-expressed network, 7 miRNAs targeted PTEN and
PTEN regulated 3 miRNAs. It is hypothesized that the 3 successors
of PTEN are regulated indirectly by the 7 predecessors through
PTEN. In addition, if the 3 successors of PTEN regulate other
genes, PTEN may regulate more genes indirectly by regulating these
3 successors. Therefore, PTEN plays an important role in the
pathogenesis and development of adenocarcinoma. There are similar
features in the associated and global networks. In addition,
hsa-miR-21 targets PTEN and PTEN regulates hsa-miR-21 in return.
Therefore, there is a self-regulating association between
hsa-miR-21 and PTEN.
 | Table I.Regulatory associations between
miRNAs and PTEN. |
Table I.
Regulatory associations between
miRNAs and PTEN.
|
| Network |
|---|
|
|
|
|---|
| Association |
Differentially-expressed | Associated | Global |
|---|
| Targets PTEN | hsa-miR-141 | hsa-miR-141 | hsa-miR-141 |
|
| hsa-miR-21 | hsa-miR-21 | hsa-miR-21 |
|
| hsa-miR-214 | hsa-miR-214 | hsa-miR-214 |
|
| hsa-miR-217 | hsa-miR-217 | hsa-miR-217 |
|
| hsa-miR-221 | hsa-miR-221 | hsa-miR-221 |
|
| hsa-miR-222 | hsa-miR-222 | hsa-miR-222 |
|
| hsa-miR-26a | hsa-miR-26a | hsa-miR-26a |
|
|
| hsa-miR-17 | hsa-miR-17 |
|
|
| hsa-miR-216a | hsa-miR-216a |
|
|
| hsa-miR-29b | hsa-miR-29b |
|
|
|
| hsa-miR-18a |
|
|
|
| hsa-miR-19a |
|
|
|
| hsa-miR-19b |
|
|
|
| hsa-miR-19b-1 |
|
|
|
| hsa-miR-19b-2 |
|
|
|
| hsa-miR-20 |
|
|
|
| hsa-miR-20a |
|
|
|
| hsa-miR-216 |
|
|
|
| hsa-miR-26a-1 |
|
|
|
| hsa-miR-26a-2 |
|
|
|
| hsa-miR-494 |
|
|
|
| hsa-miR-519a |
|
|
|
| hsa-miR-519d |
|
|
|
| hsa-miR-91 |
|
|
|
| hsa-miR-93 |
| Regulated by
PTEN | hsa-miR-21 | hsa-miR-21 | hsa-miR-21 |
|
| hsa-miR-302b | hsa-miR-302b | hsa-miR-302b |
|
| hsa-miR-22 | hsa-miR-22 | hsa-miR-22 |
|
|
| hsa-miR-25 | hsa-miR-25 |
|
|
|
| hsa-miR-19a |
|
|
|
| hsa-miR-302 |
|
|
|
| hsa-miR-302a |
|
|
|
| hsa-miR-302c |
|
|
|
| hsa-miR-302d |
|
|
|
| hsa-miR-302f |
In addition, it is known that the adjacent miRNAs in
the differentially-expressed network or associated network may
affect the pathogenesis and development of adenocarcinoma. In
addition to these miRNAs, there are 6 other miRNAs in the global
network that are not included in the differentially-expressed or
associated networks. The effect of these miRNAs on the pathogenesis
and development of adenocarcinoma remains unknown. Additional
studies investigating these miRNAs are required to increase the
understanding of adenocarcinoma. This may aid additional
understanding of the pathogenesis of adenocarcinoma.
Regulatory associations between
differentially-expressed miRNAs
The predecessor and successor nodes of the
differentially-expressed miRNAs in adenocarcinoma were extracted
from the three networks in order to analyze the regulatory
associations between the differentially-expressed miRNAs.
In total, 71 differentially-expressed miRNAs in
adenocarcinoma were identified using this method, 24 of which did
not possess adjacent nodes (data not shown). Numerous other
elements in adenocarcinoma may be regulated by 6 notable miRNAs
with 6 types of adjacent node in a three-level network. However,
only hsa-miR-143 was discussed as an example in the present
study.
In the differentially-expressed network, 2 genes,
consisting of SMAD4 and TP53, were found to regulate hsa-miR-143,
and hsa-miR-142 was found to target the FHIT and KRAS genes. In the
associated network, 6 genes were found to regulate hsa-miR-143,
which, in turn, targeted 4 genes. In the global network, 12 genes
regulated hsa-miR-143, with hsa-miR-143 targeting a total of 14
genes (Table II). Therefore, it is
hypothesized that the SMAD4 and TP53 genes regulate the FHIT and
KRAS genes indirectly, by regulating hsa-miR-143 in the
differentially-expressed network. Similar phenomena were identified
in the associated and global networks.
 | Table II.Regulatory associations between genes
and hsa-miR-143. |
Table II.
Regulatory associations between genes
and hsa-miR-143.
|
| Network |
|---|
|
|
|
|---|
| Association |
Differentially-expressed | Associated | Global |
|---|
| Regulates | SMAD4 | SMAD4 | SMAD4 |
| hsa-miR-143 | TP53 | TP53 | TP53 |
|
|
| SMAD3 | SMAD3 |
|
|
| SRC | SRC |
|
|
| TGFB1 | TGFB1 |
|
|
| TP73 | TP73 |
|
|
|
| BRD2 |
|
|
|
| CEBPB |
|
|
|
| IFNB1 |
|
|
|
| IFNG |
|
|
|
| JAG1 |
|
|
|
| KLF2 |
| Targeted by | FHIT | FHIT | FHIT |
| hsa-miR-143 | KRAS | KRAS | KRAS |
|
|
| FSCN1 | FSCN1 |
|
|
| HRAS | HRAS |
|
|
|
| COL1A1 |
|
|
|
| DNMT3A |
|
|
|
| FNDC3B |
|
|
|
| HK2 |
|
|
|
| MACC1 |
|
|
|
| MAPK12 |
|
|
|
| MAPK7 |
|
|
|
| MT-CO2 |
|
|
|
| MYO6 |
|
|
|
| SERPINE1 |
Regulatory associations between
TFs
The predecessor and successor nodes of the TFs in
adenocarcinoma were extracted from the three networks. This method
identified 27 TFs, 15 of which did not possess adjacent nodes (data
not shown). In total, 15 of the TFs possessed adjacent nodes, but
only E2F3 was discussed as an example in the present study.
E2F3 was found to demonstrate significant phenomena
in the three networks reported in Table
III. In the differentially-expressed network, 3 miRNAs,
consisting of hsa-miR-195, hsa-miR-34a and hsa-miR-34c, target the
E2F3 gene. In turn, E2F3 regulates hsa-miR-195, hsa-miR-34a and
hsa-miR-15a. In the associated network, 5 miRNAs target E2F3 and
E2F3 regulates 3 miRNAs. In the global network, 18 miRNAs were
found to target E2F3, and E2F3 was found to regulate 14 miRNAs. It
is hypothesized that certain miRNAs regulate other miRNAs
indirectly through E2F3. In addition, in the
differentially-expressed, associated and global networks, 2 miRNAs,
consisting of hsa-miR-195 and hsa-miR-34a, targeted E2F3, and E2F3
regulated these miRNAs in return. It is hypothesized that there are
self-regulating associations between hsa-miR-195 and hsa-miR-34a
and E2F3.
 | Table III.Regulatory associations between E2F3
and miRNAs. |
Table III.
Regulatory associations between E2F3
and miRNAs.
|
| Network |
|---|
|
|
|
|---|
| Association |
Differentially-expressed | Associated | Global |
|---|
| Targets E2F3 | hsa-miR-195 | hsa-miR-195 | hsa-miR-195 |
|
| hsa-miR-34a | hsa-miR-34a | hsa-miR-34a |
|
| hsa-miR-34c | hsa-miR-34c | hsa-miR-34c |
|
|
| hsa-miR-17 | hsa-miR-17 |
|
|
| hsa-miR-200b | hsa-miR-200b |
|
|
|
| hsa-miR-106b |
|
|
|
| hsa-miR-125b |
|
|
|
| hsa-miR-125b-1 |
|
|
|
| hsa-miR-125b-2 |
|
|
|
| hsa-miR-128 |
|
|
|
| hsa-miR-128-1 |
|
|
|
| hsa-miR-128-2 |
|
|
|
| hsa-miR-20 |
|
|
|
| hsa-miR-203a |
|
|
|
| hsa-miR-20a |
|
|
|
| hsa-miR-210 |
|
|
|
| hsa-miR-34 |
|
|
|
| hsa-miR-91 |
| Regulated by
E2F3 | hsa-miR-195 | hsa-miR-195 | hsa-miR-195 |
|
| hsa-miR-34a | hsa-miR-34a | hsa-miR-34a |
|
| hsa-miR-15a | hsa-miR-15a | hsa-miR-15a |
|
|
|
| hsa-let-7a |
|
|
|
| hsa-let-7a-1 |
|
|
|
| hsa-let-7a-2 |
|
|
|
| hsa-let-7a-3 |
|
|
|
| hsa-let-7i |
|
|
|
| hsa-miR-106b |
|
|
|
| hsa-miR-15b |
|
|
|
| hsa-miR-16 |
|
|
|
| hsa-miR-16-1 |
|
|
|
| hsa-miR-16-2 |
|
|
|
| hsa-miR-34 |
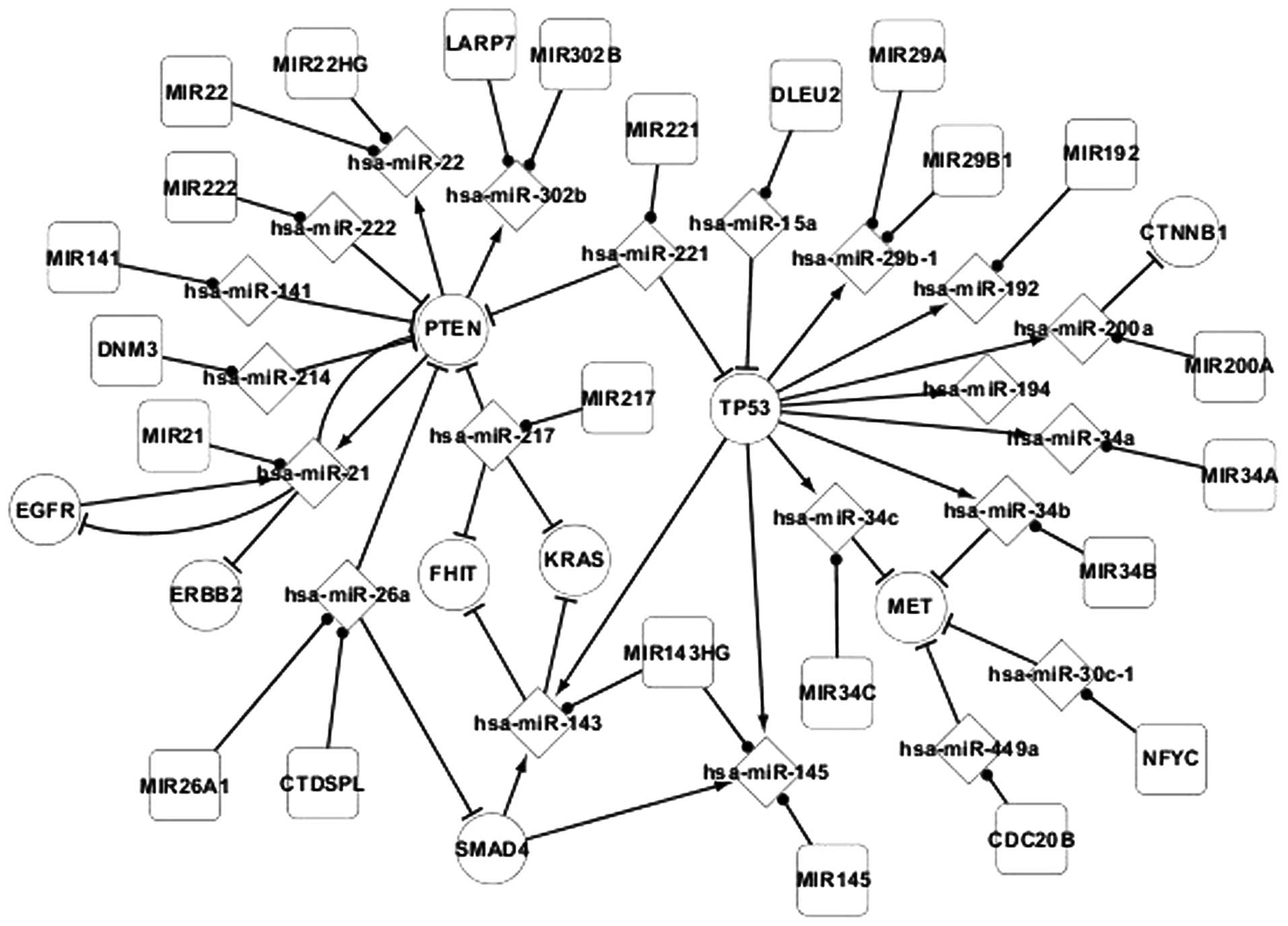
miRNA and host gene network in
adenocarcinoma
The mutation of host genes is known to possibly
affect the miRNAs in these host genes. Therefore, the regulatory
associations between host genes and miRNAs require investigation.
The host genes and the regulatory associations between the host
genes and miRNAs were extracted from the differentially-expressed
network to construct Fig. 2.
Regulatory associations were identified between the
host genes and miRNAs in adenocarcinoma (Fig. 2). miRNA is regulated by host genes or
other miRNA sequences, such as the regulation of hsa-miR-22 by
PTEN, and miRNA also targets genes, such as the targeting of PTEN
by hsa-miR-222. Host genes may code for several miRNAs, such as
MIR143HG coding for hsa-miR-143 and hsa-miR-145, and one type of
miRNA may locate to several host genes, such as hsa-miR-26a
locating to MIR26A1 and CTDSPL. In addition, there are
self-regulating associations between genes, including EGFR and
PTEN, and hsa-miR-21.
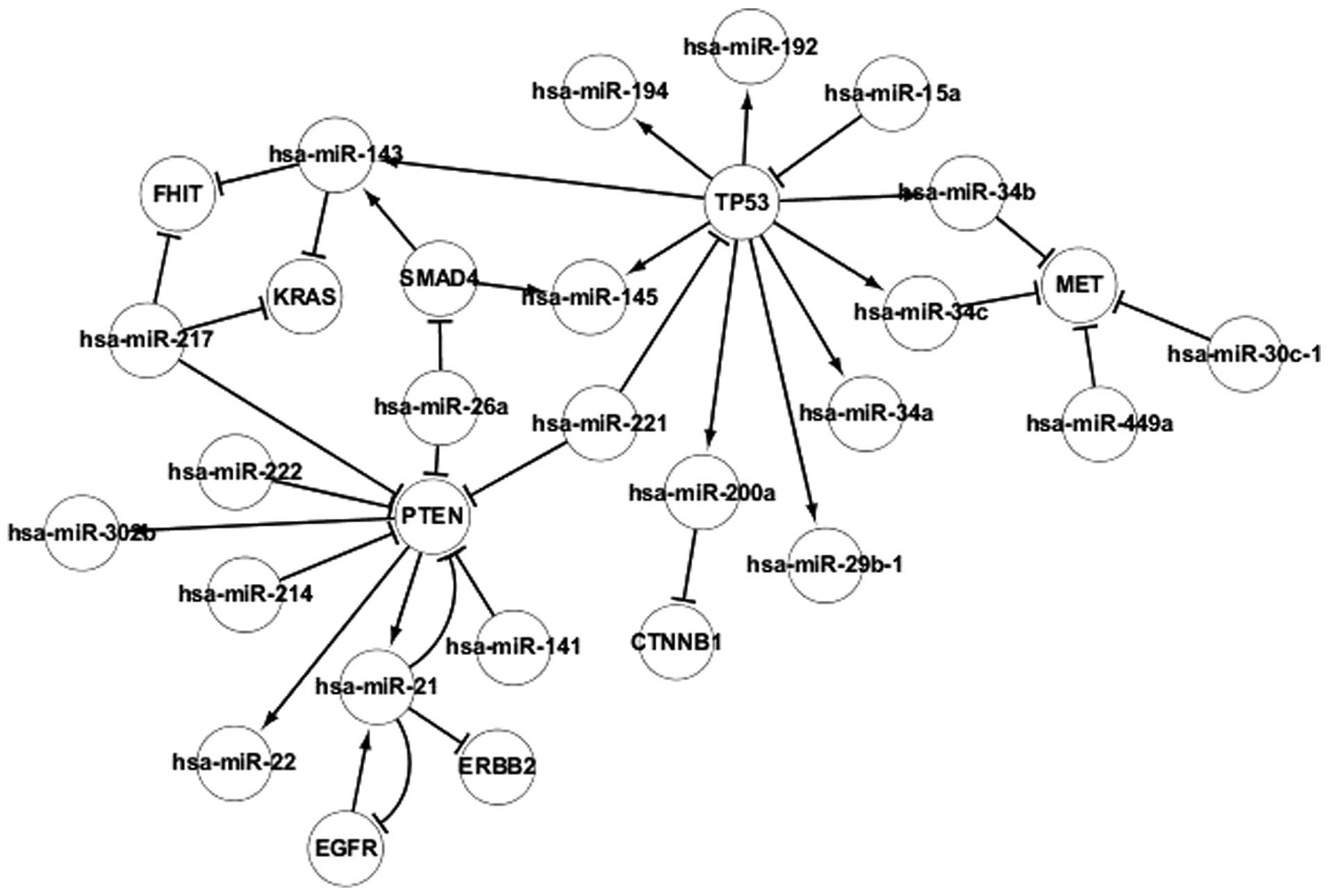
Transcriptional network of popular
TFs
The regulatory associations between TFs and
differentially-expressed miRNAs in adenocarcinoma are revealed in
Fig. 3, which contains 4 TFs,
consisting of TP53, EGFR, PTEN and SMAD4. The TFs regulate the
transcription of genes individually or with other proteins. Certain
associations were identified between TFs and miRNAs. It was found
that miRNA may be regulated by several TFs, such as hsa-miR-143
being regulated by SMAD4 and TP53, and TFs may be targeted by
several differentially-expressed miRNAs, such as hsa-miR-15a and
hsa-miR-221 targeting TP53. In addition, TFs regulate other TFs
indirectly by regulating differentially-expressed miRNAs. For
example, PTEN regulates EGFR indirectly by regulating hsa-miR-21.
miRNAs also regulate other miRNAs indirectly by regulating TFs. For
example, hsa-miR-15a regulates hsa-miR-192 indirectly by regulating
TP53. In addition, self-regulating associations were identified
between the EGFR and PTEN genes and hsa-miR-21. Therefore, PTEN and
EGFR may regulate each other. The regulatory associations between
TFs and differentially-expressed miRNAs may aid the understanding
of the pathogenesis of adenocarcinoma and may also aid the
investigation of gene therapy methods.
Discussion
Previous studies have predicted the existence of
regulatory associations between the TFs, miRNAs, and target and
host genes of miRNAs in adenocarcinoma, and these elements may
affect the development of cancer (3–7,9). However, the regulatory associations
between the elements have not been reported at present.
In the present study, the three networks were used
to collect and analyze these regulatory associations. The TFs,
miRNA, and host and target genes of miRNA in adenocarcinoma were
collected and the regulatory associations between these elements
were determined. Three level networks of adenocarcinoma were
constructed to identify key elements and pathways in
adenocarcinoma. The upstream and downstream data of the notable
elements was then extracted in order to describe the network of
adenocarcinoma more clearly.
In total, 4 key TFs, consisting of EGFR, PTEN, SMAD4
and TP53, were identified in adenocarcinoma. TP53, PTEN and SMAD4,
in particular, are extremely important in adenocarcinoma. In the
differentially-expressed network, these TFs regulate
differentially-expressed genes and the majority of the
differentially-expressed miRNAs in adenocarcinoma directly or
indirectly. This result is supported by a previous study, in which
it was concluded that TFs may regulate genes (8). The differentially-expressed network
revealed that TFs affect the pathogenesis and development of
adenocarcinoma. In addition, 14 other differentially-expressed
genes were identified. In the differentially-expressed network,
these genes are targeted by miRNAs and affect the development of
adenocarcinoma. The associated network contains numerous genes and
miRNAs that are not included in the differentially-expressed
network. These elements cannot affect adenocarcinoma as evidently
as the differentially-expressed genes and miRNAs, but they continue
to affect the pathogenesis and development of adenocarcinoma, and
require investigation.
Prior to the present study, the data regarding
adenocarcinoma was scattered across various databases, including
TransmiR (13), computational
predicted methods (42),
experimentally validated databases (15,16),
miRBase (17), the KEGG pathway
database (18), the miR2Disease
database (19), the GeneCards
database (20) and numerous relevant
studies (13,17–20,22–47).
The analysis and use of this data is challenging. In the current
study, a considerable amount of data was collected, which was
identified and used to construct three networks. The pathogenesis
of adenocarcinoma was clearly revealed and the key elements and
pathways surrounding the elements in adenocarcinoma were identified
through the three networks. It may be hypothesized that controlling
key elements and pathways appropriately to ensure genes and miRNAs
in the differentially-expressed network do not mutate may prevent
healthy individuals from developing adenocarcinoma. The regulation
of a small number of key elements in patients with adenocarcinoma,
resulting in the return of the whole network to a normal state
through the pathways between genes and miRNA, adenocarcinoma is
likely to be treated successfully. Therefore, the present study may
aid the investigation of gene therapy for adenocarcinoma and paves
the way for the successful treatment of adenocarcinoma.
In conclusion, numerous studies have identified the
regulatory associations between various elements in adenocarcinoma,
but failed to collect and analyze these regulatory associations. In
the present study, numerous data in various databases and relevant
literatures were collected to construct the three networks. Using
these networks, 4 notable TFs were identified and the important
associations between various elements were analyzed. The three
networks clearly revealed regulatory associations between various
elements in adenocarcinoma, which is important for the gene therapy
of adenocarcinoma.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 61070084 and
60905022).
Abbreviations:
|
miRNA
|
microRNA
|
|
TFs
|
transcription factors
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
TFBSs
|
transcription factor binding sites
|
References
|
1
|
Stewart BW and Wild CP: Chapter title.
Oesophageal cancer. World Cancer Report. 2014.IARC Press. 2014.
|
|
2
|
Narasimhan P, Hitti IF, Awan A, Desai M,
Kanzer BF and McDonald E: Unusual presentations of prostatic
adenocarcinoma: Lymph node metastasis. Hosp Physician. 38:43–48.
2002.
|
|
3
|
Shalgi R, Lieber D, Oren M and Pilpel Y:
Global and local architecture of the mammalian
microRNA-transcription factor regulatory network. PLoS Comput Biol.
3:e1312007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: C-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of Mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao G, Huang B, Liu Z, et al: Intronic
miR-301 feedback regulates its host gene, ska2, in A549 cells by
targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res
Commun. 396:978–982. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tran DH, Satou K, Ho TB and Pham TH:
Computational discovery of miR-TF regulatory modules in human
genome. Bioinformation. 4:371–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naeem H, Küffner R and Zimmer R: MIRTFnet:
Analysis of miRNA regulated transcription factors. PLoS One.
6:e225192011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li M, Li J, Ding X, He M and Cheng Y:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Papadopoulos GL, Reczko M, Simossis VA, et
al: The database of experimentally supported targets: A functional
update of TarBase. Nucleic Acids Res. 37:D155–D158. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu SD, Lin FM, Wu WY, et al: miRTarBase:
A database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bao J, Li Di, Wang L, et al: MicroRNA-449
and MicroRNA-34b/c function redundantly in murine testes by
targeting E2F transcription factor-retinoblastoma protein (E2F-pRb)
pathway. J Biol Chem. 287:21686–21698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Revilla-Nuin B, Parilla P, Lozano JJ, et
al: Predictive value of MicroRNAs in the progression of barrett
esophagus to adenocarcinoma in a long-term follow-up study. Ann
Surg. 257:886–893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
National Center for Biotechnology
Information: Single Nucleotide Polymorphism Database. simplewww.ncbi.nlm.gov/snpAccessed. May 10–2014
|
|
22
|
Xue Y, Tayoun AN Abou, Abo KM, et al:
MicroRNAs as diagnostic markers for pancreatic ductal
adenocarcinoma and its precursor, pancreatic intraepithelial
neoplasm. Cancer Genet. 206:217–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaduthanam S, Gade S, Meister M, et al:
Serum miR-142-3p is associated with early relapse in operable lung
adenocarcinoma patients. Lung Cancer. 80:223–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Que R, Ding G, Chen J and Cao L: Analysis
of serum exosomal microRNAs and clinicopathologic features of
patients with pancreatic adenocarcinoma. World J Surg Oncol.
11:2192013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cai B, An Y, Lv N, et al: miRNA-181b
increases the sensitivity of pancreatic ductal adenocarcinoma cells
to gemcitabine in vitro and in nude mice by targeting BCL-2. Oncol
Rep. 29:1769–1776. 2013.PubMed/NCBI
|
|
26
|
Xu FX, Su YL, Zhang H, et al: Prognostic
implications for high expression of MiR-25 in lung adenocarcinomas
of female non-smokers. Asian Pac J Cancer Prev. 15:1197–1203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang R, Zheng S, Du Y, et al: Levels of
HOXB7 and miR-337 in pancreatic ductal adenocarcinoma patients.
Diagn Pathol. 9:612014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim J, Lim NJ, Jang SG, et al: miR-592 and
miR-552 can distinguish between primary lung adenocarcinoma and
colorectal cancer metastases in the lung. Anticancer Res.
34:2297–2302. 2014.PubMed/NCBI
|
|
29
|
Wang W, Li F, Mao Y, et al: A miR-570
binding site polymorphism in the B7-H1 gene is associated with the
risk of gastric adenocarcinoma. Hum Genet. 132:641–648. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cortot AB, Younes M, Martel-Planche G, et
al: Mutation of TP53 and alteration of p14(arf) expression in EGFR-
and KRAS-mutated lung adenocarcinomas. Clin Lung Cancer.
15:124–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Heitzer E, Lax S, Lafer I, et al:
Multiplex genetic cancer testing identifies pathogenic mutations in
TP53 and CDH1 in a patient with bilateral breast and endometrial
adenocarcinoma. BMC Med Genet. 14:1292013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dulak AM, Stojanov P, Peng S, et al: Exome
and whole-genome sequencing of esophageal adenocarcinoma identifies
recurrent driver events and mutational complexity. Nat Genet.
45:478–486. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pérez-Mancera PA, Rust AG, van der Weyden
L, et al: The deubiquitinase USP9X suppresses pancreatic ductal
adenocarcinoma. Nature. 486:266–270. 2012.PubMed/NCBI
|
|
34
|
Orloff M, Peterson C, He X, et al:
Germline mutations in MSR1, ASCC1, and CTHRC1 in patients with
Barrett esophagus and esophageal adenocarcinoma. JAMA. 306:410–419.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Safran M, Dalah I, Alexander J, Rosen N,
Stein T Iny, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al:
GeneCards Version 3: The human gene integrator. Database (Oxford).
2010:baq0202010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC genome browser database: Update 2011. Nucleic Acids
Res. 39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Z, Han C, Liu J, et al: GPC5, a tumor
suppressor, is regulated by miR620 in lung adenocarcinoma. Mol Med
Rep. 9:2540–2546. 2014.PubMed/NCBI
|
|
38
|
Chen W, Qin L, Wang S, et al: CPSF4
activates telomerase reverse transcriptase and predicts poor
prognosis in human lung adenocarcinomas. Mol Oncol. 8:704–716.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Allo G, Bandarchi B, Yanagawa N, et al:
Epidermal growth factor receptor mutation-specific
immunohistochemical antibodies in lung adenocarcinoma.
Histopathology. 64:826–839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sekine S, Ogawa R, Oshiro T, et al:
Frequent lack of GNAS mutations in colorectal adenocarcinoma
associated with GNAS-mutated villous adenoma. Genes Chromosomes
Cancer. 53:366–372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen YW, Hsiao PJ, Weng CC, et al: SMAD4
loss triggers the phenotypic changes of pancreatic ductal
adenocarcinoma cells. BMC Cancer. 14:1812014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim HR, Cho BC, Shim HS, et al: Prediction
for response duration to epidermal growth factor receptor-tyrosine
kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma.
Lung Cancer. 83:374–382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luis-Ravelo D, Antón I, Zandueta C, et al:
RHOB influences lung adenocarcinoma metastasis and resistance in a
host-sensitive manner. Mol Oncol. 8:196–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mehra R, Vats P, Kalyana-Sundaram, et al:
Primary urethral clear-cell adenocarcinoma: Comprehensive analysis
by surgical pathology, cytopathology, and next-generation
sequencing. Am J Pathol. 184:584–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rondini EA, Fang H, Runge-Morris M and
Kocarek TA: Regulation of human cytosolic sulfotransferases 1C2 and
1C3 by nuclear signaling pathways in LS180 colorectal
adenocarcinoma cells. Drug Metab Dispos. 42:361–368. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Davison JM, Ellis ST, Foxwell TJ, et al:
MUC2 expression is an adverse prognostic factor in superficial
gastroesophageal adenocarcinomas. Hum Pathol. 45:540–548. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Priolli DG, Abrantes AM, Neves S, et al:
Microenvironment influence on human colon adenocarcinoma phenotypes
and matrix metalloproteinase-2, p53 and β-catenin tumor expressions
from identical monoclonal cell tumor in the orthotopic model in
athymic nude rats. Scand J Gastroenterol. 49:309–316. 2014.
View Article : Google Scholar : PubMed/NCBI
|